Abstract
Frequency of INherited Disorders database (FINDbase) (http://www.findbase.org) is a relational database, derived from the ETHNOS software, recording frequencies of causative mutations leading to inherited disorders worldwide. Database records include the population and ethnic group, the disorder name and the related gene, accompanied by links to any corresponding locus-specific mutation database, to the respective Online Mendelian Inheritance in Man entries and the mutation together with its frequency in that population. The initial information is derived from the published literature, locus-specific databases and genetic disease consortia. FINDbase offers a user-friendly query interface, providing instant access to the list and frequencies of the different mutations. Query outputs can be either in a table or graphical format, accompanied by reference(s) on the data source. Registered users from three different groups, namely administrator, national coordinator and curator, are responsible for database curation and/or data entry/correction online via a password-protected interface. Databaseaccess is free of charge and there are no registration requirements for data querying. FINDbase provides a simple, web-based system for population-based mutation data collection and retrieval and can serve not only as a valuable online tool for molecular genetic testing of inherited disorders but also as a non-profit model for sustainable database funding, in the form of a ‘database-journal’.
INTRODUCTION
As molecular genetic testing and electronic healthcare records become increasingly common features of modern medical practice, there is a need to integrate information in genetic databases to establish a detailed understanding of how genome sequence differences impact on human health. To date, there are several depositories that fall under the banner of ‘mutation databases’, which can be divided into three main categories: central (or core) databases, such as Online Mendelian Inheritance in Man [OMIM, http://www3.ncbi.nlm.nih.gov/omim; (1)], the Human Gene Mutation Database [http://www.hgmd.org; (2)], locus-specific databases (LSDBs), a large group with over 570 members [http://www.centralmutations.org; reviewed in Ref. (3)] and National/Ethnic Mutation databases [NEMDBs, (4)].
The recent emergence of National and Ethnic Mutation Databases (NEMDBs) is justified by the fact that the spectrum of mutations observed for any gene or disease will oftendiffer not only between population groups across the planet but also between distinct ethnic groups within a geographical region. NEMDBs provide data that can be used to e.g. stratify national molecular diagnostic services, study human demographic history, admixture patterns and gene/mutation flow (5,6).
We have previously described the development of specialized software, namely ETHNOS (7), which has facilitated the construction of the Hellenic, Cypriot, Iranian, Lebanese and Serbian NEMDBs (7–9). However, this software could not handle advanced data querying and storing of large datasets. Here, we report the construction of Frequency of INherited Disorders database (FINDbase), a relational database based on an upgraded version of the ETHNOS software, capable to accommodate large datasets pertaining to frequencies of causative mutations, leading to inherited disorders in various populations and ethnic groups worldwide.
DATABASE DESCRIPTION
Primary data sources and database contents
The initial information data sources in FINDbase were acquired from the European Union FP5 Cystic Fibrosis Thematic Network Consortium data (www.cfnetwork.be), also reported by the World Health Organization (http://www.who.int/genomics/publications/en/HGN_WB_04.02_report.pdf; ‘The molecular epidemiology of cystic fibrosis’) and partially published elsewhere (10), and population-specific mutation frequency data derived from two LSDBs, namely the HbVardatabase of human hemoglobin variants and thalassemia mutations [http://globin.bx.psu.edu/hbvar; (11–14)] and the PAHdb phenylalanine hydroxylase locus knowledgebase [http://www.pahdb.mcgill.ca; (15,16)]. In addition, original and review articles retrieved by manual searching of all articles listed in the PubMed literature database (http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=PubMed), international conference proceedings and personal communications by research groups also consist part of FINDbase data source. The information source (publications, conference proceedings, etc.) is displayed by selecting the ‘Submissions’ option, located in the ‘Overview’ section. In case of multiple articles on a single genetic disorder in a certain population group, calculation of mutation frequencies is based on a single and most representative study, involving sufficient numbers of patients and controls. Estimation of mutation frequencies based on multiple reports has the inherent danger of including redundant cases, which can alter the calculated frequencies. By only including number of chromosomes and not sensitive personal data of their carriers, the important issue of anonymity is adequately preserved.
