Abstract
We demonstrate that naturally occurring C14 and C16-specific acyl-acyl carrier protein (ACP) desaturases from plants can complement the unsaturated fatty acid (UFA) auxotrophy of an Escherichia coli fabA/fadR mutant. Under the same growth conditions, C18-specific Δ9-stearoyl (18:0)-ACP desaturases are unable to complement the UFA auxotrophy. This difference most likely results from the presence of sufficient substrate pools of C14 and C16 acyl-ACPs but a relative lack of C18 acyl-ACP pools in E. coli to support the activities of the plant fatty acid desaturase. Based on this, a substrate-dependent selection system was devised with the use of the E. coli UFA auxotroph to isolate mutants of the castor Δ9-18:0-ACP desaturase that display enhanced specificity for C14 and C16 acyl-ACPs. Using this selection system, a number of desaturase variants with altered substrate specificities were isolated from pools of randomized mutants. These included several G188L mutant isolates, which displayed a 15-fold increase in specific activity with 16:0-ACP relative to the wild-type castor Δ9-18:0-ACP desaturase. Expression of this mutant in Arabidopsis thaliana resulted in the accumulation of unusual monounsaturated fatty acids to amounts of >25% of the seed oil. The bacterial selection system described here thus provides a rapid means of isolating variant fatty acid desaturase activities for modification of seed oil composition.
Acyl-acyl carrier protein (ACP)-desaturases (DES) are a family of soluble enzymes that are associated with the synthesis of monounsaturated fatty acids in plants (1). These enzymes catalyze the insertion of a double bond into saturated fatty acids bound to ACP in the plastids of plant cells, and thus serve as one of the primary determinants of the unsaturated fatty acid content of plant membranes and seed storage oils. The most widely occurring member of this family is the Δ9-stearoyl (18:0‡)-ACP DES, which introduces the double bond of oleic acid (1). In addition, a number of other acyl-ACP DES have been identified that are associated with the synthesis of unusual monounsaturated fatty acids and have altered substrate and regio-specifities relative to the Δ9-18:0-ACP DES. Variant acyl-ACP DES identified to date include the Δ4-palmitoyl (16:0)-ACP DES from Umbelliferae seed (2, 3), the Δ6-16:0-ACP DES from black-eyed Susan vine (Thunbergia alata) seed (4), the Δ9-myristoyl (14:0)-ACP DES from geranium (Pelargonium xhortorum) trichomes (5), and Δ9-16:0-ACP DES from cat's claw (Doxantha unguis-cati) (6) and milkweed (Asclepias syriaca) (7) seeds. These enzymes share ≥60% amino acid sequence identity with the Δ9-18:0-ACP DES.
This collection of structurally related, but functionally diverged, enzymes provides a tool for understanding how acyl-ACP DES recognize the chain length of substrates and position the placement of double bonds. Characterization of these properties also has been greatly aided by information from the crystal structure of the castor Δ9-18:0-ACP DES (8). From the three-dimensional structure of this enzyme, a hydrophobic channel was identified that likely corresponds to the binding pocket for fatty acid substrates bound to ACP (8, 9). Results from modeling studies with acyl-ACP DES sequences predict that amino acids lining the lower portion of the hydrophobic channel set constraints on the chain lengths of fatty acid substrates (8, 9). These residues thus represent prime targets for the rational modification of substrate specificities. This has been successfully demonstrated in mutagenesis experiments with the castor Δ9-18:0-ACP DES (9). In these studies, replacement of leucine 118§ and proline 179 with the bulkier phenylalanine and isoleucine, respectively, resulted in the conversion of the castor Δ9-18:0-ACP DES into an enzyme that was most active with 16:0-ACP (9).
To further characterize residues that influence the substrate specificity of acyl-ACP DES, experiments were undertaken to establish a selection system to identify mutants of the Δ9-18:0-ACP DES with increased activity for fatty acid substrates with <18 carbon atoms. As described here, we have established such a system that is based on the ability of acyl-ACP DES to function in vivo in Escherichia coli to produce monounsaturated fatty acids (5,10). This property is combined with the fact that pools of acyl-ACPs in E. coli are enriched in 14:0- and 16:0-ACP but are deficient in 18:0-ACP (11). Based on this, we demonstrate that variant acyl-ACP DES with substrate preference for 14:0- and 16:0-ACP are able to produce sufficient monounsaturated fatty acids to complement an E. coli unsaturated fatty acid (UFA) auxotroph. In contrast, wild-type Δ9-18:0-ACP DES display little or no ability to complement the UFA auxotrophy. This scheme provides a tool for rapidly selecting mutants of the Δ9-18:0-ACP DES that have increased activity with 14:0- and 16:0-ACP substrates.
Ultimately, mutant acyl-ACP DES with altered substrate specificities may be useful for producing monounsaturated fatty acids for nutritional and industrial applications. In this regard, we also show that the expression of a mutant form of the Δ9-18:0-ACP DES obtained from our E. coli selection system can result in large alterations in the monounsaturated fatty acid profile of Arabidopsis seeds.
Materials and Methods
Complementation of an E. coli UFA Auxotroph.
The MH13 mutant of E. coli K12 (12) was used to characterize the activity of acyl-ACP DES. This strain (kindly provided by John Cronan, University of Illinois, Urbana, IL) is fadR∷Tn5 of strain DC308 (13) constructed by phage P1 transduction from strain RS3069 (14). This cell line requires exogenous unsaturated fatty acids for growth at all temperatures because of a temperature-sensitive lesion in fabA and transposon disruption of fadR.
