Abstract
Epstein–Barr virus (EBV) establishes a life-long persistent infection in most of the human population. In the peripheral blood, EBV is restricted to memory B cells that are resting and express limited genetic information. We have proposed that these memory cells are the site of long-term persistent infection. We now show that memory cells in the tonsil express the genes for EBV nuclear antigen 1 (EBNA1) (from the Qp promoter), latent membrane protein 1 (LMP1), and LMP2a but do not express EBNA2 or the EBNA3s. This pattern of latent gene expression has only been seen previously in EBV-associated tumors such as nasopharyngeal carcinoma, Hodgkin's disease (HD), and T/NK lymphomas. Normal circulating memory B cells frequently reenter secondary lymphoid tissue, where they receive signals essential for their survival. Specifically they require signals from antigen-specific T helper cells and from antigen itself. LMP1 and LMP2 are known to be able to generate these signals in a ligand-independent fashion. We suggest, therefore, that the transcription pattern we have found in latently infected, tonsillar, memory B cells is used because it allows for the expression of LMP1, LMP2a, and EBNA1 in the absence of the immunogenic and growth-promoting EBNA2 and EBNA3 molecules. LMP1 and LMP2a are produced to provide the surrogate rescue and survival signals needed to allow latently infected memory cells to persist, and EBNA1 is produced to allow replication of the viral episome.
Epstein–Barr virus (EBV) establishes a life-long, persistent, latent infection of B cells in >90% of the human population (reviewed in ref. 1). This infection is usually benign but occasionally is associated with lymphoma or carcinoma. Currently, very little is known about how EBV sustains a persistent infection in vivo. Restricted, latency-associated, viral gene transcription programs have been discovered in tumors and tumor-derived cell lines, but it has yet to be demonstrated that these programs are part of the normal biology of viral persistence in vivo (reviewed in ref. 1).
Recently, a form of latent infection was described for infected cells in the peripheral circulation of healthy carriers. In this case no viral latent genes are expressed (2), with the possible exception of latent membrane protein 2a (LMP2a) (3–5). The latently infected cells are remarkably specific in cellular phenotype, being restricted to resting memory B cells (6–8, 48). Based on these observations we proposed that life-long, persistent infection by EBV is maintained within the B cell memory compartment of the peripheral blood (6, 9, 10) and proposed that this form of infection be referred to as the latency or true latency program.
We reasoned that if EBV persists in a transcriptionally quiescent state in peripheral memory B cells, then there must be a mechanism to ensure the long-term survival of these latently infected memory cells. Normal memory B cells in the peripheral blood are also quiescent; however, their survival is absolutely dependent on two signals that the cells receive when they enter secondary lymphoid tissue. The first is supplied by antigen-specific T helper cells (11), and the second is transmitted through the antigen receptor (12, 13). The EBV encoded latent membrane proteins, LMP1 and LMP2a, are capable of delivering these two signals in a constitutive and ligand-independent fashion (14, 15). We hypothesized that latently infected, memory B cells recirculating into lymph nodes may express LMP1 and LMP2a to generate the signals necessary to ensure the long-term survival of the cells in the memory compartment. There are two known transcription programs that could allow for such expression of LMP1 and LMP2a. The first and best characterized occurs when EBV infects normal B cells in vitro (reviewed in ref. 16). The infected B cells become proliferating activated lymphoblasts that express all of the known latent proteins, including six nuclear antigens (Epstein–Barr virus nuclear antigens, EBNAs) and the LMPs. We refer to this as the growth latency program. With this transcription program, expression of all of the latent genes depends on the viral transcription factor EBNA2 (17–19). This is an unlikely program to be used for the maintenance of persistent infection because it may lead to unregulated growth of the latently infected cells, and it is unclear how a memory cell expressing this program could turn it off and enter into a resting state. Furthermore, certain latent proteins, especially EBNA2 and the EBNA3s, encode potent epitopes that are recognized by cytotoxic T cells (20). Cells expressing these epitopes would be rapidly destroyed.
The other program that allows for the expression of LMP1 and LMP2 is the restricted form of latency, mentioned above, that is found in the EBV-associated tumors, including Hodgkin's disease (HD) (21–24), nasopharyngeal carcinoma (25–27), and EBV-associated T/NK lymphomas (28, 29). In these tumors latent gene expression is limited to the expression of EBNA1, LMP1, and LMP2. EBNA2 and the EBNA3s are not expressed. Because EBNA2 is absent, EBNA1, which is essential for the maintenance of the viral episome (30), cannot be made from the promoters used in the growth program. Instead it is expressed from a unique, EBNA2-independent promoter called Qp (31, 32). Because this type of transcription pattern allows EBNA1 expression without the highly immunogenic EBNA2 and EBNA3 proteins, it has been proposed that limited transcription programs, involving EBNA1 from Qp, might be expressed in cells responsible for long-term persistence in the blood (1, 33, 34). However, EBNA1 expression from Qp in the blood has only been detected by one group, and then only in a few healthy carriers (5). Other groups have failed to reproduce this observation (3, 8).