FINDbase is the richest NEMDB among those currently available content-wise (4) with a total of 3 379 records and contains information on 31 different inherited disorders studied in 83 population groups, resulting from 1 223 causative mutations in 24 genomic regions (October 2006 release). Automated summary listings on FINDbase contents can be generated by selecting the respective options (‘Records’, ‘Disorders’, ‘Populations’, ‘Mutations’ and ‘Genes’ respectively) in the menu, located at the left side of the screen (see also Supplementary Figure).
DATABASE DESIGN, IMPLEMENTATION AND ACCESS
FINDbase was developed to facilitate easy creation and maintenance of fully web-based population-specific databases, is platform-independent and uses PHP and MySQL (http://www.mysql.com, MySQL AB, Uppsala, Sweden), open source software. At present, data are stored in a single data table, although a future upgrade includes splitting the data in multiple tables, corresponding to each population group documented in FINDbase (see below). In other words, FINDbase will ultimately be a collection of many NEMDBs, operating under the same software. Database design follows all content criteria and Human Genome Variation Society (HGVS; http://www.hgvs.org) recommendations. Finally, FINDbase design follows certain database guidelines in order to conform to quality (17).
FINDbase is a freely available online resource, which can be accessed on the World Wide Web at the URL address: http://www.findbase.org. Detailed instructions for both using and querying the database are also available from the same site (‘User guide’). There has been no claim of ownership of the information stored in this database by anyone involved in this initiative. However, this compilation and representation of it are subject to copyright and usage principles to ensure that FINDbase and its contents remains freely available to all interested individuals.
REGISTRATION AND DATA ENTRY
In FINDbase, data entry and modification is only possible for registered users. There are three levels of registered users, as outlined below in hierarchical order:
Administrators have full access rights to all database's functionalities and contents.
National coordinators have data entry and modification rights, registration and account activation for advisors/curators and data re-allocation to another advisor/curator. National coordinators are responsible for managing the overall construction and maintenance of a NEMDB, contributing to FINDbase. Their role is also to promote the usage of their NEMDB in their population and to promote investigations on those genetic disorders, which mutation spectrum is not yet known in their countries.
Advisors and curators have data entry and modification rights only for those data entered by him/herself and under no circumstances can alter data entered by another advisor/curator. Advisors and curators are responsible for data entry in the NEMDB, for which they are registered. If an advisor/curator wish to end his involvement with FINDbase, their data are allocated to another curator who will then be responsible for their curation.
National coordinator or advisor/curator accounts can be requested in the corresponding (‘Register’) section. The request is then automatically sent to the administrator and national coordinator of that particular NEMDB respectively. The account is subject to eligibility criteria, such as active involvement in the area of Human Genetics, and thorough knowledge of the genetic basis of inherited disorders in a particular population.
The data entry page can be found in the ‘Curators’ section. Once logged in, the registered user connects with Publication data editor, for further guidance through the data entry procedure. Each entry contains empty fields in a Table format, where the registered user enters the genetic disorder, the gene, accompanied by the respective OMIM ID, the mutation in the correct (official) nomenclature, the number of chromosomes or mutation frequencies. Population name is automatically set, depending on the population/NEMDB for which the curator is registered, and, where available, the entry of the ethnic group or geographical region is required.
QUERYING THE DATABASE
FINDbase provides a browsing/filtering interface to the underlying data allowing one to formulate queries in order to better explore the variety and depth of information recorded therein. Such queries can be useful in a clinical setting to reach a diagnosis or to better coordinate mutation screening. Stored data can be queried via the Search page in two different ways, namely automated summary listings, sorted by Disorder, Gene or Mutation and user-defined queries, by Population or Mutation. FINDbase does not yet provide a function to formulate ad hoc queries.
