Abstract
TransportDB (http://www.membranetransport.org/) is a comprehensive database resource of information on cytoplasmic membrane transporters and outer membrane channels in organisms whose complete genome sequences are available. The complete set of membrane transport systems and outer membrane channels of each organism are annotated based on a series of experimental and bioinformatic evidence and classified into different types and families according to their mode of transport, bioenergetics, molecular phylogeny and substrate specificities. User-friendly web interfaces are designed for easy access, query and download of the data. Features of the TransportDB website include text-based and BLAST search tools against known transporter and outer membrane channel proteins; comparison of transporter and outer membrane channel contents from different organisms; known 3D structures of transporters, and phylogenetic trees of transporter families. On individual protein pages, users can find detailed functional annotation, supporting bioinformatic evidence, protein/DNA sequences, publications and cross-referenced external online resource links. TransportDB has now been in existence for over 10 years and continues to be regularly updated with new evidence and data from newly sequenced genomes, as well as having new features added periodically.
INTRODUCTION
Membrane transporters are a large group of proteins that span the cell membrane and form an intricate system of pumps and channels through which they deliver essential nutrients, eject waste products and assist the cell to sense environmental conditions. Transporters represent a large and diverse group of proteins that differ in membrane topology, energy coupling mechanism and substrate specificities. They play indispensable roles in the fundamental cellular processes of all organisms (1).
With the advent of the genomics era, comprehensive genome-wide bioinformatic comparisons of predicted membrane transporters across a range of organisms in all three domains of life have become possible. Previously, we have reported a series of comparative analyses of transport systems in a collection of prokaryotic and eukaryotic organisms (2–4). We started a web portal showing our bioinformatic prediction of transporters in sequenced genomes back in 1996, and have had a continual web presence since then. The current incarnation of TransportDB dates back to 2002, when we moved to a relational database structure and greatly enhanced the available features (5). The aim of TransportDB is to present the comprehensive transporter profiles of each sequenced prokaryotic and eukaryotic organisms, as well as to provide comparative and phylogenetic tools to view, search, compare and download the transporter data in an easy-to-navigate format. We describe in this paper the data content and web features of TransportDB, with a focus on the recent additions and improvements.
DATABASE STRUCTURE AND CONTENT
TransportDB uses a relational database to store all the data associated with membrane transporters and outer membrane channels. It was built specifically to hold many different genomes and to allow cross-genome queries and comparisons. TransportDB database consists of 20 tables and is implemented in MySQL (http://www.mysql.com/). Data stored in TransportDB database can be accessed using Structured Query Language (SQL). Users can search TransportDB via a web interface which facilitates building a custom query without interacting directly with the database. Examples include searches for transporter class, family, protein and substrate. The relational database format also allows users to select a subset of organisms of their interest and compare the overall transporter features as well as each individual transporter family.
TransportDB adopts a ‘3-tier’ database architecture. The top tier of TransportDB is the user web interface. It sends the requests for data, formats the output of the query and displays it on the web. The Application Programmer Interface (API), or middle tier, connects the database and retrieves datasets by explicit keys (e.g. the name of a transporter gene) using a single query. We adopt PHP (http://www.php.net/), a widely used server-side scripting language, as our API. The database itself is the bottom tier. This architecture enables the web application to utilize SQL to query the database, while not limiting the top tier to any specific database system.
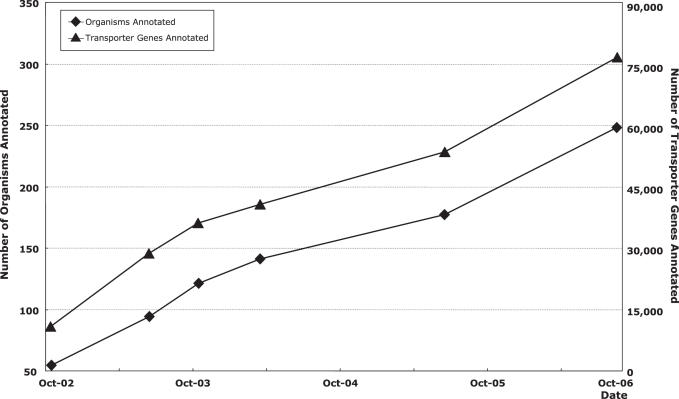
TransportDB stores the complete array of predicted transporters and outer membrane channels from various prokaryotic and eukaryotic organisms, with detailed information and supporting evidence for each protein. Figure 1 shows the evolution of data collection in TransportDB. When we moved to a relational database format in 2002, we had bioinformatic analyses of 54 organisms and over 10 000 transporter genes (5). During the past 4 years, a very considerable number of organisms have been added to the database and the total number of annotated transporter genes has increased by a factor of seven. Currently, TransportDB contains data from 248 organisms, including 197 bacteria, 24 archaea and 27 eukaryota. This collection of organisms represents a broad phylogenetic diversity. A total of 71 659 transport proteins and 4790 outer membrane channels have been annotated and assigned to 183 families according to the TC classification system (1,6). These families are further classified into different types according to their transport mode and energy coupling mechanism: seven families of primary active transporter that couple the transport process with a primary energy source (e.g. ATP hydrolysis); 83 families of secondary transporter that utilize an ion or solute electrochemical gradient; 33 families of energy-independent channels; two families of group translocators which modify their substrates during transport; 38 families of porins/outer membrane channels that are prevalent in the outer membrane of Gram-negative bacteria and certain eukaryotic organelles; and 11 families with an unknown transport mechanism. Transporters are unevenly distributed among these families: some are very large superfamilies with thousands of members, such as the ABC superfamily (7) (32 099 proteins annotated) and the MFS superfamily (8) (7942 proteins), both of which are widely distributed across prokaryotic and eukaryotic species; some families, however, only exist in a very limited phylogenetic spectrum and/or are present in only limited numbers.