Based on the arguments detailed above, we reasoned that memory cells expressing a restricted pattern of latency, involving EBNA1 from Qp plus LMP1 and LMP2, should be present in the lymph node, not the peripheral blood. To test this idea we have used reverse transcription–PCR (RT-PCR) to analyze the state of viral gene expression in memory B cells within the lymphoid tissue. Specifically, we have performed RT-PCR for EBNA2/EBNA3s versus EBNA1 from Qp to distinguish the growth program from the restricted forms of latency found in the tumors, combined with RT-PCR for LMP1 and LMP2. We chose the tonsil for our studies because there are well-defined, specific markers for identifying and isolating functional subsets of tonsillar B cells (35, 36), and the tonsil is a known site of viral infection and persistence (37). We found that highly purified memory cells from the tonsil express EBNA1 from Qp, LMP1, and LMP2a in the absence of detectable EBNA2 and EBNA3s. This is the first report of an infected B cell subset consistently producing EBNA1 from Qp. We suggest that this transcription program is part of the normal biology of EBV infection, and memory cells expressing this program represent an essential stage in the long-term maintenance of the virus in the memory compartment.
Materials and Methods
Sample Preparation.
Tonsils, obtained from patients undergoing routine tonsillectomies at the Massachusetts General Hospital, were minced in 1× PBS/0.5% BSA, and the resulting suspension was passed through silk screen to remove any connective tissue. The lymphocytes were isolated by Ficoll-Hypaque (Amersham Pharmacia) centrifugation, washed two times with 1× PBS/0.5% BSA, and resuspended at 2 × 107 cells/ml.
Lymphocyte Fractionation.
Naive (IgD+) B cells were isolated by positive selection, using the magnetic bead cell sorting (MACS) (Miltenyi Biotec, Auburn, CA) system as described previously (6). The memory B cells (IgD− CD10−) were isolated by a combination of MACS and fluorescence-activated cell sorting (FACS). Lymphocytes were incubated with biotinylated antibodies to IgD (0.060 μg/ml; Southern Biotechnology Associates) and CD3 (0.03 μg/ml; PharMingen) and fractionated by using the MACS system (Miltenyi Biotec). The negatively selected cells (CD3−, IgD−) were stained with phycoerythrin (PE)-coupled αCD10 (1:50; PharMingen) and FITC-coupled αIgD (1:500), and the CD10− IgD− B cells were sorted with a FACSort. Samples of the isolated cells were stained with the indicated markers for FACS reanalysis to confirm purity (Fig. 1). All separations and manipulations were performed at 4°C.
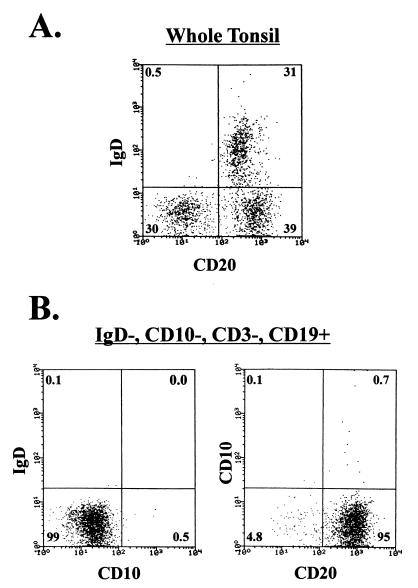
Figure 1.
FACS analysis of the purified memory population. Whole tonsillar lymphocytes were depleted of IgD+ naive B cells and CD3+ T cells by MACS. The depletions were 99% efficient. The resulting population was stained for CD10 and IgD expression, and the memory B cells (IgD−, CD10−) were separated from the germinal center B cells (IgD−, CD10+) by FACS. (A) Unfractionated tonsil cells stained with FITC anti-CD20 (a pan-B cell marker) and PE-coupled anti-IgD. (B) FACS reanalysis of the purified populations with FITC anti-CD10 and PE-coupled anti-IgD (Left) or FITC anti-CD20 and PE-coupled anti-CD10 (Right). Note that the purified population is 95% pure B cells, based on restaining for CD20, and contains only 0.1% IgD+ (naive) and 0.5–0.8% CD10+ (germinal center) B cells.