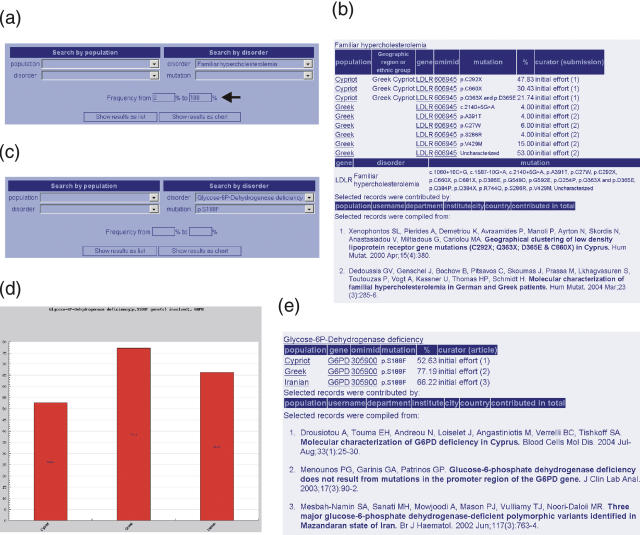
Two characteristic sample queries are shown in Figures 1 and 2. First of all, a user can query for all the different inherited disorders found in a particular population, such as all the Hungarian population. In this case, the user needs either to click on Hungary's capital from the world map (Figure 1a) in the Home page or to select for the ‘Hungarian’ population from the population list in the Home (Supplementary Figure) or Search page (Figure 1b). Query results are then provided in a summary list, displaying the disorder, the gene found mutated and the mutations found in the population in question. If a genetic disorder is further specified for the selected population, the query returns a detailed list of all relevant mutations together with their frequencies and data source in a table of graphical format. In the latter case, a chart is automatically created with the corresponding mutation frequencies shown in the bars (Figure 1c and d), while mutation frequencies from different ethnic groups or geographical regions are displayed in different colors (Figure 1d). The disorder and gene names and OMIM ID are hyperlinked to further information in the OMIM central database. The Search page also allows the user to insert the desired frequency range (Figures 1d and 2a).
Figure 1.
Querying FINDbase for frequencies of mutations leading to inherited disorders in a certain population. (a) The world map in the Home page, from where the user can quickly select the desired population to query upon by clicking on a country's capital (in this case, Hungary). A communication box automatically appears containing the corresponding national flag and the number of disorders currently documented in FINDbase. (b) Construction of the query ‘Find all inherited disorders in the Hungarian population’. Only parts of the Search page are shown. The user needs either to click on Hungary's capital from the world map (as previously described) or to select for the ‘Hungarian’ population from the population list, located either in the Home page [next to the world map (Supplementary Figure)] or at the left part of the Search page. Query results are in a table format, indicating the disorder, the gene found mutated and the mutations in their official nomenclature. (c) Construction of the query ‘Find all mutations leading to β-thalassemia in the Hungarian population’. By selecting ‘Hungarian’ and ‘Beta-thalassemia’ from the population and disorder's list respectively, the query returns a list of all β-thalassemia mutations found in the Hungarian population together with their frequencies and data source in a table (by selecting the ‘Show results as table’ button) or graphical format (by selecting the ‘Show results as chart’ button). The disorder and gene names and OMIM ID are hyperlinked to further information in the OMIM central database. (d) Sample query ‘Find all mutations leading to Cystic Fibrosis in the Austrian population with a frequency range of 1% and above’. By selecting ‘Austrian’ and ‘Cystic Fibrosis’ from the population and disorder's list respectively and specifying the desired frequency range in the corresponding boxes (indicated with an arrow), the query returns a list of all CFTR mutations with a frequency range of 1% and above found in the Austrian population together with their data source in a table or graphical format. In the latter case, the different CFTR mutation frequencies found in different geographical regions are differentially displayed, with the corresponding mutation frequencies shown in the bars.
Figure 2.
Querying FINDbase for frequencies of mutations leading to a certain inherited disorder in different populations. (a) Construction of the query ‘Find frequent (between 2 and 100%) familial hypercholesterolemia mutations in all populations’. (b) The user needs to select for the ‘Familiar hypercholesterolemia’ disorder from the disorder's menu, located either in the Home page [next to the world map (Supplementary Figure)] or at the right part of the Search page. Query results are in a table format, indicating the populations and ethnic groups (if applicable) where LDLR mutations are found. Information is accompanied by the data source. (c) Construction of the query ‘Find the frequency of the p.S188F mutation leading to Glucose 6P-Dehydrogenase (G6PD) deficiency in all populations’. By selecting ‘Glucose 6P-Dehydrogenase deficiency’ from the disorder's list and ‘p.S188F’ from the mutation's list, the query returns a list of populations in which the p.S188F mutation is found together with the corresponding frequencies. Query output can be in a graphical (d) or table format (e), always accompanied by the data source.