To provide additional reductant for in vivo acyl-ACP DES activity, a chloramphenicol-resistant expression plasmid containing the Anabaena vegetative ferredoxin gene (15) was introduced into the E. coli MH13 cell line. Anabaena ferredoxin was moved as an XbaI/EcoRI fragment from pET9d (15) into the corresponding sites of pLac3d (10) to generate the plasmid pLacAnFd. (pLac3d is equivalent to pET3d, except that the T7 RNA polymerase promoter has been replaced with a lac promoter.) A BglII/HindIII fragment from pLacAnFd then was inserted into the BamHI/HindIII sites of pACYC184 (16). The resulting plasmid was introduced into E. coli MH13 to generate the host cell line E. coli MH13/pAnFd.
The mature coding sequences of acyl-ACP DES in pET expression vectors as previously described (2, 4, 5, 7, 17, 18) were introduced into pLac3a or pLac3d vectors and transformed into E. coli MH13/pAnFd cells. Cells were grown on LB plates that contained ampicillin (100 μg/ml), chloramphenicol (35 μg/ml), and kanamycin (40 μg/ml). For permissive growth, plates were supplemented with oleic acid and Tergitol Nonidet P-40 (Sigma) at final concentrations of 250 μg/ml and 0.2% (vol/vol), respectively. Media used to test for complementation contained 0.4 mM isopropyl β-d-thiogalactoside (IPTG) but lacked UFA. Cultures were maintained at 30°C for 40 h.
Fatty Acid Analyses of E. coli Expressing Acyl-ACP DES.
Cultures were initiated from single colonies of MH13/pAnFd cells expressing plant acyl-ACP DES in vector pLac3a or pLac3d. Cells were grown at 30°C to OD600 ≈ 0.3–0.5 in 25 ml of LB media with chloroamphenicol, kanamycin, ampicillin, and IPTG at concentrations described above. Cultures were collected by centrifugation, and lipids were extracted as described (19) and dried under N2. Fatty acid methyl esters were prepared by transesterification of the lipid extract with sodium methoxide (20) and subsequently analyzed with a Hewlett–Packard 6890 GC fitted with a 30-m × 0.25 mm (i.d.) HP-INNOWAX (Hewlett–Packard) column. The oven temperature was from 170°C (25 min hold) to 185°C at 3.5°C/min.
The double-bond positions of monounsaturated fatty acid methyl esters were determined by GC-MS of dimethyl disulfide derivatives (21).
Saturation Mutagenesis at Selected Codons of the Castor Δ9-18:0-ACP DES cDNA.
Saturation mutagenesis was performed at codons for amino acids 114, 118, 179, or 188 of the castor Δ9-18:0-ACP DES by replacing the target codon by NNN with the use of overlap extension PCR (22). The mature castor Δ9-18:0-ACP DES was used as the template, and amplification reactions were conducted by using Pfu polymerase (Stratagene). Saturating mutations were introduced at the codon for amino acid 114 by initially conducting two separate amplification reactions by using the following oligonucleotide primer combinations: reaction 1, T7 primer and 5′-GAACTCCATCCAAGGTATTCAGNNNTGTTTGATAAGTGGGAAGGGC-3′ (primer A); and reaction 2, 5′-CTGAATACCTTGGATGGAGTTC-3′ (primer B) and 5′-GCAAAAGCCAAAACGGTACCATCAGGATCA-3′ (primer C). Agarose gel-purified reaction products were combined and amplified together with the T7 primer and primer C. The resulting product was digested with XbaI and BamHI and ligated in place of the corresponding portion of the coding sequence for the mature wild-type castor Δ9-18:0-ACP DES in pLac3d. Similar methodology was used to perform saturation mutagenesis at codon 118, except that primers A and B were replaced with the respective primers: 5′-GTTTCATCCCGAACTCCATCNNNGGTATTCAGCATTGTTTGATAAG-3′ and 5′-GATGGAGTTCGGGATGAAAC-3′.
Saturation mutagenesis at codon 179 was performed as described above, except that primers A, B, and C were replaced with the respective primers: 5′-GTATGGACTGTTTTCTGTCCGNNNATCCATTCCTGAACCAATCAA-3′, 5′-CGGACAGAAAACAGTCCATACC-3′, and 5′-TCGTCCTGCACTTTGATCACCTACAGCTTCACTTG-3′. The second-round PCR product was digested with NcoI and BclI and ligated into the NcoI/BamHI sites of pLac3d. Saturated mutations at codon 188 were introduced as described above for codon 179, except that primers A, B, and C were replaced with the respective primers: 5′-CCTGGAATGATGTATAGATGAANNNAAGGTATGGACTGTTTTCTGTC-3′, 5′-TTCATCTATACATCATTCCAGG-3′, and 5′-TCGTCCTGCACTTTGATCACCTACAGCTTCACTTG-3′.
Plasmid libraries containing saturating mutations at codons for amino acids 114, 118, 179, or 188 of the castor Δ9-18:0-ACP DES were amplified in E. coli DH5α cells. Seven randomly chosen colonies from cells transformed with the codon 188 library were sequenced to confirm the efficacy of the positional randomization. The sequences of the seven clones at codon 188 encoded threonine (ACC), serine (AGC, TCC, TCT), arginine (CGC), and a stop codon (TGA).
Saturation mutagenesis at both positions 114 and 188 was achieved by insertion of the XbaI/BamHI-digested PCR product from the codon 114 library in place of the corresponding portion of clones from the codon 188 library.