Because of the lack of biotinylated antibodies for the markers IgA, IgM, and IgG, populations selected for expression of these markers were obtained by FACS separation. For analysis of memory B cells expressing different isotypes, IgD− CD19+ B cells were isolated by MACS as described above, and the cells were costained with anti-CD20-FITC (DAKO) and PE-coupled anti-IgA, IgM, or IgG (Southern Biotechnology Associates). Relevant populations were sorted on a FACStar Plus (Becton Dickinson) or a MoFlo (Cytomation, Fort Collins, CO).
RT-PCR.
RNA was purified from 5 × 106 cells with Trizol (GIBCO) as described by the manufacturer. If necessary, lymphocytes from an EBV-negative tonsil were added to samples to bring the cell number up to 5 × 106. cDNA was prepared as described previously (6), except that the 20-μl cDNA mixture was not ethanol precipitated, but brought up to 100 μl with HPLC H2O and used directly. PCR was performed on the synthesized cDNA for EBNA-1(Q-K) (EBNA-1 from the Qp promoter), EBNA-2, LMP-1, and LMP-2a. The reaction was carried out in a final volume of 50 μl of reaction mix, which consisted of 50 mM KCl, 20 mM Tris (pH 8.4), 2.5 mM MgCl2, 0.2 mM dNTPs, and 20 pM each of the amplimers (final concentrations). The exception was LMP1, for which 3.0 mM MgCl2 was used. The amplimers were as follows: EBNA1(Q-K), 5′-TGGCCCCTCGTCAGACATGATT-3′ and 5′-AGCGTGCGCTACCGGAT-3′ (gift of Sam Speck); EBNA-2, 5′-CATAGAAGAAGAAGAGGATGAAGA-3′ and 5′-GTAGGGATTCGAGGGAATTACTGA-3′ (4); LMP-1, 5′-TTGGTAGTACTCCTACTGATGATCACC-3′ and 5′-AGTAGATCCAGATACCTAAGACAA GT-3′ (4); LMP-2a, 5′-ATGACTCATCTCAACACATA-3′ and 5′-CATGTTAGGCAAATTGCAAA (4). Master mixes containing the above conditions were aliquoted to 200-μl Microamp reaction tubes, and 20 μl of the cDNA suspension described above was added. Given that the cDNA synthesis reaction was in 100 μl, it was possible to perform RT-PCR for all four latent genes described above from one cDNA pot. Reactions were incubated at 95°C for 5 min, and 1 unit of Taq DNA polymerase (Perkin–Elmer) was added to each tube. The tubes were loaded into a Geneamp 9600 thermocycler, and the following conditions were run: for EBNA-1(Q-K) and EBNA-2, 95°C for 15 s, 62°C for 30 s, and 72°C for 30 s, repeated for 40 cycles; LMP-1, 95°C for 15 s, 65°C for 30 s, and 72°C for 30 s, repeated for 40 cycles; LMP-2a, 95°C for 15 s, 55°C for 30 s, and 72°C for 1 min, repeated for 40 cycles. All PCRs were concluded with a 5-min incubation at 72°C to complete the extension of all synthesized products. PCR products were visualized by Southern blotting as described above. Blots were probed by using PCR product derived from the IB4 or RAEL cell lines.
Limiting Dilution DNA PCR.
To determine the absolute number of infected cells in each population, a limiting dilution DNA PCR analysis was performed as described previously (2, 38). Isolated populations were serially diluted, and replicates (usually eight) of each cell number were aliquoted at the desired number into a 96-well V-bottom microtiter plate (Nunclon; Nalge Nunc International, Roskilde, Denmark). Serial dilutions were never performed on cell extracts or isolated DNA. The cells were pelleted; the supernatant was aspirated; 10 μl of a lysis solution containing 0.45% Tween-20, 0.45% Nonidet P-40, 2 mM MgCl2, 50 mM KCl, 10 mM Tris (pH 8.3), and 0.5 mg/ml proteinase K was added to each well; and the plate was incubated for at least 2 h at 55°C. After incubation, the plate was centrifuged quickly to remove condensation from the lid of the plate. PCR was performed in a final volume of 50 μl per reaction. Five microliters of cell lysate was added to each PCR reaction. The PCR and Southern blotting conditions used to detect the PCR products were as described previously (7). The DNA PCR can detect the presence of a single viral genome in as many as 106 uninfected cells. Poisson statistics were used to calculate the frequency of EBV-infected cells.
Results
EBV Is Present in Tonsillar Memory B Cells.
