Abstract
The neuron-restrictive silencer factor (NRSF; also known as REST for repressor element-1 silencing transcription factor) is a transcriptional repressor of multiple neuronal genes, but little is known about its function in vivo. NRSF is normally down-regulated upon neuronal differentiation. Constitutive expression of NRSF in the developing spinal cord of chicken embryos caused repression of two endogenous target genes, N-tubulin and Ng-CAM, but did not prevent overt neurogenesis. Nevertheless, commissural neurons that differentiated while constitutively expressing NRSF showed a significantly increased frequency of axon guidance errors. These data suggest that down-regulation of NRSF is necessary for the proper development of at least some classes of neurons in vivo.
The determination and differentiation of neurons is governed by a complex transcriptional program involving both positive- and negative-acting factors. Although a great deal has been learned recently about positive transcriptional determinants of the neuronal fate, much less is known about negative regulators (1–3). In fact, the only transcription factor currently known to inhibit neurogenesis in vivo is Hes1, a basic helix–loop–helix factor that is a downstream target of Notch signaling (4–6). Hes1 acts early in the transcriptional cascade to inhibit basic helix–loop–helix neuronal determination genes, however, rather than as a direct negative regulator of genes characteristic of terminal neuronal differentiation.
The neuron-restrictive silencer factor (NRSF; also known as REST for repressor element-1 silencing transcription factor) is a zinc-finger transcription factor isolated based on its ability to repress transcription from the promoters of neuron-specific, terminal differentiation (“structural”) genes in nonneuronal cells (7, 8). Upwards of 30 different neuronal genes have been shown to contain a consensus 21-bp NRSF-binding site (the NRSE) (9). Such genes encode a broad range of molecules involved in neuronal development and function, including ion channels (10), neurotransmitter receptors (11–16) and their synthesizing enzymes (17), neurotrophins (18), synaptic vesicle proteins (19, 20), adhesion molecules (21, 22), growth-associated (23), and cytoskeletal proteins (9, 24).
Expression studies have indicated that NRSF mRNA is present at much higher levels in nonneuronal than in neuronal cells and tissues. Furthermore, in the developing nervous system, NRSF is expressed at higher levels in the ventricular zone than in the marginal zone (7, 8, 24). These data have suggested that NRSF might function in part to repress neuronal gene expression in both nonneuronal tissues and neural precursors. They also imply that down-regulation of NRSF during neurogenesis may be important for proper neuronal differentiation. At apparent odds with this latter idea is the observation that NRSF is expressed at low levels by most neurons in the adult brain (25). Whether this low level has any functional significance, however, is not yet clear; indeed, neurons (but not nonneuronal cells) have recently been shown to express a splice variant of NRSF (25) that acts as a dominant inhibitor of NRSF function in neuronal cell lines (26).
Despite the panoply of potential NRSF targets, its suggestive pattern of expression and a wealth of in vitro and biochemical data, it has been difficult to establish the precise functional role of NRSF in vivo. NRSF/Rest mutant mouse embryos begin to degenerate at embryonic day 9.0–9.5 of gestation (24), indicating an essential function in early development but precluding a clear analysis of its role in central nervous system neuronal differentiation, which mostly occurs after this stage. However, mosaic expression of a dominant-negative form of NRSF in the developing chick spinal cord resulted in precocious expression of some neuronal target genes in neural progenitors (24). Although frank neuronal differentiation within the ventricular zone was not observed, these experiments provided in vivo evidence that expression of NRSF in neural progenitors is functionally important to prevent premature initiation of neuronal gene expression during development.
An important question not addressed by these previous studies is whether down-regulation of NRSF expression during neurogenesis is required for proper neuronal differentiation. To address this question we have constitutively expressed NRSF in differentiating neuroblasts and their progeny in vivo. This perturbation leads to a down-regulation of certain endogenous NRSF target genes, which are, conversely, up-regulated by loss-of-function manipulations (24). To assess the consequence of this gain-of-function manipulation more generally for the proper development of neuronal morphology and connectivity, we have used a tauGFP marker to trace the axonal projections of the cells in which sustained expression of NRSF was achieved. Although in general NRSF-overexpressing cells develop an apparently normal neuronal morphology, significantly more commissural neurons exhibit axon pathfinding errors than in controls. These results provide evidence that down-regulation of NRSF during neurogenesis is necessary for proper development of an important aspect of the neuronal phenotype by at least some classes of neurons, in vivo.
Materials and Methods
Plasmid Construction.
Expression constructs harboring a full-length (3.25-kb) murine NRSF cDNA (C. J. Schoenherr and D.J.A., unpublished work) were made in the nonintegrating plasmid vector pCS2+ (27, 28) and the replication-competent retroviral vector RCASBP(B) (29). These constructs were designed to include as little of the 5′ and 3′ untranslated regions as possible: 140 bp of 5′ untranslated region and 40 bp of 3′ untranslated region. Nuclear-localized β-galactosidase was expressed from the construct pCS2 + n-βgal (27, 28). TauGFP was a derivative of mGFP6 (30). Details of the vector construction procedures are available on request.