Figure 1.
The evolution of TransportDB data storage. The rhomboid points represent number of organisms annotated over 4 years. The triangle points show the number of transporter genes annotated.
DATABASE ACCESS AND WEB FEATURES
TransportDB is accessible online at http://www.membranetransport.org/.
There are several ways for users to access data stored in TransportDB. Users can browse the database by selecting the organism from drop-down boxes on the left of the web page, or by clicking the links from the ‘Organism List’ page (Figure 2). All the transport proteins are listed in a tabular format with predicted substrate or function. A hierarchical top-down structure was deployed for easy data access, which arranges transporters in the orders of kingdom (bacteria, archaea or eukaryota), organism, transporter type, transporter family/subfamily and transport protein. Each transport protein is presented in separate web pages where users can find detailed information such as transporter substrate/function annotation, TC classification, transmembrane segment prediction by TMHMM (9), genomic locus information, protein/DNA sequence, etc. Evidence types associated with functional annotation are included, such as hidden Markov models [Pfam (10) and TIGRfam (11)], BLAST (12) and COG (13) data. Cross-referenced links to external database are also provided, including Entrez Gene (14), TIGR's Comprehensive Microbial Resource (CMR) (15), MIPS (16), EcoCyc (17) and PubMed. A keyword-based text search is available for users to search by criteria such as transporter type, transporter family, transporter protein name or substrate. The results are grouped by transporter family and organism. Each result contains links to individual family and protein pages. The protein and DNA sequences in TransportDB are readily available for BLAST search. Users can submit a single peptide or nucleotide sequence in the ‘Blast’ section. The output of the BLAST search includes transporter family information (TC number, family name) in addition to the standard features.
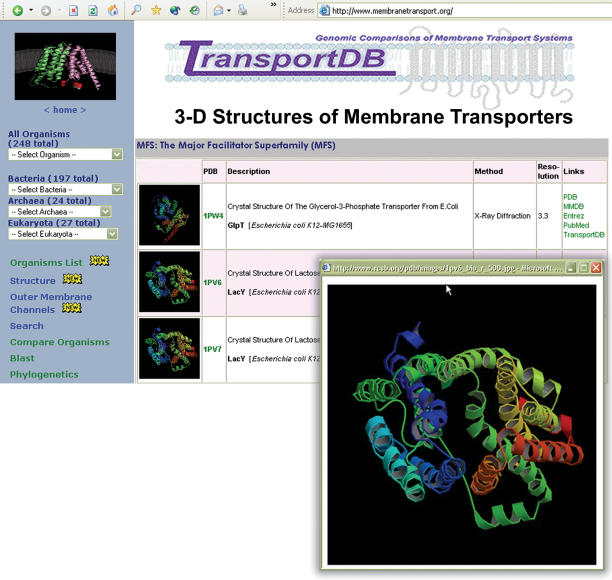
Figure 2.
Graphic illustrations of the TransportDB web interface describing 3D structures of membrane transporters. These structures are listed in a tabular format and arranged by transporter families. Information on structure description, method and resolution are included. Cross-referenced links to PDB, PDB_TM, MMDB, Entrez Gene and PubMed are also provided.