We isolated memory B cells from whole tonsils by negative selection. T cells, naive B cells, and germinal center B cells were depleted with anti-CD3, anti-IgD, and anti-CD10, respectively. A FACS analysis of the final isolated, IgD−, CD10− memory population is shown in Fig. 1. The purified memory population consisted of 95% pure CD20+ B cells contaminated with <1% CD10+ (germinal center) or IgD+ (naive) B cells. Table 1 shows the frequency of virus-infected cells within the memory (IgD−, CD10−, CD20+) population compared with total B cells (isolated by positive selection for CD19 expression) and naive B cells (isolated by positive selection for IgD expression) (6). The results for all five tonsils show that there are significant numbers of infected memory cells, ranging from 1 to 22/105 B cells, which cannot be accounted for by contamination from other populations. We have previously shown that all IgD−, CD20+ cells in tonsils are latently infected (6), and no viral replication is ongoing in this population. Therefore, we conclude that there are latently infected, memory B cells in the tonsil.
Table 1.
Frequency of EBV-infected cells in the tonsillar memory B cell compartment
| No. of infected cells per
107 total cells
| |||
|---|---|---|---|
| Tonsil | Total B | Naive | Memory |
| 1 | >400 | 350 | 215 |
| 2 | 100 | 40 | 180 |
| 3 | 1,500 | 450 | 2,200 |
| 4 | 60 | 30 | 140 |
| 5 | 150 | 75 | 100 |
Tonsils 1, 2, 3, and 4 are tonsils 1, 2, 4, and 5 in Fig. 3.
EBV Latent Gene Expression in Tonsillar Memory B Cells.
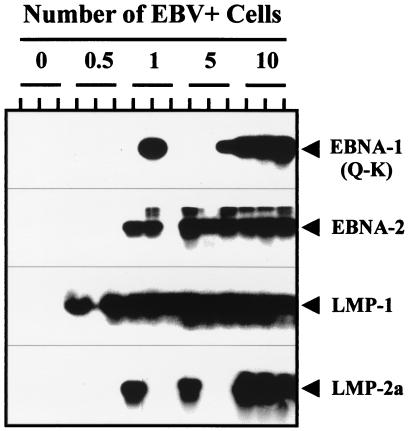
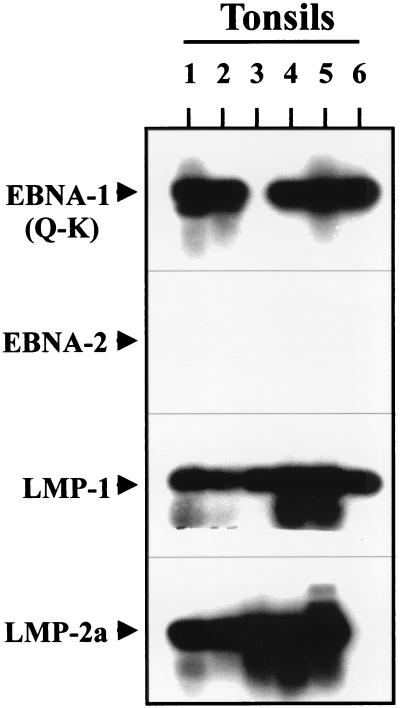
To assess latent gene expression in the purified memory cells, we used RT-PCR to test for expression of the indicator genes that distinguish the tumor-associated pattern of gene expression [EBNA2−, EBNA1(Q-K)+, LMP1+, LMP2+] from the in vitro lymphoblastoid form of latency [EBNA2+, EBNA1(Q-K)−, LMP1+, LMP2+]. Sensitivity controls for the RT-PCR assay are shown in Fig. 2. It is apparent that all four assays can detect as little as a single cell from a tissue culture cell line. Because the transcript copy numbers in the cell lines or in vivo are not known, we cannot be sure of the absolute sensitivity of the assays. Nevertheless, the results shown indicate that the assays are highly sensitive and provide a control for experiment-to-experiment comparisons. The results of the RT-PCR analysis for memory cells from six different tonsils are shown in Fig. 3. EBNA1(Q-K) was detected in 5/6, LMP1 in 6/6, and LMP2 in 5/6 of the tonsils tested, whereas EBNA2 was never detected. In a separate set of experiments, using assays that routinely detected expression in a single EBV-positive cell from a cell line, we were unable to detect expression of EBNA 3a, b, or c in the IgD− cells from five separate tonsils (not shown).
Figure 2.
Sensitivity controls for the RT-PCR analysis. EBV-positive cells from a cell line were spiked into 5 × 106 EBV-negative tonsillar lymphocytes at the numbers indicated before RNA extraction and cDNA synthesis. Each number of cells was tested independently in triplicate. The cell lines used were IB4 for EBNA2, LMP1, and LMP2a and RAEL for EBNA1(Q-K).
Figure 3.
RT-PCR analysis for EBV latent gene expression in tonsillar memory B cells. RT-PCR analysis for the latent genes EBNA-1(Q-K), EBNA-2, LMP-1, and LMP-2a was performed on 106 purified memory B cells from six independent tonsils. For details see Materials and Methods. The migration points for the PCR products are indicated by arrows.