Electroporation.
White Leghorn chicken eggs were incubated at 38°C until stages 12–15 of development (31), or about 16–25 somites, embryonic day 2. Supercoiled plasmid for electroporation was used at a concentration of 1 or 2 mg/ml in TE (10 mM Tris/1 mM EDTA, pH 7.5) with 0.025% Fast Green (Sigma). Embryos were injected with DNA solution into the lumen of the neural tube and then subjected to electroporation using a BTX ElectroSquarePorator, BTX Genetrodes, and accompanying equipment (Genetronics). The electrodes were spaced 6–8 mm apart and positioned such that the DNA was driven into the cells on only one side of the neural tube. Embryos were pulsed five times for 50 ms each at 25 V. Eggs were then reincubated for 2 or 3 days before the embryos were fixed for 2 h at 4°C in 4% paraformaldehyde in PBS. Embryos were then washed in PBS, cryoprotected with 30% sucrose, mounted in OCT (Tissue-Tek), and sectioned at 20 μm at lower cervical and thoracic levels.
Immunohistochemistry.
Antibody staining of sectioned tissue was carried out by incubating sections overnight at 4°C with the following primary antibodies at the indicated dilutions. Mouse monoclonal antibodies to: murine NRSF (12C11; ref. 24), undiluted; neuron-specific tubulin (TuJ1, BAbCo), 1:1000; NgCAM (8D9, Developmental Studies Hybridoma Bank), 1:10; cyn-1 (32), 1:5; and neurofilament (NN18, Sigma) 1:250. Rabbit polyclonal antibodies to: β-galactosidase (5′–3′), 1:500; GFP (CLONTECH), 1:1000; SCG10 (33), 1:1000. Samples were developed using Alexa dye-conjugated secondary antibodies (488 and 568, Molecular Probes) and visualized using a Zeiss Axioskop fluorescence microscope.
Quantitative Analysis of Axon Guidance Errors.
The number of stray tauGFP+ axons in the ventrolateral quadrant of the spinal cord contralateral to the electroporated side was counted in a blinded manner on coded photographs of sections from each of 11 NRSF-IRES-tauGFP (NTG)-expressing and 7 tauGFP (TG)-expressing embryos collected from two separate electroporation sessions. A total of 38 randomly selected sections from NTG embryos and 40 sections from TG embryos were scored. The average number of stray axons per section was calculated for each embryo, and the mean of these average values was compared between NTG- and TG-expressing embryos. Statistical significance was computed using a two-tailed Student's t test.
Results
Repression of Endogenous Target Genes by Overexpression of NRSF.
An important step in neurogenesis is the induction of neuronal, terminal differentiation (or “structural”) gene expression. In the developing spinal cord, neural progenitors in the ventricular zone normally express low levels of class III β-tubulin (or N-tubulin) (Fig. 1B, arrowhead), and newly born neurons in the marginal zone quickly up-regulate it to high levels (Fig. 1B, open arrow). Expression of the neuron-glial cell adhesion molecule (Ng-CAM) is also up-regulated during neuronal differentiation, but unlike N-tubulin, it is not detectable until the cells have migrated to the marginal zone (Fig. 1D, open arrow). Importantly, N-tubulin and NgCAM are both NRSF direct target genes (9) that have been previously shown to be prematurely up-regulated by inhibition of NRSF function in vivo (24).
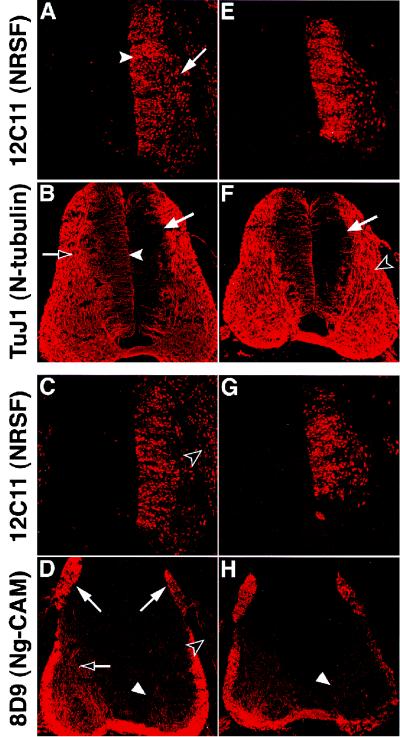
Figure 1.