To select mutants that would complement the UFA auxotrophy of E. coli MH13/pAnFd cells, the plasmid libraries were introduced into this cell line by electroporation. Transformed cells then were plated on LB media containing IPTG, kanamycin, ampicillin, and chloramphenicol, without the addition of UFA, and incubated at 30°C for up to 48 h. In addition, in some experiments, complementation was conducted in liquid culture. In these experiments, transformed E. coli MH13/pAnFd cells were grown in the same LB media with shaking (250 rpm) at 30°C and harvested at OD600 ≈ 0.25. To increase the proportion of mutants with enhanced specificity for 14:0- and 16:0-ACP, plasmid DNA isolated from these cells was used to retransform E. coli MH13/pAnFd. The cells were again grown at 30°C and harvested at OD600 ≈ 0.25. Plasmid DNA isolated from colonies selected on plates or from cells selected in liquid culture was transformed into E. coli DH5α cells and grown with ampicillin selection. The sequences of the selected mutants of the castor Δ9-18:0-ACP DES in pLac3d were determined to identify the substitution.
Characterization of the Substrate Specificity of Mutants of the Castor Δ9-18:0-ACP DES.
The coding sequences of mutants of the castor Δ9-18:0-ACP DES selected in bacterial complementation experiments were transferred from the vector pLac3d to the vector pET9d. Recombinant protein then was generated by expression of the resulting plasmids in E. coli BL21 (DE3) cells and enriched to 90–95% purity by cation exchange chromatography as described (6, 9).
Mutant castor Δ9-18:0-ACP DES were assayed as described (6, 9) with [1-14C]14:0-, 16:0-, or 18:0-ACP as substrates using recombinant spinach ACP-I (23, 24). Methyl esters of fatty acids were analyzed by argentation TLC, and radioactivity in products was quantified as described (6, 9).
Transgenic Expression of the Wild-Type Castor Δ9-18:0-ACP DES and G188L Mutant in Arabidopsis thaliana Seeds.
The coding sequences of the mature wild-type castor Δ9-18:0-ACP DES and G188L mutant were amplified by PCR with the use of Pfu polymerase (Stratagene) and the primers 5′-TTTCCATGGCCTCTACCCTCAAGTCTG-3′ and 5′-TTTTCTAGACTACAGCTTCACTTGCCTATC-3′. The resulting DES ORF was linked by a NcoI site with sequence for the plastid transit peptide of the coriander Δ4-16:0-ACP DES in pBluescript SK(−) (2). The resulting chimeric coding sequences were moved as EcoRV/XbaI fragments into the SmaI/XbaI sites of the binary expression vector pDN2. This vector is derived from pDN (25) by introduction of the SmaI/SacI portion of the pBluescript SK(−) polylinker. Vector pDN2 and the resulting plamids containing the coding sequences of either the wild-type DES or the G188L mutant were introduced into the fab1 mutant or wild-type A. thaliana (Columbia) by means of Agrobacterium tumefaciens-mediated transformation by vacuum infiltration (26). The resulting transgenic plants were selected for kanamycin resistance. Introduction of the desired transgenes was confirmed by sequence analysis of PCR products amplified with DES-specific primers from genomic DNA of transgenic plants.
The fatty acid content of single seeds from T1 plants was determined by gas chromatography. Fatty acid methyl esters were prepared by homogenization of single seeds in 10 μl of trimethylsulfoniumhydroxide/methanol (27) in a 100-μl autosampler vial. After a 15-min reaction at room temperature, samples were dried, resuspended in 35 μl of hexane, and analyzed with a Hewlett–Packard 5890 GC fitted with a 30-m × 320-μm (i.d.) Omegawax 320 column (Supelco). The oven temperature was from 185°C (3 min hold) to 200°C at a rate of 2°C/min and then to 220 (2 min hold) at a rate of 7°C/min.
Results
Complementation of an E. coli UFA Auxotrophy by the Expression of Plant 14:0- and 16:0-ACP DES.
We previously have demonstrated that plant 14:0 and 16:0-ACP DES can use in vivo pools of acyl-ACPs in E. coli to produce monounsaturated fatty acids (5, 9). In contrast, it has been reported that the Δ9-18:0-ACP DES does not generate any detectable monounsaturated fatty acids when expressed in E. coli (28). These apparently contradictory findings are likely related to the relative pool sizes of acyl-ACPs. Fatty acid analyses of acyl-ACPs of E. coli have shown that these pools are enriched in 14:0 and 16:0 but contain little or no detectable 18:0 (11). Therefore, E. coli cells likely do not contain sufficient pools of 18:0-ACP to support the activity of the Δ9-18:0-ACP DES.
We examined activity of plant acyl-ACP DES in E. coli further using an unsaturated fatty acid auxotrophic strain. Similar strains have been used to isolate an acyl-lipid desaturase by complementation (29). For these experiments, E. coli MH13 cells were used, which require exogenous UFA for growth at all temperatures because of lesions in the fabA and fadR loci. The fabA gene product, β-hydroxydecanoyl-ACP dehydratase, introduces a cis double bond in acyl chains during fatty acid synthesis, and fadR encodes a transcriptional activator of fabA and other fatty acid biosynthetic genes. An inducible expression plasmid for Anabaena vegetative ferredoxin (Fd) was introduced into MH13 cells to provide additional reductant to support the in vivo activity of plant acyl-ACP DES. The resulting cell line was designated MH13/pAnFd.