Quantitative Analysis of the Expressed Latent Genes.
For three tonsils, where positive signals were obtained for EBNA1(Q-K), LMP1, and LMP2, we performed serial dilutions of the memory cells before RNA purification for RT-PCR analysis of the four indicator genes. The details of one such experiment are shown in Fig. 4. EBNA2 was never detected at any of the dilutions tested, but the signals for EBNA1(Q-K), LMP1, and LMP2 were detected in as few as 105 cells. By comparing this end point to the measured frequency of infected cells in the population (2 in 105), it was possible to estimate that no more than two infected cells were required to detect the latent genes (tonsil 2 in Table 2). The results from all three tonsils tested are summarized in Table 2. Taken together, these experiments demonstrate that 20–100% of the infected memory cells are expressing EBNA1(Q-K), LMP1, and LMP2. These results exclude the possibility that the RT-PCR signals we are detecting are caused by spurious contamination with some other B cell subset. Therefore, all three genes are expressed in a large fraction of the infected memory cells, and they are probably expressed together in the same cell. If we assume, based on the cell line controls, that we can detect a single cell expressing EBNA2 transcripts, then we may conclude that less than 2% of the infected memory cells in vivo express EBNA2. This is exactly the phenotype of the EBV-associated tumors and represents the first demonstration that this phenotype can occur in a normal B cell population.
Figure 4.
RT-PCR analysis of titrated memory cells. Memory cells were isolated, and RT-PCR analysis for EBNA-1(Q-K), EBNA-2, LMP-1, and LMP-2a was performed. The number of cells tested for each dilution is shown. For each dilution tested the number of cells was brought up to 5 × 106 by the addition of EBV-negative tonsillar lymphocytes before RNA extraction and cDNA synthesis. EBV-negative tonsillar lymphocytes (5 × 106) were also used as the negative control. The migration points for the PCR products are indicated by arrows.
Table 2.
Fraction of EBV-infected memory cells expressing EBNA1(Q-K), LMP1, and LMP2
| Tonsil | 1 | 2* | 3* |
| Frequency of infected cells per 105 memory cells | 10 | 2 | 20 |
| No. of infected cells required to detect LMP1 | 1 | ≤2 | ≤2 |
| No. of infected cells required to detect LMP2 | ≤5 | ≤2 | ≤2 |
| No. of infected cells required to detect EBNA1(Q-K) | ≤5 | ≤2 | 2–20 |
| No. of infected cells required to detect EBNA2† | >100 | >40 | >200 |
The number of infected cells required to detect the latent genes was estimated from the minimum number of cells required to detect a signal in titrations of the type shown in Fig. 2 for tonsil 2. The estimated number of infected cells for each cell number tested in an RT-PCR reaction was derived from the frequencies measured with the limiting dilution DNA PCR method.
EBNA2 was never detected in any samples.
The IgD−, CD3−, CD10− B Cells Express Surface Ig.
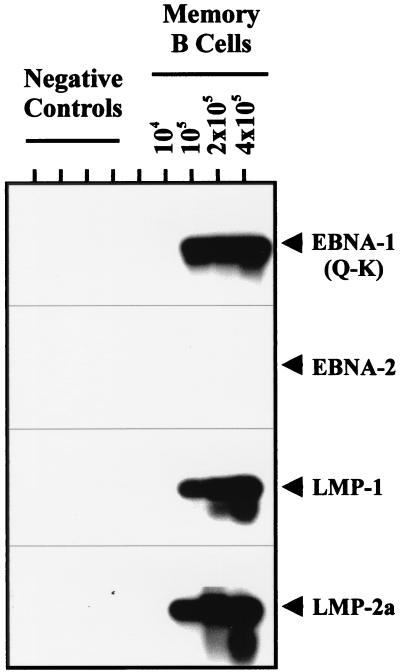
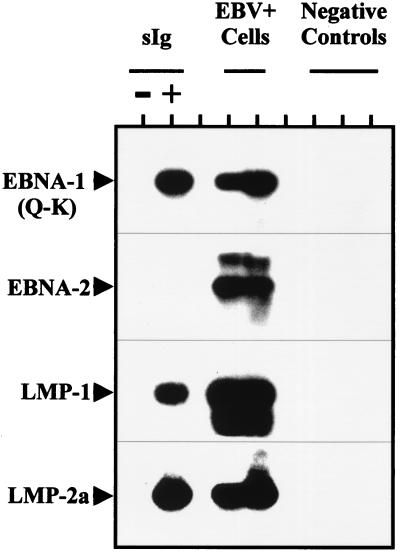
Most of our experiments were performed with memory cells defined as IgD−, CD3−, CD10− B Cells. However, it was possible that the infected cells reside in an aberrant sIg-negative subset that copurified with the memory cells. To test this hypothesis we have positively selected the memory population for the expression of sIg, using antibodies that recognize the IgA, IgG, and IgM isotypes. In these experiments both the DNA PCR (not shown) and RT-PCR signals (Fig. 5) fractionated with the sIg-positive cells, indicating that the latently infected B cells were bona fide memory cells.