Overexpression of NRSF results in repression of direct neuronal target genes. A and B, C and D, E and F, and G and H illustrate pairs of adjacent sections stained with antibodies to mouse NRSF (A, C, E, and G), N-tubulin (B and F), and Ng-CAM (D and H). A–D represent sections from one embryo and E–H represent sections from another embryo. Exogenous NRSF is expressed throughout the ventricular (A, arrowhead) and marginal (A, arrow) zones on the electroporated (right) side of both embryos (see also C, E, and G). N-tubulin expression is repressed by NRSF in the ventricular zone (B and F, arrows; cf. B, arrowhead) but not in the marginal zone (F, open arrowhead) on the electroporated (right) but not the contralateral (left) side of the spinal cord (cf. A vs. B, E vs. F). Ng-CAM expression is reduced in axons of commissural neurons in the marginal zone (D and H, arrowheads; cf. D, open arrow), sensory axons in the DREZ (D, right vs. left arrows), and neurons of the DRG (D, open arrowhead). Levels of Ng-CAM expression also appear lower in the ventral funiculus of the spinal cord on the electroporated side relative to controls (D and H, white arrowheads), because much of this staining represents the axons of ipsilateral commissural neurons located more dorsally, in which NRSF expression is high (C and G). In addition, NRSF may also repress Ng-CAM expression in neurons differentiating within the ventrolateral quadrant (see also Fig. 2 B and D).
To assess the effect of sustained NRSF expression on the induction of neuronal gene expression in vivo, we electroporated chick embryos with a full-length mouse NRSF cDNA expressed from a replication-competent retroviral vector, RCASBP(B) (29). Although the 3.43-kb NRSF insert exceeds the theoretical packaging limit of 2.4 kb (29) for the generation of infectious retrovirus, we found that expression of murine NRSF from this vector persisted longer than was observed using a nonintegrating expression vector (Fig. 1 A, C, E, and G, and data not shown). This apparent difference may reflect the integration of proviral DNA into the host chromosome.
After unilateral electroporation of the RCAS-NRSF construct into stage 12–13 chicken embryos and further incubation for 3 days, widespread expression of murine NRSF, detected with a mouse-specific monoclonal antibody (12C11), was observed on the electroporated but not the contralateral side of the spinal cord within both the ventricular (Fig. 1A, arrowhead) and marginal (Fig. 1A, arrow) zones. The presence of NRSF-expressing cells in the marginal zone suggested that sustained expression of NRSF does not prevent the emigration of newly generated neurons from the ventricular zone to their normal site of differentiation. However, we cannot exclude that those cells expressing murine NRSF in the marginal zone migrated from the ventricular zone before elevated levels of exogenous NRSF expression were achieved.
Immunostaining of adjacent sections with antibodies to either N-tubulin or Ng-CAM revealed a down-regulation of these markers caused by misregulation of NRSF. Overexpression of NRSF within the ventricular zone resulted in repression of the low level of N-tubulin expression normally seen in these neural progenitors (Fig. 1 B and F, arrows) when compared with the control contralateral side. No obvious decrease in N-tubulin expression was observed in the marginal zone, however (Fig. 1F, open arrowhead). In contrast, decreased expression of Ng-CAM (which is normally not expressed in the ventricular zone) was detected in the marginal zone in the axons of ventrally projecting commissural neurons (Fig. 1 D and H, arrowheads). Electroporation of a negative control construct expressing nuclear-localized β-galactosidase revealed no difference between control and electroporated sides in the expression of either N-tubulin or Ng-CAM (data not shown).
No differences in the expression of other neuronal genes were detected at this time point (data not shown). For example, apparently equal numbers of cells expressing the pan-neuronal cytoplasmic antigen cyn-1 (32) were detected on the NRSF-overexpressing and control sides of the neural tube. In addition, two other putative direct targets of NRSF, the growth-associated protein, SCG10 (23), and middle molecular weight neurofilament (9), did not exhibit obviously decreased levels of expression in NRSF-electroporated embryos.
Alterations in Axon Pathfinding in NRSF-Overexpressing Neurons.
The sustained expression of NRSF could have affected the levels of many other neuronal target genes for which appropriate antibodies are not available. We therefore sought to assess more generally the consequence of sustained NRSF expression for the development of proper neuronal morphology and connectivity. To this end, we adapted a system (34) for tracing the cellular morphology and axonal projections of the cells that constitutively expressed NRSF. An expression cassette (“NTG”) was constructed in which the murine NRSF coding sequence was followed by an internal ribosome entry site (IRES), followed by a sequence encoding a fusion of the microtubule-associated protein tau with the green fluorescent protein (tauGFP). This design permitted visualization of the cell body morphology and processes of cells expressing exogenous NRSF, using an antibody to GFP. As a control, tauGFP was expressed alone (TG). To evaluate axonal trajectories at early stages of development we examined embryos at two rather than 3 days after electroporation. This allowed use of a nonintegrating plasmid, pCS2+ (28), which is expressed more rapidly (35, 36) and at higher levels than the retroviral vector. Repression of N-tubulin and Ng-CAM expression by murine NRSF was seen with this plasmid expression system at the earlier time point as well (Fig. 2 A and B, arrow and data not shown).