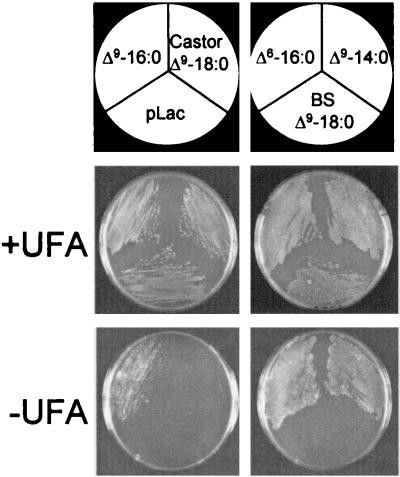
Experiments were designed to compare the ability of different plant acyl-ACP DES to complement the UFA auxotrophy of MH13/pAnFd cells. For these studies, MH13/pAnFd cells were transformed with plasmids containing the coding sequences of acyl-ACP DES behind a lac promoter. When maintained at 30°C, IPTG-induced cells expressing three different 14:0- and 16:0-ACP DES (geranium Δ9-14:0-, milkweed Δ9-16:0-, and black-eyed Susan vine Δ6-16:0-ACP DES) were able to grow in the absence of UFA (Fig. 1). In contrast, induced cells expressing Δ9-18:0-ACP DES from two different plant species (castor and black-eyed Susan vine) displayed little or no ability to grow at 30°C on plates lacking UFA. Expression of Δ9-18:0-ACP DES in these cells was confirmed by Western blot analysis of extracts from induced cultures maintained with UFA supplementation (results not shown). The inability of the Δ9-18:0-ACP DES to complement the UFA auxotrophy of MH13/pAnFd cells is thus consistent with the lack of sufficient 18:0-ACP substrate pools in E. coli.
Figure 1.
Complementation of UFA auxotrophy of E. coli MH13/pAnFd cells by expression of plant 14:0- and 16:0-ACP DES. E. coli MH13/pAnFd cells were transformed with the expression vector pLac3d (pLac) alone or with pLac3d containing the coding sequences of the milkweed Δ9-16:0-ACP DES (Δ9-16:0), the black-eyed Susan Δ6-16:0-ACP DES (Δ6-16:0), the geranium Δ9-14:0-ACP DES (Δ9-14:0), the castor Δ9-18:0-ACP DES (Castor Δ9-18:0), or the black-eyed Susan Δ9-18:0-ACP DES (BS Δ9-18:0). Transformed cells were grown at 30°C for 40 h either under permissive conditions on media containing oleic acid (+UFA) or under selective conditions on media lacking oleic acid (−UFA) as described in Materials and Methods.
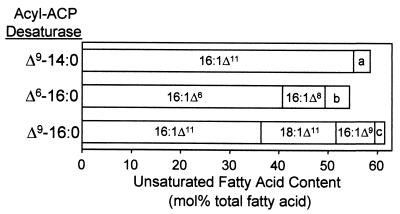
Interestingly, MH13/pAnFd cells expressing the 14:0- and 16:0-ACP DES accumulated unusual monounsaturated fatty acids to >50 mol % of the total fatty acids (Fig. 2). The monounsaturated isomers detected were those expected from the in vitro activities of these enzymes with 14:0- or 16:0-ACP and from subsequent fatty acid chain elongation.
Figure 2.
UFA content of E. coli MH13/pAnFd cells expressing the geranium Δ9-14:0-ACP desaturase (Δ9-14:0), the black-eyed Susan Δ6-16:0-ACP desaturase (Δ6-16:0), or the milkweed Δ9-16:0-ACP desaturase (Δ9-16:0). Cultures were initiated from single colonies and grown at 30°C to OD600 ≈ 0.3–0.5 with IPTG induction and antibiotic selection as described in Materials and Methods. Double bond positions were determined by GC-MS of dimethyl disulfide derivatives of fatty acid methyl esters (21). Minor unsaturated fatty acid components are indicated by a, b, and c: a includes ≤3.5 mol% of 14:1Δ9, 16:1Δ9, and 18:1Δ13; b includes ≤5 mol% of 18:1Δ8; and c includes ≤2 mol% 18:1Δ9 and 18:1Δ13.
Substrate-Dependent Selection of Mutants of the Castor Δ9-18:0-ACP DES.
The above results demonstrated the ability of 14:0- and 16:0-ACP DES to complement the UFA auxotrophy of E. coli MH13/pAnFd cells but an inability of 18:0-ACP DES to complement this auxotrophy. This observation suggested the possibility of using MH13/pAnFd cells as the basis for a selection system to identify mutants of the Δ9-18:0-ACP DES with altered substrate specificity. In this regard, we hypothesized that mutations in the Δ9-18:0-ACP DES that increase the specific activity of this enzyme with 14:0- and 16:0-ACP substrates should yield activities that are able to complement the UFA auxotrophy of E. coli MH13/pAnFd cells.
The ability of E. coli MH13/pAnFd cells to serve as a substrate-dependent selection system was tested by transforming this cell line with pools of mutant cDNAs of the castor Δ9-18:0-ACP DES and subsequently characterizing the in vitro activities of the recombinant DES that complement the UFA auxotrophy. For these experiments, expression libraries of the castor Δ9-18:0-ACP DES cDNA were generated that were saturated for mutations at codons for amino acids that line the bottom of the substrate binding pocket based on the crystal structure of this enzyme (8, 9). The libraries constructed included those saturated for mutations at codons corresponding to residues 114, 118, 188, and 179 individually as well as at codons for both residues 114 and 188. E. coli MH13/pAnFd cells were transformed with these libraries and maintained in media lacking UFA.