The relational database format allows easy manipulation of the data stored in TransportDB. An overview page is accessible for each organism, summarizing its complete transporter content, including transporter types and individual transporter families, and their statistics. Users can choose any two or more organisms from the ‘Compare Organisms’ section to compare their transport gene complement. All these results are generated on the fly to reflect the most recent updates. In the ‘Phylogenetics’ section, users can view the pre-computed neighbor-joining trees for each of the transporter families through an ATV java applet (18). This enables users to access the up-to-date phylogenetic trees of every transporter family, and to manipulate the trees to display subtrees, zoom in and out, or collapse subtrees to single nodes, etc. Transport proteins in each family are also available for download in FASTA or multiple sequence alignment formats.
RECENT FEATURE ENHANCEMENT
In addition to bioinformatic predictions, we have recently begun to comprehensively track experimental evidence for transporter gene function based on the primary literature. We retrieved from Entrez (19) all related publications on transport proteins in TransportDB by e-utilities (20), which submitted queries containing gene name and organism to NCBI server and returned the related literature. The publications on genomic sequencing and massive gene expression studies were manually excluded. A total of 13 936 PubMed entries were retrieved which covers 3862 transporters and outer membrane channels. The abstracts of all these literature as well as links to PubMed are accessible at the individual transporter protein pages.
A new section has been added to TransportDB describing experimentally determined 3D structures of membrane transporters from crystallization or NMR-based studies. The data to populate this new section were derived by searching our entire collection of transport proteins against the protein data bank (PDB) (21). A total of 273 structures were retrieved, representing 98 transporters (multiple structures are available for some transporters). On the TransportDB website, these structures are listed in a tabular format and arranged by transporter families (Figure 2). Information on structure description, method and resolution are included. Cross-referenced links to PDB, PDB_TM (22), MMDB (23), Entrez Gene and PubMed are also provided. Membrane transporters represent 3–12% of total proteins of various organisms (2–4). Currently there are more than 39 000 structures deposited in the PDB. Membrane transporters are highly underrepresented and consist of <1% of all structures. This lack of representation reflects the difficulties in the purification and crystallization of transporter proteins due to their hydrophobic nature and solubility only in the presence of detergents.
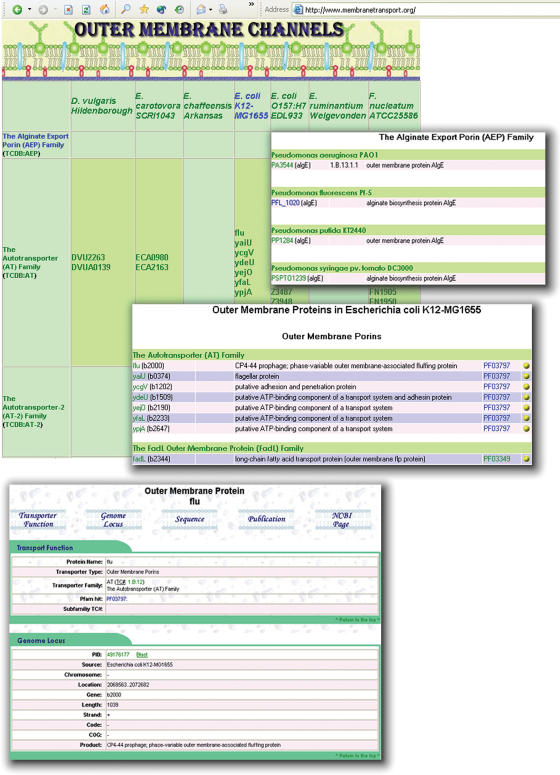
Another recently added section to TransportDB describes outer membrane channels. Gram-negative bacteria and certain eukaryotic organelles, such as mitochondria and peroxisomes, are characteristically surrounded by an outer membrane that shows little permeability for hydrophilic solutes. Outer membrane proteins form nonspecific diffusion channels across the outer membrane to allow the influx of nutrients as well as the extrusion of wastes (24). A total of 4664 outer membrane channels from 142 organisms are currently annotated in TransportDB and classified into 38 families according to the TC classification. These proteins are listed in a tabular format in the ‘Outer Membrane Channels’ section (Figure 3). Each outer membrane channel has an individual page showing supporting bioinformatic evidence, protein/DNA sequences, publications and cross-referenced external links. Users can also pull out a list of outer membrane channels from a specific organism, or a list of proteins from a specific family in all the organisms.
Figure 3.
Graphic illustrations of the TransportDB web interface describing outer membrane channels. Proteins are presented in a tabular format. Each outer membrane channel has individual pages showing supporting bioinformatic evidence, protein/DNA sequence, publications and cross-referenced external links, etc. Users can also pull out a list of outer membrane channels from a specific organism, or a list of proteins from a specific family in all organisms.