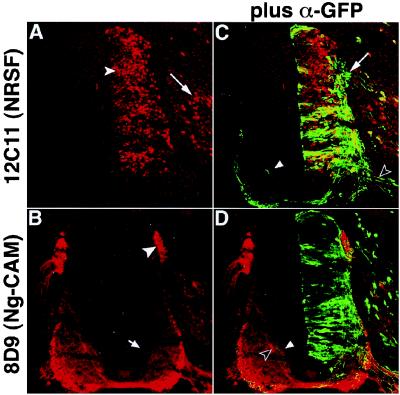
Figure 2.
Morphology of NRSF-misexpressing neurons identified by coexpression of tauGFP. Embryos were electroporated with the NTG expression construct that encodes an NRSF-IRES-tauGFP cassette. A and C show a single section double-stained with antibodies to NRSF (A) and GFP (C) visualized to display either NRSF expression alone (A) or both NRSF and tauGFP expression (C). B and D show an adjacent section double-stained with antibodies to Ng-CAM (B) and GFP (D), visualized in a similar manner. Many cells expressing exogenous NRSF show GFP staining (C); cells expressing GFP but not NRSF are rare. NRSF-overexpressing sensory neurons in the DRG (A, arrow) send processes into the dorsal spinal cord (C, arrow; see also Fig. 3E, arrow), whereas motor neurons extend processes out of the ventral root (C, open arrowhead). Expression of NRSF from this nonintegrating expression plasmid also represses Ng-CAM expression in commissural (B, arrow) and sensory (B, arrowhead) axons, as observed using the retroviral vector (cf. Fig. 1). Inappropriate projection of the axons of NRSF-overexpressing commissural neurons is found within the contralateral commissural axon tract (C and D, arrowhead, green process), in close association with Ng-CAM positive commissural axons that have not yet crossed the midline (D, open arrowhead).
The NTG plasmid was electroporated into stage 13–14 chicken embryos, and incubation of the eggs continued for 2 more days. Immunostaining for NRSF revealed abundant expression of the transgene in the ventricular and marginal zones on the electroporated side of the spinal cord (Fig. 2A, arrowhead), as well as in sensory neurons of the DRG (Fig. 2A, arrow). Double staining with antibodies to NRSF and GFP indicated that the vast majority of cells expressing one protein also expressed the other (Fig. 2C). Some cells expressing NRSF at high levels showed low-level GFP expression, but GFP-expressing cells that had very low levels of NRSF were rare.
Visualization of the tauGFP fusion revealed that NRSF overexpression did not prevent cells from attaining a neuronal morphology. Sensory neurons expressing exogenous NRSF sent axons into the spinal cord from the DRG (Figs. 2C and 3E, arrows), and motor axons appeared to project normally out of the spinal cord through the ventral root (Fig. 2C, open arrowhead). The axons of NRSF-overexpressing commissural neurons, however (whose cell bodies are located in the dorsolateral spinal cord; Fig. 3C, open arrowhead), showed abnormalities in an important aspect of their trajectory. These axons grew normally toward the floor plate and were comparably fasciculated as those in control embryos expressing tauGFP alone (TG; Fig. 3 C and D, arrows). Many of the NTG-expressing commissural axons also crossed the ventral commissure beneath the floor plate (Fig. 3A, open arrowhead). A number of these axons, however, failed to immediately turn anteriorly into the ventral or lateral funiculi as expected (37–39). Instead, they remained at the same rostrocaudal level and projected dorsally, either within the neural tube (Fig. 2 C and D, arrowheads and Fig. 3 A, C, and E, arrowheads) or along the outer margin of the spinal cord (data not shown). In some cases, decussated NTG-expressing axons appeared to take a trajectory parallel to that taken by contralateral Ng-CAM+ commissural axons (Fig. 2D, arrowheads).
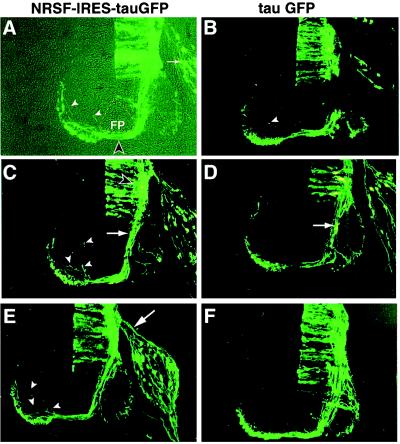
Figure 3.