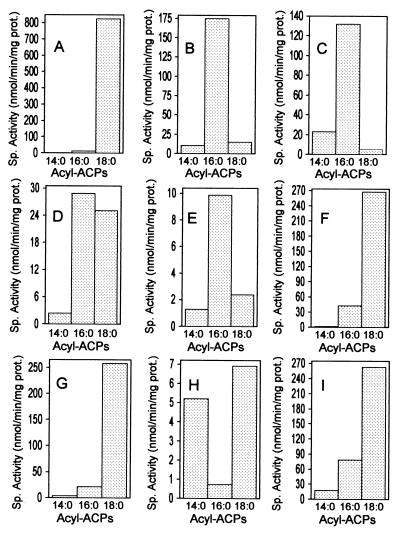
Plasmids containing the coding sequence for mutant castor Δ9-18:0-ACP DES were isolated from cells that were able to grow under the nonpermissive growth conditions. These then were used for the generation of purified recombinant enzyme for in vitro assay of activity with acyl-ACPs of different chain lengths. All of the characterized mutants found to complement the UFA auxotrophy of the E. coli MH13/pAnFd cells displayed increases in their relative activity with 14:0- and/or 16:0-ACP substrates compared with the wild-type castor Δ9-18:0-ACP DES (Fig. 3A). Of the libraries tested, the one containing random mutations at the codon for residue 188 yielded the most extreme phenotype after selection in the E. coli UFA auxotroph (Fig. 3B). Of more than 15 selected clones analyzed, all encoded a leucine at this position, and all of the six possible codons for this amino acid were represented among the analyzed clones. The replacement of a glycine by a leucine at residue 188 resulted in an enzyme that is 10-fold more active with 14:0-ACP and 15-fold more active with 16:0-ACP than the wild-type DES (Fig. 3B). This mutant was also more than 50-fold less active with 18:0-ACP relative to the wild-type enzyme. A related mutant M114I/G188L also was identified from selection with a library saturated for mutations at codons for both amino acids 188 and 114. This mutant displayed further reduction in the activity with 18:0-ACP (160-fold lower than the wild-type enzyme) (Fig. 3C). Furthermore, the M114I/G188L mutant was 29-fold more active with 14:0-ACP and 12-fold more active with 16:0-ACP than the wild-type Δ9-18:0-ACP DES. In addition to 18:1Δ9, approximately 10% and 40% of the products formed by mutants G188L and M114I/G188L, respectively, with 18:0-ACP were identified as the Δ10 isomer of 18:1. A similar dual regiospecificity with 18:0-ACP was previously noted for the L118F/P179I mutant of the castor Δ9-18:0-ACP DES (9).
Figure 3.
Substrate specificity profiles of the wild-type castor Δ9-18:0-ACP DES (A) and mutants of this enzyme obtained from substrate-dependent selection by E. coli MH13/pAnFd cells (B–I). Shown are the specific activities of purified recombinant enzymes with 14:0-, 16:0-, and 18:0-ACP. Mutants of the castor Δ9-18:0-ACP DES were selected from libraries containing random mutations at codons for amino acids 114, 118, 179, or 188 as well as both amino acids 114 and 188 as described in the text. Activities are shown for the wild-type DES (A) as well as mutants G188L (B), M114I/G188L (C), L118W (D), L118Y (E), L118F (F), M114I (G), M114F (H), and P179I (I).
Of more than 20 clones analyzed, complementation of the UFA auxotrophy of E. coli MH13/pAnFd cells by enzymes saturated for mutations at residue 118 resulted in the selection of those containing only aromatic side chains in place of leucine at this position (Fig. 3 D–F). Mutants containing either the bulkier tyrosine or tryptophan at amino acid 118 were most active with 16:0-ACP (Fig. 3 D and E), although the specific activity with this substrate was more than 6- to 12-fold lower than that observed with the G188L mutant. It should be noted that the L118Y mutant was prone to precipitation during purification and purified as two peaks using cation exchange chromatography. The apparent instability of this enzyme during purification may account for its relatively low in vitro activity (Fig. 3E). Mutants containing a phenylalanine substitution at residue 118 were most active with 18:0-ACP; however, the specific activity with 16:0-ACP was nearly 4-fold greater than that of the wild-type enzyme (Fig. 3F).
The enzymes selected from a library saturated for mutations at the codon 114 varied depending on whether complementation was performed in liquid culture or directly on plates lacking UFA (see Materials and Methods). The predominant enzyme selected by complementation in liquid culture contained an isoleucine at amino acid 114 in place of the methionine found in the wild-type enzyme. This mutant, although most active in vitro with 18:0-ACP, was 2-fold more active with 16:0-ACP and nearly 5-fold more active with 14:0-ACP than the wild-type castor Δ9-18:0-ACP DES (Fig. 3G). In contrast, the only mutants that were identified by complementation directly on plates lacking UFA were those that contained either tyrosine or phenylalanine at residue 114. Enzymes with either amino acid substitution displayed qualitatively and quantitatively similar in vitro activity. This substrate specificity profile of the mutant enzymes was characterized by a 6-fold increase in specific activity with 14:0-ACP relative to the wild-type enzyme (Fig. 3H). Interestingly, the M114F and M114Y mutants were more active with 14:0-ACP than with 16:0-ACP. Consistent with this, >50% of the fatty acids of cells expressing these mutants were monoenes originating from the Δ9 desaturation of 14:0-ACP (e.g., 14:1Δ9, 16:1Δ11, and 18:1Δ13).