Figure 5.
RT-PCR analysis of tonsillar lymphocytes fractionated on the basis of sIg expression. Purified memory cells were labeled with FITC-coupled anti-IgG, IgM, and IgA, and cells from the positive and negative fractions were isolated by FACS. RT-PCR was performed on 106 cells of each population, as described in Fig. 3 and Materials and Methods. The positive controls were five cells from an EBV-positive cell line brought up to 5 × 106 by the addition of EBV-negative tonsillar lymphocytes before RNA extraction and cDNA synthesis. EBV-negative tonsillar lymphocytes (5 × 106) also were used as the negative control.
Discussion
We have discovered that tonsillar memory B cells express a restricted pattern of latent gene transcripts (EBNA1(Q-K)+, EBNA2−, EBNA3−, LMP1+, LMP2+) that resembles that described previously only in the EBV-associated tumors. Previous studies have used RT-PCR to detect the EBNA1 transcript arising from the Qp promoter, to test for restricted forms of EBV latency in the peripheral blood. One study reported that these transcripts were present in the blood of a few healthy carriers (5), but these studies have not been confirmed by our (8) or other (3) laboratories. By comparison, we have found these transcripts in memory cells from 80% of tonsils tested. Furthermore, the RT-PCR signals we obtained were strong and unequivocal and titered out to the cell number expected from the known frequency of virus infected cells in the memory populations. Because our RT-PCR includes the ORF for the protein, we can conclude that we are detecting bona fide transcripts and not the aborted Qp transcripts reported to be present in all EBV-infected cells (39).
It has been hypothesized that the expression of EBNA1(Q-K) alone would characterize the site of long-term persistence (1, 33, 34) because EBNA1 is not recognized by cytotoxic T lymphocytes (40). Our data suggest instead that it is the tumor-associated transcription pattern that is associated with long-term persistence. The function of EBNA1(Q-K) is most likely to allow EBNA1 expression in situations where growth and or survival of the latently infected cells depends, not on the EBNA2-driven growth program used in immortalized lymphoblasts, but on the expression of LMP1 and LMP2a alone. We propose that LMP1 and LMP2 are expressed on tonsillar memory B cells to provide the surrogate T cell help and B cell receptor signals that memory cells require for long-term survival. If this hypothesis is correct, then memory cells persistently infected with EBV may not be true antigen-dependent memory cells, but cells that have been signaled into the long-term memory compartment through the actions of the latent proteins LMP1 and LMP2.
Previous studies have used in situ histochemistry to identify latent gene expression in tonsils. However, these studies are usually performed on tonsils from patients with infectious mononucleosis (37, 41), because the frequency of infected cells is much higher in these individuals than in healthy carriers. Unfortunately it is not clear whether infectious mononucleosis is representative of persistent infection or represents a pathological and, therefore, atypical type of infection. The frequency of infected cells in the tonsils of healthy carriers, such as we have studied here, is around 1 in 105 (see Table 1), which is too low to be analyzed meaningfully by in situ studies. Our studies, detecting viral DNA and gene expression by PCR in large numbers of highly purified B cell subsets, provide an alternative experimental approach to the difficult problem of defining EBV latent gene expression in vivo in healthy carriers. Although our experiments do not address the issue of gene expression at the level of the protein, it is nevertheless clear that we have demonstrated a tight regulation in the expression of EBV latent genes in tonsillar memory cells at the level of mRNA.
The expression of the EBNA1(Q-K)+, LMP1+, EBNA2−, EBNA3− program in virtually every EBV-associated tumor, with the exception of those found in immunosuppressed patients, suggests that this transcription program may be the preferred program that the virus uses in latently infected cells. For this reason we propose that this form of latency be called the default program. In support of this conclusion we have observed that fractionation of tonsillar memory cells into resting and activated populations, based on cell surface markers, reveals that EBNA1(Q-K) transcripts are only found in the activated population (data not shown). By comparison LMP2 transcripts are detected in both populations, and LMP2 is the only latent gene found to be expressed in the resting memory population, the same result as found in peripheral blood. Because the EBNA2 and EBNA3 molecules, found in the growth program and in tumors of immunosuppressed patients (42, 43), are the primary targets of the cellular immune response (20), the default program may in part represent a mechanism of immune evasion for both the normal infected memory B cells and the tumors.