Constitutive expression of NRSF in commissural neurons causes axon guidance errors. Sections stained with antibody to GFP are illustrated from embryos electroporated with the NTG construct encoding NRSF (A, C, and E) or the control TG construct encoding tauGFP alone (tauGFP; B, D, and F). In A both bright field and epifluorescence images are superimposed. The floorplate (FP) is outlined by the dashed line and the midline decussation of GFP-expressing axons is indicated by the open arrowhead. Solid arrowheads indicate stray axons in the contralateral quadrant in this as well as the other panels. Arrow indicates sensory neurons in the DRG. (C and D) Commissural axons (open arrowhead) exhibit a similar extent of fasciculation on the ipsilateral side of NRSF-expressing (C) and control (D) embryos (arrows), but stray axons are visible in the NRSF-expressing embryo (C, arrowheads). Such stray axons are occasionally seen in control embryos as well (B, arrowhead); see Fig. 4 for quantification. Arrow in E indicates sensory axons growing toward the dorsal spinal cord.
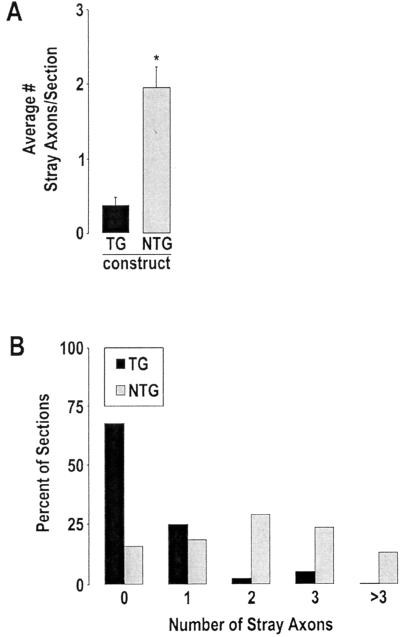
Such pathfinding errors by decussated commissural axons were significantly less frequent in embryos electroporated with the control TG construct (Fig. 3 B, arrowhead, and D and F). Quantification (see Materials and Methods) of the number of stray axons on the contralateral side in an average of three sections per embryo from each of 11 NTG-transfected and 7 TG-transfected embryos (from two independent experiments) indicated that the mean frequency of stray axons per section was 4- to 5-fold higher in NTG-expressing (1.95 ± .29 errors/section; mean ± SEM) than in TG-expressing (0.38 ± 0.12 errors per section) embryos (Fig. 4A, P < 0.001). A histogram of these data also showed that the percentage of sections containing two or more stray axons was significantly higher for NTG- than for TG-expressing embryos (Fig. 4B). Thus, constitutive expression of NRSF caused an increased frequency of axon guidance errors by developing commissural axons.
Figure 4.
Misexpression of NRSF causes a statistically significant increase in axon guidance errors by commissural axons. (A) The average number of stray axons per section is over 5-fold higher in NRSF-expressing (NTG) than in control (TG) embryos (P < 0.001; see Materials and Methods for details). (B) Alternate representation of the data illustrating a histogram of the percentage of sections analyzed that contain stray commissural axons on the contralateral side. The data are derived from analysis of approximately 40 sections from 11 NTG- and 7 TG-expressing embryos obtained in two independent experiments.
Discussion
In this report we describe the effects of misregulating NRSF expression in vivo in the developing nervous system of chicken embryos. Exogenous NRSF was constitutively expressed in the ventricular zone of the neural tube, resulting in higher-than-normal levels of expression in neural progenitors and in sustained expression in their progeny, where the endogenous gene is normally down-regulated. The pattern of defects observed suggests that down-regulation of endogenous NRSF in differentiating neuronal precursors is necessary for both proper induction of neuronal gene expression and for correct axon pathfinding.
NRSF Overexpression Represses Endogenous Neuronal Target Genes.
Two neuronal genes that are direct targets of NRSF (9, 21) were down-regulated by constitutive expression of NRSF in the developing spinal cord. N-tubulin, which is normally expressed at low levels in neural progenitors and is highly up-regulated in differentiating neurons, was found to be repressed to almost undetectable levels in those regions of the ventricular zone containing the highest levels of exogenous NRSF. Ng-CAM, which is normally only detectable in differentiated neurons, was repressed by NRSF in the marginal zone of the spinal cord. Interestingly, Ng-CAM and N-tubulin were both de-repressed by expression of a dominant-negative form of NRSF in chick embryos (24). The symmetry of these loss- and gain-of-function molecular phenotypes provides strong evidence that NRSF is important in the negative regulation of these target genes in vivo. The failure to detect repression of N-tubulin in the marginal zone may reflect its expression in neurons that had differentiated before high levels of exogenous NRSF expression were achieved. Alternatively, the high baseline level of N-tubulin expression in this region may obscure down-regulation of the marker in the subset of neurons that overexpress NRSF.
Overexpression of NRSF caused repression of some but not all of the target genes that we examined. The varying responsiveness of NRSF target genes to perturbation of NRSF expression levels by both loss- (24) and gain-of-function manipulations (this study) in vivo is consistent with other studies. These studies have indicated that the relative contribution of NRSF to the transcriptional regulation of a given reporter gene depends on what other regulatory elements are present in the promoter (12, 17), as well as on the cellular context (18, 21, 22, 24, 40, 41). Thus, both inhibition and increase of NRSF function leads to the conclusion that the relative importance of NRSF for maintaining repression is gene- as well as cell-type-dependent (see ref. 24 for detailed discussion).