Variants selected from the codon 179 library, although not examined as extensively as those from libraries described above, included mutant P179I that displayed a 7-fold and 20-fold increase in specific activity with 16:0-ACP and 14:0-ACP, respectively, compared with the wild-type castor Δ9-18:0-ACP DES (Fig. 3I).
Expression of the G188L Mutant in A. thaliana Seeds.
The results detailed above demonstrate the ability to identify in vitro activities with the E. coli-based mutant selection system. Additional studies were performed to assess whether the selected mutants could be used to generate altered unsaturated fatty acid compositions in seed oils of plants. These experiments focused on the G188L mutant of the castor Δ9-18:0-ACP DES (Fig. 3B), which displayed the highest in vitro specific activity of the selected mutants with a substrate other than 18:0-ACP. To ensure proper organellar targeting and appropriately timed expression, the ORF of a G188L mutant was linked to the plastid transit peptide of the coriander Δ4-16:0-ACP DES and placed behind the seed-specific napin promoter in a plant expression vector. The resulting expression constructs were introduced into A. thaliana by means of Agrobacterium-mediated transformation. In addition to wild-type plants, the fab1 mutant of Arabidopsis was used as a background for expression of the recombinant DES. The fab1 mutant contains a lesion in the gene for β-ketoacyl-ACP synthase II (30), which catalyzes the initial condensation reaction in the elongation of 16:0-ACP to 18:0-ACP. As a result, these plants have an elevated palmitic acid content (30), presumably because of an increase in the size of plastidic 16:0-ACP pools. Given that the G188L mutant is most active with 16:0-ACP (Fig. 3B), Arabidopsis fab1 plants thus were considered to be an ideal host for this variant.
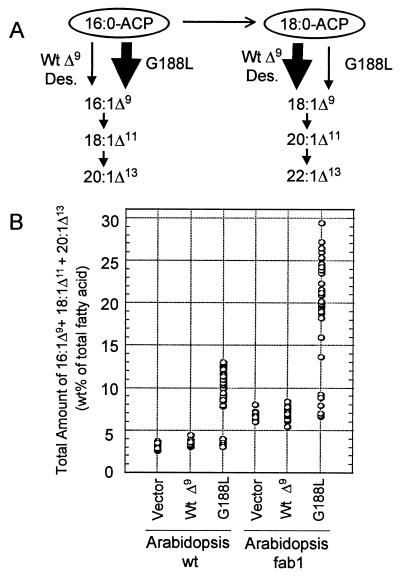
As shown in Fig. 4A, it was anticipated that the expression of the G188L mutant would result in enhanced metabolic flux into monounsaturated fatty acids derived from the Δ9 desaturation of 16:0-ACP. This would lead to increased amounts of not only 16:1Δ9, but also its elongation products 18:1Δ11 and 20:1Δ13, all of which are considered to be “unusual” fatty acids in plants. Consistent with this, seeds from 14 of 17 T1 lines transformed with the G188L mutant in wild-type Arabidopsis displayed 2- to 4-fold increases in amounts of these fatty acids relative to seeds from vector control plants. A similar enrichment in amounts of 16:1Δ9, 18:1Δ11, and 20:1Δ13 also was observed in seeds from six of nine T1 lines transformed with the G188L mutant in the Arabidopsis fab1 background. In contrast, seeds from plants transformed with the wild-type castor Δ9-18:0-ACP DES in either Arabidopsis background displayed little or no alteration in total amounts of 16:1Δ9, 18:1Δ11, and 20:1Δ13 relative to seeds from vector control plants.
Figure 4.
Diversion of fatty acid flux into unusual monounsaturated fatty acids derived from the Δ9 desaturation of 16:0-ACP by expression of the G188L mutant of the castor Δ9-18:0-ACP DES. Consistent with its in vitro activity (see Fig. 3B), expression of the G188L mutant in transgenic Arabidopsis seeds results in preferential use of 16:0-ACP pools to produce the unusual plant fatty acids 16:1Δ9, 18:1Δ11, and 20:1Δ13, as illustrated in A. This contrasts with the wild-type Δ9-18:0-ACP DES, which preferentially uses pools of 18:0-ACP to generate 18:1Δ9 and its elongation products 20:1Δ11 and 22:1Δ13. (B) The total amount of 16:1Δ9, 18:1Δ11, and 20:1Δ13 in single segregating seeds from a representative T1 plant transformed with expression constructs for either the wild-type castor Δ9-18:0-ACP DES (Wt Δ9), the G188L mutant (G188L), or the vector alone (Vector) in a wild-type or fab1 Arabidopsis background. Each data point represents the fatty acid composition of a single seed, and for each of the transformation events shown, 25–35 single seeds were analyzed.