The discovery that the default program is used by tonsillar memory cells is particularly interesting in the context of HD. The demonstration of hypermutation within the Ig genes of HD tumor cells led to the suggestion that HD is derived from a B cell that was post-germinal center in origin (44). The data presented here are consistent with that idea and raise the possibility that EBV-positive HD is a tumor of latently infected memory cells that are resident in secondary lymphoid tissue and normally express the default transcription program. If these cells genuinely represent an essential intermediate state in the maintenance of latently infected memory cells, then they would be expected to turn off latent gene expression as they exit the cell cycle and leave the lymph nodes to enter the peripheral circulation. However, if the cells acquired a mutation that prevented them from undergoing this differentiation step, they would not exit the cell cycle and the result could be constitutive expression of LMP1 and LMP2. It is already well established that constitutive, unregulated expression of LMP1 can be oncogenic in experimental systems (45–47).
In conclusion, our experiments provide evidence that the default latency program, previously found in EBV-associated tumors, is also used in the maintenance of persistent EBV infection within the B cell memory compartment.
Acknowledgments
We thank Allen Parmalee for excellent flow cytometry and Cheryl Greene for providing the tonsils. Our work is supported by Public Health Service Grants AI 18757 and CA 65883.
Abbreviations
- EBV
Epstein–Barr virus
- EBNA
Epstein–Barr virus nuclear antigen
- MACS
magnetic bead cell sorting
- LMP
latent membrane protein
- FACS
fluorescence-activated cell sorting
- HD
Hodgkin's disease
- RT-PCR
reverse transcription–PCR
- PE
phycoerythrin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200366597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200366597
References
- 1.Rickinson A B, Kieff E. In: Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Raven; 1996. pp. 2397–2446. [Google Scholar]
- 2.Babcock G J, Decker L L, Freeman R B, Thorley-Lawson D A. J Exp Med. 1999;190:567–576. doi: 10.1084/jem.190.4.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F, Zou J Z, di, R. L, Winberg G, Hu L F, Klein E, Klein G, Ernberg I. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu L, Rowe D T. J Virol. 1992;66:3715–3724. doi: 10.1128/jvi.66.6.3715-3724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tierney R J, Steven N, Young L S, Rickinson A B. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babcock G J, Decker L L, Volk M, Thorley-Lawson D A. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 7.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 8.Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. J Virol. 1997;71:4882–4891. doi: 10.1128/jvi.71.7.4882-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorley-Lawson D A, Babcock G J. Life Sci. 1999;65:1433–1453. doi: 10.1016/s0024-3205(99)00214-3. [DOI] [PubMed] [Google Scholar]
- 10.Thorley-Lawson D A. Epstein–Barr Virus Rep. 1999;6:158–163. [Google Scholar]
- 11.Gray D, Siepmann K, van Essen D, Poudrier J, Wykes M, Jainandunsing S, Bergthorsdottir S, Dullforce P. Immunol Rev. 1996;150:45–61. doi: 10.1111/j.1600-065x.1996.tb00695.x. [DOI] [PubMed] [Google Scholar]
- 12.Gray D, Skarvall H. Nature (London) 1988;336:70–73. doi: 10.1038/336070a0. [DOI] [PubMed] [Google Scholar]
- 13.Lam K P, Kuhn R, Rajewsky K. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 14.Caldwell R G, Wilson J B, Anderson S J, Longnecker R. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 15.Gires O, Zimber-Strobl U, Gonnella R, Ueffing M, Marschall G, Zeidler R, Pich D, Hammerschmidt W. EMBO J. 1997;16:6131–6140. doi: 10.1093/emboj/16.20.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieff E. In: Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. New York: Raven; 1996. pp. 2343–2396. [Google Scholar]