Relationship of NRSF to Transcriptional Inhibitors of Neurogenesis.
Although constitutive expression of NRSF down-regulated a subset of neuronal genes, it did not appear to prevent overt neurogenesis: many NRSF-expressing cells could be observed in the mantle layer, and they coexpressed neuronal markers such as SCG10, neurofilament, and cyn-1. In contrast, overexpression of another negative regulator, Hes1 (4), in neural progenitors of the forebrain prevented cells from migrating out of the ventricular zone (5), and in both forebrain and retina prevented overt neuronal differentiation (5, 6).
The apparently different biological activities of NRSF/Rest and Hes genes in gain-of-function experiments may reflect differences between the levels of the neurogenic regulatory hierarchy at which these genes act. The only known direct targets of Hes1 are two transcription factors: Hes1 itself (42) and the neuronal differentiation gene, hASH1 (43). Furthermore, Hes1 is not only able to repress the transcription of positive regulators of neurogenesis such as ASH genes, but because of its helix–loop–helix structure, it is also able to directly inhibit their function by heterodimerization. Therefore, Hes1 acts by several mechanisms to inhibit regulatory factors that function at early steps in the genetic hierarchy controlling neurogenesis. The known targets of NRSF, by contrast, appear primarily to be structural rather than regulatory genes (9). The inability of NRSF to inhibit neurogenesis may thus be explained by the fact that it does not repress early-acting positive regulators of the neurogenic differentiation program. Consistent with this, misexpression of NRSF did not prevent expression of the basic helix–loop–helix neuronal determination gene neurogenin2 (44) in the spinal cord (data not shown).
Overexpression of NRSF in Neurons Perturbs Axon Pathfinding.
Overexpression of NRSF in spinal cord commissural neurons caused axon pathfinding defects. The axons of some of these neurons did not turn anteriorly into the longitudinal axis after crossing the floor plate, as they normally do (37–39). Rather, they continued in the same rostrocaudal segment of the spinal cord and projected dorsally. Such aberrant projections were also detected in control embryos expressing the TG construct, but at a significantly lower frequency. Interestingly, loss-of-function genetic perturbations of axon guidance molecules often appear to stabilize aberrant projections, which occur transiently during normal development but are subsequently corrected (45). The effect of NRSF misexpression on axon guidance is therefore similar to the kind of phenotype that would be expected from down-regulating multiple axon guidance components. The fact that guidance errors are detected only after commissural axons have crossed the midline may reflect the fact that proper guidance of such decussated axons requires changes in the expression levels of several cell surface proteins (ref. 46; reviewed in refs. 47 and 48). Perhaps forced expression of NRSF inhibits the postdecussational induction of some of these guidance factors.
Constitutive expression of NRSF caused a down-regulation of Ng-CAM expression in commissural axons. In earlier experiments, injection of function-blocking antibodies against Ng-CAM into the neural tube of chick embryos was shown to cause defasciculation of commissural axons before decussation (49). In contrast, there was no apparent defasciculation of commissural axons on the ipsilateral side of NRSF-misexpressing embryos, relative to controls. There is not necessarily any inconsistency between these observations. It is not uncommon for axon guidance phenotypes to be more severe when guidance molecules are perturbed immunologically rather than genetically (e.g., see refs. 45 and 49–51). This may reflect a lesser degree of specificity of the immune reagents, either because of antibody cross-reactivity or secondary effects on other guidance molecules associated with the primary antigenic target. Alternatively, the antibodies may have achieved a more complete inhibition of Ng-CAM function than was caused by transcriptional down-regulation of Ng-CAM expression.
A more likely explanation, however, is that inhibition of Ng-CAM function yields a different phenotype than does NRSF misexpression, because there are potentially dozens of other target genes in addition to Ng-CAM that may be down-regulated by forced expression of NRSF (52). If these target genes include those encoding other guidance molecules that normally function in opposition to (or whose function is negatively regulated by) Ng-CAM itself (e.g., see ref. 53), then the down-regulation of these other molecules by NRSF could counteract or modify its effect to down-regulate Ng-CAM.
The potential multiplicity of NRSF targets may also explain why NRSF caused guidance errors by decussated commissural axons on the contralateral side, whereas injection of Ng-CAM antibodies did not (49). However, it is important to note that the antibody injection was a bilateral manipulation, whereas NRSF was misexpressed only on one side of the spinal cord. Therefore, in the present experiments decussated commissural axons misexpressing NRSF should encounter normal levels of Ng-CAM expression on the contralateral side, whereas in the antibody injection experiments Ng-CAM function would be neutralized on both sides of the spinal cord. It would be interesting to test the effects of unilateral neutralization of Ng-CAM function on commissural axon guidance. However, we wish to emphasize that we feel it is unlikely that the axon guidance defects caused by NRSF misexpression can be explained solely by its effect on Ng-CAM expression. An understanding of the molecular mechanism whereby NRSF misexpression affects axon guidance will require a more systematic analysis of the target genes that are negatively regulated by NRSF in commissural neurons.