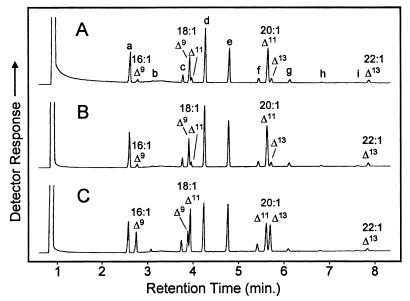
Analyses of single seeds from T1 plants indicated that wild-type Arabidopsis transformed with the G188L mutant accumulated 16:1Δ9, 18:1Δ11, and 20:1Δ13 to amounts as high as 13% of the total fatty acids of the seed oil (Fig. 4B). In contrast, these fatty acids accounted for 3–4% of the total fatty acids in seeds of wild-type Arabidopsis transformed with the expression vector alone or with the wild-type castor Δ9-18:0-ACP DES. The background level of these fatty acids presumably arises from the low, but detectable, activity of the wild-type castor Δ9-18:0-ACP DES with 16:0-ACP (Fig. 3A). In addition, seeds from the Arabidopsis fab1 background transformed with the G188L mutant accumulated the unusual monounsaturated fatty acids to amounts as high as 25–29% of the total fatty acids, which was nearly 4-fold higher than that detected in control plants (Figs. 4B and 5). Overall, these results demonstrate that the altered in vitro activity displayed by the G188L mutant also is revealed in the seed oil of transgenic plants expressing this enzyme.
Figure 5.
Gas chromatographic analyses of fatty acid methyl esters prepared from single seeds of Arabidopsis fab1 plants transformed with the expression vector (A), the wild-type castor Δ9-18:0-ACP DES (B), or the G188L mutant of the castor Δ9-18:0-ACP DES (C). Peaks corresponding to the monounsaturated fatty acid components are labeled. The additional peaks correspond to the following fatty acids: a, 16:0; b, 16:2Δ9,12; c, 18:0; d, 18:2Δ9,12; e, 18:3Δ9,12,15; f, 20:0; g, 20:2Δ11,14; h, 20:3Δ11,14,17; i, 22:0.
Discussion
We have demonstrated the ability of an E. coli UFA auxotroph to serve as an efficient selection system for the isolation of variants of the Δ9-18:0-ACP DES with enhanced activity for C14 and C16 fatty acid substrates. The mutagenesis-based complementation approach described is a significant advantage over rational design, because mutations that result in subtle changes in the position of substrates in the active site often lead to large and undesired changes in kinetic parameters that cannot be predicted with confidence from crystallographic models. Thus, an alternative approach to rational design is to use the structural model as a guide to select positions likely to affect the function of the enzyme and to use saturation mutagenesis at these locations to introduce the maximum local structural variation. The selection system then is used to discriminate between the different enzymes based on their enzymatic properties.
Using this approach, four amino acids that line the bottom of the substrate binding pocket of the castor Δ9-18:0-ACP DES based on the crystallographic model (8, 9) were targeted for saturation mutagenesis. Subsequent selection by complementation of the E. coli UFA auxotroph resulted in the identification of a large number of catalytically competent variants with altered substrate specificity. The selected mutants included several that displayed strong substrate preference for 16:0- rather than 18:0-ACP. For example, the ratio of specific activity with 18:0-ACP vs. 16:0-ACP was converted from 70:1 displayed by the wild-type enzyme to 1:25 in the case of the M114I/G188L double mutant. In general, the amino acid substitutions in the selected DES variants contained bulkier side chains than those of the wild-type enzyme, which is consistent with a reduced ability of the substrate binding pocket to accommodate the longer carbon chain of 18:0-ACP.
The selection system described offers several additional features for structural studies of acyl-ACP DES. For example, amino acid residues that affect substrate binding indirectly cannot be readily predicted from the crystal structure of the castor Δ9-18:0-ACP DES. These residues can instead be identified by random mutagenesis in conjunction with a substrate-dependent selection such as that described in this study. Another advantage of the auxotroph complementation system is that it not only selects for enzymes that are capable of desaturating shorter chain fatty acids, but it also can be used to identify the most catalytically competent enzymes that are active under physiologically relevant substrate concentrations. This contrasts with rational design studies conducted without selection, which typically require a “trial and error” approach to identify variant enzymes that display adequate solubility and stability and also possess the desired catalytic properties.
Selection under physiological substrate concentrations likely explains why the Kms measured for the variant DES from this study are similar to those of the wild-type enzyme. Although technical difficulties make precise values difficult to obtain, we estimate that the Kms for all of the mutants investigated are below 1 μM, and not substantially different from the 0.4 μM Km of the wild-type enzyme with 18:0-ACP measured under our assay conditions. For example, the double mutant M114I/G188L was found to have Kms of 0.6 μM and 0.7 μM with 16:0- and 18:0-ACP, respectively (data not shown), despite the fact that the specific activity for 18:0-ACP is reduced by 160-fold with respect to the wild-type enzyme. Thus, because the Kms are similar, the changes in substrate specificity (kcat/Km) likely results in large part from increases in kcat.
In addition to understanding the structure–function relationships of DES, a goal of these studies was to engineer enzymes that are capable of producing large amounts of novel monounsaturated fatty acids in a transgenic oilseed crop. Such fatty acids have utility in industrial applications such as the production of nylon precursors (31). As described, expression of a G188L mutant of the castor Δ9-18:0-ACP DES in Arabidopsis fab1 plants resulted in the accumulation of the unusual monoenes 16:1Δ9, 18:1Δ11, and 20:1Δ13 to amounts approaching 30% of the total fatty acids of the seed oil. Such a marked change in the unsaturated fatty acid composition of seed oil has yet to be achieved by the transgenic expression of any naturally occurring acyl-ACP DES variants (2). In addition to our findings, it recently has been shown that rationally designed acyl-ACP thioesterases can be used to alter the saturated fatty acid content of canola seeds (32). Together, these results demonstrate that mutagenesis of fatty acid biosynthetic and modification enzymes is a viable design strategy for the metabolic engineering of oilseed composition.