FUTURE PERSPECTIVES
In summary, TransportDB was developed as a relational database for the comprehensive representation of cytoplasmic membrane transport systems. This is the only active database in the field dedicated to the comprehensive and comparative study of membrane transporters and outer membrane channels in different organisms with fully sequenced genomes. We are continuing to expand the TransportDB database to incorporate data from newly published genomes. TransportDB will be routinely updated with new annotation information and with data from newly sequenced organisms.
New enhancements that we are focusing on for the short- to medium-term future include the following: (i) Make our transporter annotation pipeline available to the public through our web portal so that they may customize it for their genome annotation efforts. Over the past 4 years, we have undertaken transporter annotation of over 60 unpublished genomes from custom requests from a broad range of different research groups (these data are not released to public until the publication of the relevant genome paper or until the genome data has been deposited into the public databases). We believe that providing access to our annotation pipeline through the web will serve to fulfill the increasing demands of such efforts. (ii) Add additional bioinformatic analyses to the transporter annotation pipeline, such as examining the genomic context of candidate genes, and increased use of phylogenetic approaches. (iii) Move to using a controlled vocabulary, a carefully selected list of words and phrases, for membrane transporter substrate prediction, so that they may be retrieved by searches more efficiently. The controlled vocabulary for substrate prediction can also facilitate substrate specificity comparisons and aid the automatic derivation of transport reaction equations for metabolic modeling and flux balance analysis for different sequenced genomes. As a starting point, we plan to use the hierarchical compound lists that are defined in the MetaCyc database (25).
Acknowledgments
We would like to thank all the past contributors to TransportDB. We would also like to thank Dr Martin Wu and Dr Dongying Wu for their assistance on the construction of the phylogenetic trees and for their helpful feedback on other aspects of TransportDB website. Funding to pay the Open Access publication charges for this article was provided by TIGR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Saier M.H., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 2000;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paulsen I.T., Sliwinski M.K., Saier M.H., Jr Microbial genome analyses: global comparisons of transport capabilities based on phylogenies, bioenergetics and substrate specificities. J. Mol. Biol. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 3.Paulsen I.T., Nguyen L., Sliwinski M.K., Rabus R., Saier M.H., Jr Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 4.Ren Q., Paulsen I.T. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 2005;1:190–201. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren Q., Kang K.H., Paulsen I.T. TransportDB: a relational database of cellular membrane transport systems. Nucleic Acids Res. 2004;32:D284–D288. doi: 10.1093/nar/gkh016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saier M.H., Jr, Tran C.V., Barabote R.D. TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomii K., Kanehisa M. A comparative analysis of ABC transporters in complete microbial genomes. Genome Res. 1998;8:1048–1059. doi: 10.1101/gr.8.10.1048. [DOI] [PubMed] [Google Scholar]
- 8.Pao S.S., Paulsen I.T., Saier M.H., Jr Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 10.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Tatusov R.L., Natale D.A., Garkavtsev I.V., Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33:D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson J.D., Umayam L.A., Dickinson T., Hickey E.K., White O. The comprehensive microbial resource. Nucleic Acids Res. 2001;29:123–125. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mewes H.W., Frishman D., Mayer K.F., Munsterkotter M., Noubibou O., Pagel P., Rattei T., Oesterheld M., Ruepp A., Stumpflen V. MIPS: analysis and annotation of proteins from whole genomes in 2005. Nucleic Acids Res. 2006;34:D169–D172. doi: 10.1093/nar/gkj148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zmasek C.M., Eddy S.R. ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics. 2001;17:383–384. doi: 10.1093/bioinformatics/17.4.383. [DOI] [PubMed] [Google Scholar]
- 19.Schuler G.D., Epstein J.A., Ohkawa H., Kans J.A. Entrez: molecular biology database and retrieval system. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 20.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Church D.M., DiCuccio M., Edgar R., Federhen S., Helmberg W., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tusnady G.E., Dosztanyi Z., Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Anderson J.B., DeWeese-Scott C., Fedorova N.D., Geer L.Y., He S., Hurwitz D.I., Jackson J.D., Jacobs A.R., Lanczycki C.J., et al. MMDB: Entrez's 3D-structure database. Nucleic Acids Res. 2003;31:474–477. doi: 10.1093/nar/gkg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caspi R., Foerster H., Fulcher C.A., Hopkinson R., Ingraham J., Kaipa P., Krummenacker M., Paley S., Pick J., Rhee S.Y., et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2006;34:D511–D516. doi: 10.1093/nar/gkj128. [DOI] [PMC free article] [PubMed] [Google Scholar]