The present results provide the first in vivo gain-of-function data for NRSF. They indicate that the down-regulation of endogenous NRSF expression that normally occurs during neurogenesis may be important for those aspects of neural development that require a fine-tuning of gene expression, such as axon guidance. By extension, the persistent expression of NRSF mRNA observed in adult neurons (25) could reflect a role to quantitatively modulate the expression of genes underlying neuronal plasticity (54). Consistent with this idea, the NRSE has been found to be important in preventing overactivation of the BDNF promoter in response to kainic acid-induced seizures in adult mice (18). The fact that only a subset of its potential target genes are down-regulated by NRSF misexpression in neurons may, therefore, reflect mechanisms that normally allow endogenous NRSF to be used to selectively regulate certain phenotypic properties in differentiated neurons while preventing deleterious repression of its many other targets.
Acknowledgments
We thank the Developmental Studies Hybridoma Bank for monoclonal antibodies, Jim Haseloff and Emma Dormand for IRES-tauGFP cassettes, Hieu Phan for scoring sections, Gabe Miller and Pat White for help with statistical analysis and preparation of figures, and Kai Zinn and Jane Dodd for critical comments on the manuscript. This work was supported by National Institutes of Health Grant NS23476. D.J.A. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- NRSF
neuron-restrictive silencer factor
- NTG
NRSF-IRES-tauGFP
- TG
tauGFP
References
- 1.Beatus P, Lendahl U. J Neurosci Res. 1998;54:125–136. doi: 10.1002/(SICI)1097-4547(19981015)54:2<125::AID-JNR1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 2.Kageyama R, Nakanishi S. Curr Opin Genet Dev. 1997;7:659–665. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- 3.Lee J E. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 4.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 5.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomita K, Ishibashi M, Nakahara K, Ang S L, Nakanishi S, Guillemot F, Kageyama R. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- 7.Schoenherr C J, Anderson D J. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 8.Chong J A, Tapia-Ramirez J, Kim S, Toledo-Aral J J, Zheng Y, Boutros M C, Altshuller Y M, Frohman M A, Kraner S D, Mandel G. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 9.Schoenherr C J, Paquette A J, Anderson D J. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraner S D, Chong J A, Tsay H J, Mandel G. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 11.Wood I C, Roopra A, Buckley N J. J Biol Chem. 1996;271:14221–14225. doi: 10.1074/jbc.271.24.14221. [DOI] [PubMed] [Google Scholar]
- 12.Mieda M, Haga T, Saffen D W. J Biol Chem. 1997;272:5854–5860. doi: 10.1074/jbc.272.9.5854. [DOI] [PubMed] [Google Scholar]
- 13.Bessis A, Salmon A M, Zoli M, Le Novere N, Picciotto M, Changeux J P. Neuroscience. 1995;69:807–819. doi: 10.1016/0306-4522(95)00303-z. [DOI] [PubMed] [Google Scholar]
- 14.Myers S J, Peters J, Huang Y, Comer M B, Barthel F, Dingledine R. J Neurosci. 1998;18:6723–6739. doi: 10.1523/JNEUROSCI.18-17-06723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai G, Norton D D, Prenger M S, Kusiak J W. J Biol Chem. 1998;273:1086–1091. doi: 10.1074/jbc.273.2.1086. [DOI] [PubMed] [Google Scholar]
- 16.Mu W, Burt D R. Brain Res Mol Brain Res. 1999;67:137–147. doi: 10.1016/s0169-328x(99)00049-2. [DOI] [PubMed] [Google Scholar]
- 17.Lonnerberg P, Schoenherr C J, Anderson D J, Ibanez C F. J Biol Chem. 1996;271:33358–33365. doi: 10.1074/jbc.271.52.33358. [DOI] [PubMed] [Google Scholar]
- 18.Timmusk T, Palm K, Lendahl U, Metsis M. J Biol Chem. 1999;274:1078–1084. [PubMed] [Google Scholar]
- 19.Li L, Suzuki T, Mori N, Greengard P. Proc Natl Acad Sci USA. 1993;90:1460–1464. doi: 10.1073/pnas.90.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoch S, Cibelli G, Thiel G. J Biol Chem. 1996;271:3317–3323. doi: 10.1074/jbc.271.6.3317. [DOI] [PubMed] [Google Scholar]
- 21.Kallunki P, Jenkinson S, Edelman G M, Jones F S. J Biol Chem. 1995;270:21291–21298. doi: 10.1074/jbc.270.36.21291. [DOI] [PubMed] [Google Scholar]