- 17.Jin X W, Speck S H. J Virol. 1992;66:2846–2852. doi: 10.1128/jvi.66.5.2846-2852.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimber-Strobl U, Suentzenich K O, Laux G, Eick D, Cordier M, Calender A, Billaud M, Lenoir G M, Bornkamm G W. J Virol. 1991;65:415–423. doi: 10.1128/jvi.65.1.415-423.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Tsang S F, Kurilla M G, Cohen J I, Kieff E. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanna R, Burrows S R, Moss D J. Microbiol Rev. 1995;59:387–405. doi: 10.1128/mr.59.3.387-405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oudejans J J, Dukers D F, Jiwa N M, van den Brule A J, Grasser F A, de Bruin P C, Horstman A, Vos W, van Gorp J, Middeldorp J M, Meijer C J. J Clin Pathol. 1996;49:897–902. doi: 10.1136/jcp.49.11.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deacon E M, Pallesen G, Niedobitek G, Crocker J, Brooks L, Rickinson A B, Young L S. J Exp Med. 1993;177:339–349. doi: 10.1084/jem.177.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbst H, Dallenbach F, Hummel M, Niedobitek G, Pileri S, Muller L N, Stein H. Proc Natl Acad Sci USA. 1991;88:4766–4770. doi: 10.1073/pnas.88.11.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niedobitek G, Kremmer E, Herbst H, Whitehead L, Dawson C W, Niedobitek E, von Ostau C, Rooney N, Grasser F A, Young L S. Blood. 1997;90:1664–1672. [PubMed] [Google Scholar]
- 25.Young L S, Dawson C W, Clark D, Rupani H, Busson P, Tursz T, Johnson A, Rickinson A B. J Gen Virol. 1988;69:1051–1065. doi: 10.1099/0022-1317-69-5-1051. [DOI] [PubMed] [Google Scholar]
- 26.Brooks L, Yao Q Y, Rickinson A B, Young L S. J Virol. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahraeus R, Fu H L, Ernberg I, Finke J, Rowe M, Klein G, Falk K, Nilsson E, Yadav M, Busson P, et al. Int J Cancer. 1988;42:329–338. doi: 10.1002/ijc.2910420305. [DOI] [PubMed] [Google Scholar]
- 28.van Gorp J, Brink A, Oudejans J J, van den Brule A J, van den Tweel J G, Jiwa N M, de Bruin P C, Meijer C J. J Clin Pathol. 1996;49:72–76. doi: 10.1136/jcp.49.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang A K, Tao Q, Srivastava G, Ho F C. Int J Cancer. 1996;68:285–290. doi: 10.1002/(SICI)1097-0215(19961104)68:3<285::AID-IJC3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 30.Yates J L, Warren N, Sugden B. Nature (London) 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 31.Tsai C N, Liu S T, Chang Y S. DNA Cell Biol. 1995;14:767–776. doi: 10.1089/dna.1995.14.767. [DOI] [PubMed] [Google Scholar]
- 32.Schaefer B C, Strominger J L, Speck S H. Proc Natl Acad Sci USA. 1995;92:10565–10569. doi: 10.1073/pnas.92.23.10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein G. Cell. 1994;77:791–793. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 34.Masucci M G, Ernberg I. Trends Microbiol. 1994;2:125–130. doi: 10.1016/0966-842x(94)90599-1. [DOI] [PubMed] [Google Scholar]
- 35.Pascual V, Liu Y J, Magalski A, de Bouteiller O, Banchereau J, Capra J D. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y J, Arpin C. Immunol Rev. 1997;156:111–126. doi: 10.1111/j.1600-065x.1997.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 37.Niedobitek G, Hamilton D S, Herbst H, Finn T, Vetner M, Pallesen G, Stein H. Hum Pathol. 1989;20:796–799. doi: 10.1016/0046-8177(89)90075-0. [DOI] [PubMed] [Google Scholar]
- 38.Khan G, Miyashita E M, Yang B, Babcock G J, Thorley-Lawson D A. Immunity. 1996;5:173–179. doi: 10.1016/s1074-7613(00)80493-8. [DOI] [PubMed] [Google Scholar]
- 39.Tao Q, Robertson K D, Manns A, Hildesheim A, Ambinder R F. J Virol. 1998;72:7075–7083. doi: 10.1128/jvi.72.9.7075-7083.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald M P, Klein G, Kurilla M G, Masucci M G. Nature (London) 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 41.Anagnostopoulos I, Hummel M, Kreschel C, Stein H. Blood. 1995;85:744–750. [PubMed] [Google Scholar]
- 42.Young L, Alfieri C, Hennessy K, Evans H, O'Hara C, Anderson K C, Ritz J, Shapiro R S, Rickinson A, Kieff E, et al. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 43.Thomas J A, Hotchin N A, Allday M J, Amlot P, Rose M, Yacoub M, Crawford D H. Transplantation. 1990;49:944–953. doi: 10.1097/00007890-199005000-00022. [DOI] [PubMed] [Google Scholar]
- 44.Kuppers R, Rajewsky K. Annu Rev Immunol. 1998;16:471–493. doi: 10.1146/annurev.immunol.16.1.471. [DOI] [PubMed] [Google Scholar]
- 45.Moorthy R K, Thorley-Lawson D A. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baichwal V R, Sugden B. Oncogene. 1988;2:461–467. [PubMed] [Google Scholar]
- 47.Wang D, Liebowitz D, Kieff E. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 48.Joseph A M, Babcock G J, Thorley-Lawson D A. J Immunol. 2000;165:2975–2981. doi: 10.4049/jimmunol.165.6.2975. [DOI] [PubMed] [Google Scholar]