FINDbase data can be also queried by disorder and refined by mutation. In this case, the user needs to select from the disorder's menu, located either in the Home page [next to the world map (Supplementary Figure)] or at the right part of the search page (Figure 2a) and refine the search by specifying the mutation name (Figure 2c). Query results are again in a table (Figure 2b and e) or graphical format (Figure 2d), indicating the populations and ethnic groups (if applicable) where this disorder is found.
CONCLUSIONS AND FUTURE PROSPECTS
NEMDBs are comprehensive sources of information on the extant genetic heterogeneity of different populations. National and ethnic-specific mutant alleles, identified from diagnostic laboratories, should be ideally stored in such databases to become freely accessible to all individuals, researchers and laboratories involved in molecular genetic testing in an either national or even international scale. Combining relevant information from many population groups and with numerous links for the documented genetic disorders, genes and LSDBs, FINDbase has the potential to expand significantly to become a reference source of information on the incidence of inherited disorders in various populations.
FINDbase is being upgraded: (i) to enrich the existing data collection, (ii) to upgrade ETHNOS software, (iii) to optimize database content and quality management. Upgrade is estimated to last ∼2 years, while funding for maintenance and upgrade is secured for a 5-year period from EC projects and private sources. The latter is encouraging since opportunities for funding database projects are difficult to secure (18).
We are currently in the process of collecting population-specific data from the published literature by manual or automated means, i.e. data mining software. Also, being part of two European Commission consortia increases not only the numbers of FINDbase end-users but also the number of potential data contributors. In June 2006, we have started dispatching invitation letters to consortia members to contribute population-specific mutation frequency data in those cases where existing data were either not representative, e.g. based on a small population size, or really outdated. So far, we have seen a very satisfactory response and we hope that more users will positively react to our call. We also plan to provide database pages in a language other than English, a feature, currently not available in any NEMDB (4), which will appeal the database to non-specialist users, such as patients and family members.
As far as further upgrading of the ETHNOS software is concerned, we are currently working on establishing separate NEMDBs per population (source databases), hosted either in the main FINDbase server or in a remote location (see also Database design, implementation and access), which will intercommunicate with FINDbase [central (warehouse) database] bi-directionally. The latter is facilitated by the fact that both the source and central databases operate under the same software, which not only contributes towards NEMDB uniformity (4) but also allows for maintaining updated versions of the linked NEMDBs using a very simple server task tool. This will also allow local (national) curators to maintain a curator's page that will differ from NEMDB to NEMDB and could redirect the end-user to other websites, containing information that may not appear in FINDbase itself, URLs from local human genetic societies, conferences of interest, etc. Our preliminary data show that such operation is feasible and we anticipate that the existing NEMDBs, currently based on the previous ETHNOS (version1.0) flat-file database format [http://www.goldenhelix.org; (7–9)] will migrate to the SQL-based ETHNOS software in the beginning of 2007.
Last, but not least, database content and quality management is one of the key elements for success of database projects. So far, FINDbase control is maintained locally, but as data influx will continue to rise, it would seem logical for its control and decision making to pass to a multinational consortium of experts from the field of human molecular genetics to form FINDbase steering committee (4). Also, in order to provide incentives to potential contributors and researchers to submit their data to FINDbase and at the same time to prevent population frequency data from being lost or kept unpublished, FINDbase would act as a specialized ‘database-journal’, exclusively dedicated to document the molecular basis of inherited disorders in different populations. The main advantage of such an approach is that wherein all data gets immediately submitted and, once accepted, deposited into an Internet-accessible structured depository, like FINDbase, all of which will be interconnected [see also Ref. (18)]. FINDbase steering committee can serve as the editorial board of such database-journal, evaluating the quality of the data submitted, i.e. conforming to a number of pre-determined requirements (e.g. minimum population size tested, etc), while each submission, once entered into FINDbase, would be registered under a unique PubMed ID, providing authors with a certain degree of credit for their contribution. Such approach has been previously proposed by the HGVS for LSDBs (19). We believe that this innovative approach, if adopted, could lead the way and can, thereby, produce a non-profit model for sustainable funding for such an entity.