- 22.Kallunki P, Edelman G M, Jones F S. J Cell Biol. 1997;138:1343–1354. doi: 10.1083/jcb.138.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori N, Schoenherr C, Vandenbergh D J, Anderson D J. Neuron. 1992;9:45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z F, Paquette A J, Anderson D J. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 25.Palm K, Belluardo N, Metsis M, Timmusk T. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimojo M, Paquette A J, Anderson D J, Hersh L B. Mol Cell Biol. 1999;19:6788–6795. doi: 10.1128/mcb.19.10.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rupp R A, Snider L, Weintraub H. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 28.Turner D L, Weintraub H. Genes Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- 29.Morgan B A, Fekete D M. Methods Cell Biol. 1996;51:185–218. doi: 10.1016/s0091-679x(08)60629-9. [DOI] [PubMed] [Google Scholar]
- 30.Schuldt A J, Adams J H, Davidson C M, Micklem D R, Haseloff J, Johnston D S, Brand A H. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamburger V, Hamilton H L. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 32.Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell T M, Briscoe J. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- 33.Stein R, Mori N, Matthews K, Lo L C, Anderson D J. Neuron. 1988;1:463–476. doi: 10.1016/0896-6273(88)90177-8. [DOI] [PubMed] [Google Scholar]
- 34.Mombaerts P, Wang F, Dulac C, Chao S K, Nemes A, Mendelsohn M, Edmondson J, Axel R. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- 35.Muramatsu T, Mizutani Y, Ohmori Y, Okumura J. Biochem Biophys Res Commun. 1997;230:376–380. doi: 10.1006/bbrc.1996.5882. [DOI] [PubMed] [Google Scholar]
- 36.Briscoe J, Pierani A, Jessell T M, Ericson J. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 37.Bovolenta P, Dodd J. Development (Cambridge, UK) 1990;109:435–447. doi: 10.1242/dev.109.2.435. [DOI] [PubMed] [Google Scholar]
- 38.Yaginuma H, Homma S, Kunzi R, Oppenheim R W. J Comp Neurol. 1991;304:78–102. doi: 10.1002/cne.903040107. [DOI] [PubMed] [Google Scholar]
- 39.Silos-Santiago I, Snider W. J Comp Neurol. 1992;325:514–526. doi: 10.1002/cne.903250405. [DOI] [PubMed] [Google Scholar]
- 40.Bessis A, Champtiaux N, Chatelin L, Changeux J P. Proc Natl Acad Sci USA. 1997;94:5906–5911. doi: 10.1073/pnas.94.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kallunki P, Edelman G M, Jones F S. Proc Natl Acad Sci USA. 1998;95:3233–3238. doi: 10.1073/pnas.95.6.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 43.Chen H, Thiagalingam A, Chopra H, Borges M W, Feder J N, Nelkin B D, Baylin S B, Ball D W. Proc Natl Acad Sci USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer L, Ma Q, Anderson D J. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 45.Harrelson A L, Goodman C S. Science. 1988;242:700–708. doi: 10.1126/science.3187519. [DOI] [PubMed] [Google Scholar]
- 46.Dodd J, Morton S B, Karagogeos D, Yamamoto M, Jessell T M. Neuron. 1988;1:105–116. doi: 10.1016/0896-6273(88)90194-8. [DOI] [PubMed] [Google Scholar]
- 47.Flanagan J, Van Vactor D V. Cell. 1998;92:429–432. doi: 10.1016/s0092-8674(00)80935-6. [DOI] [PubMed] [Google Scholar]
- 48.Stoeckli E, Landmesser L. Curr Opin Neurobiol. 1998;8:73–79. doi: 10.1016/s0959-4388(98)80010-x. [DOI] [PubMed] [Google Scholar]
- 49.Stoeckli E T, Landmesser L T. Neuron. 1995;14:1165–1179. doi: 10.1016/0896-6273(95)90264-3. [DOI] [PubMed] [Google Scholar]
- 50.Lin D M, Fetter R D, Kopczynski C, Grenningloh G, Goodman C S. Neuron. 1994;13:1055–1069. doi: 10.1016/0896-6273(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 51.Dahme M, Bartsch U, Martini R, Anliker B, Schachner M, Mantei N. Nat Genet. 1997;17:346–349. doi: 10.1038/ng1197-346. [DOI] [PubMed] [Google Scholar]
- 52.Schoenherr C J, Paquette A J, Anderson D J. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunz S, Spirig M, Ginsburg C, Buchstaller A, Berger P, Lanz R, Rader C, Vogt L, Kunz B, Sonderegger P. J Cell Biol. 1998;143:1673–1690. doi: 10.1083/jcb.143.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abel T, Martin K C, Bartsch D, Kandel E R. Science. 1998;279:338–341. doi: 10.1126/science.279.5349.338. [DOI] [PubMed] [Google Scholar]