Acknowledgments
We thank Jim Jardine and Sarah Hall for assistance with Arabidopsis plants, Dr. Jane Setlow for editorial assistance, Dr. John Cronan for providing the E. coli MH13 cell line, and the Office of Basic Energy Sciences of the U.S. Department of Energy for financial support.
Abbreviations
- ACP
acyl carrier protein
- DES
desaturase
- UFA
unsaturated fatty acid
- IPTG
isopropyl β-d-thiogalactoside
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Fatty acid nomenclature: X:Y indicates that the fatty acid contains X numbers of carbon atoms and Y numbers of double bonds; Δz indicates that a double bond is positioned at the zth carbon atom from the carboxyl terminus.
Amino acid numbering corresponds to the sequence of the mature castor Δ9-18:0-ACP DES (see ref. 8).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210276297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210276297
References
- 1.Shanklin J, Cahoon E B. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- 2.Cahoon E B, Shanklin J, Ohlrogge J B. Proc Natl Acad Sci USA. 1992;89:11184–11188. doi: 10.1073/pnas.89.23.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahoon E B, Ohlrogge J B. Plant Physiol. 1994;104:827–844. doi: 10.1104/pp.104.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahoon E B, Cranmer A M, Shanklin J, Ohlrogge J B. J Biol Chem. 1994;269:27519–27526. [PubMed] [Google Scholar]
- 5.Schultz D J, Cahoon E B, Shanklin J, Craig R, Cox-Foster D L, Mumma R O, Medford J I. Proc Natl Acad Sci USA. 1996;93:8771–8775. doi: 10.1073/pnas.93.16.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cahoon E B, Shah S, Shanklin J, Browse J. Plant Physiol. 1998;117:593–598. doi: 10.1104/pp.117.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahoon E B, Coughlan S J, Shanklin J. Plant Mol Biol. 1997;33:1105–1110. doi: 10.1023/a:1005821007291. [DOI] [PubMed] [Google Scholar]
- 8.Lindqvist Y, Huang W, Schneider G, Shanklin J. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 9.Cahoon E B, Lindqvist Y, Schneider G, Shanklin J. Proc Natl Acad Sci USA. 1997;94:4872–4877. doi: 10.1073/pnas.94.10.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cahoon E B, Mills L A, Shanklin J. J Bacteriol. 1996;178:936–939. doi: 10.1128/jb.178.3.936-939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohlrogge J, Savage L, Jaworski J, Voelker T, Post-Beittenmiller D. Arch Biochem Biophys. 1995;317:185–190. doi: 10.1006/abbi.1995.1152. [DOI] [PubMed] [Google Scholar]
- 12.Henry M F. Ph.D. thesis. Urbana-Champaign: University of Illinois; 1992. [Google Scholar]
- 13.Clark D P, de Mendoza D, Polacco M P, Cronan J E., Jr Biochemistry. 1983;22:5897–5902. doi: 10.1021/bi00294a032. [DOI] [PubMed] [Google Scholar]
- 14.Simons R W, Egan P A, Chute H T, Nunn W D. J Bacteriol. 1980;142:621–632. doi: 10.1128/jb.142.2.621-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng H, Westler W M, Xia B, Oh B-H, Markley J L. Arch Biochem Biophys. 1995;316:619–634. doi: 10.1006/abbi.1995.1082. [DOI] [PubMed] [Google Scholar]
- 16.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shanklin J, Somerville C. Proc Natl Acad Sci USA. 1991;88:2510–2514. doi: 10.1073/pnas.88.6.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cahoon E B, Becker C K, Shanklin J, Ohlrogge J B. Plant Physiol. 1994;106:807–808. doi: 10.1104/pp.106.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 20.Roughan G, Nishida I. Arch Biochem Biophys. 1990;276:38–46. doi: 10.1016/0003-9861(90)90007-l. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto K, Shibahara A, Nakayama T, Kajimoto G. Chem Phys Lipids. 1991;60:39–50. [Google Scholar]
- 22.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Rock C O, Garwin J L. J Biol Chem. 1979;254:7123–7128. [PubMed] [Google Scholar]
- 24.Beremand P D, Hannapel D J, Guerra D J, Kuhn D N, Ohlrogge J B. Arch Biochem Biophys. 1987;256:90–100. doi: 10.1016/0003-9861(87)90428-0. [DOI] [PubMed] [Google Scholar]
- 25.Broun P, Shanklin J, Whittle E, Somerville C. Science. 1998;282:1315–1317. doi: 10.1126/science.282.5392.1315. [DOI] [PubMed] [Google Scholar]
- 26.Bechtold N, Pelletier G. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 27.Butte W, Eilers J, Kirsch M. Anal Lett. 1982;15:841–850. [Google Scholar]
- 28.Thompson G A, Scherer D E, Foxall-Van Aken S, Kenny J W, Young H L, Shintani D K, Kridl J C, Knauf V C. Proc Natl Acad Sci USA. 1991;88:2578–2582. doi: 10.1073/pnas.88.6.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguilar P S, Cronan J E, Jr, de Mendoza D. J Bacteriol. 1998;180:2194–2200. doi: 10.1128/jb.180.8.2194-2200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J, James D W, Jr, Dooner H K, Browse J. Plant Physiol. 1994;106:143–150. doi: 10.1104/pp.106.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohlrogge J B. Plant Physiol. 1994;104:821–826. doi: 10.1104/pp.104.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Facciotti M T, Bertain P B, Yuan L. Nat Biotechnol. 1999;17:593–597. doi: 10.1038/9909. [DOI] [PubMed] [Google Scholar]