FINDbase would be also linked to other online repositories, such as the WayStation (http://www.centralmutations.org) as part of the Human Variome Project (20) and to complement efforts with GeneTests-GeneClinics [http://www.geneclinics.org; (21)] and OrphaNet (http://www.orpha.net), which are useful online resources for the various molecular genetic laboratories and tests provided in the United States and Europe respectively.
Finally, it is known from our previous experience that the user input is fundamental for improving the overall database quality and data accuracy. Database users frequently contact the administrators in order to report missing information and pinpoint inconsistencies and/or erroneous entries. Therefore, we urge FINDbase users to frequently communicate their opinions and notify the administrators and national coordinators for errors or for incomplete information. This will certainly contribute towards keeping the database as complete and up-to-date as possible.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
Most of our work has been supported by the Ithanet FP6 Collaboration Action (026539), the EuroGenTest FP6 Network of Excellence (512148), corporate funding from Asclepion Genetics (Switzerland) to GPP, by VZFNM 00064203(6112), Snip2Chip and Micro2DNA projects to MMJr and by a Fondazione per la ricerca sulla Fibrosi Cistica-Onlus (Italy) project (FFC#7-2004) to FR. We particularly thank Prof. Richard Cotton and the HGVS for their continuous encouragement and our national collaborators, who could not be listed in full due to space constraints, for provision of their individual CFTR mutation distribution datasets. Finally, we are indebted to all FINDbase users worldwide for their valuable comments and suggestions, which helped us to keep the information as updated and complete as possible and also contributed to the continuous improvement of the database profile and contents. Funding to pay the Open Access publication charges for this article was provided by EuroGenTest FP6 Network of Excellence (512148).
Conflict of interest statement. None declared.
REFERENCES
- 1.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 2.Hamosh A., Scott A.F., Amberger J., Bocchini C., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claustres M., Horaitis O., Vanevski M., Cotton R.G. Time for a unified system of mutation description and reporting: a review of locus-specific mutation databases. Genome Res. 2002;12:680–688. doi: 10.1101/gr.217702. [DOI] [PubMed] [Google Scholar]
- 4.Patrinos G.P. National and ethnic mutation databases: documenting populations' genography. Hum. Mutat. 2006;27:879–887. doi: 10.1002/humu.20376. [DOI] [PubMed] [Google Scholar]
- 5.Scriver C.R. Human genetics: lessons from Quebec populations. Annu. Rev. Genomics Hum. Genet. 2001;2:69–101. doi: 10.1146/annurev.genom.2.1.69. [DOI] [PubMed] [Google Scholar]
- 6.Laberge A.M., Michaud J., Richter A., Lemyre E., Lambert M., Brais B., Mitchell G.A. Population history and its impact on medical genetics in Quebec. Clin. Genet. 2005;68:287–301. doi: 10.1111/j.1399-0004.2005.00497.x. [DOI] [PubMed] [Google Scholar]
- 7.Patrinos G.P., van Baal S., Petersen M.B., Papadakis M.N. The Hellenic National Mutation database: A prototype database for inherited disorders in the Hellenic population. Hum. Mutat. 2005;25:327–333. doi: 10.1002/humu.20157. [DOI] [PubMed] [Google Scholar]
- 8.Kleanthous M., Patsalis P.C., Drousiotou A., Motazacker M., Christodoulou K., Cariolou M., Baysal E., Khrizi K., Pourfarzad F., Moghimi B., et al. The Cypriot and Iranian National Mutation Frequency databases. Hum. Mutat. 2006;27:598–599. doi: 10.1002/humu.9422. [DOI] [PubMed] [Google Scholar]
- 9.Megarbane A., Chouery E., van Baal S., Patrinos G.P. The Lebanese National Mutation Frequency database. Eur. J. Hum. Genet. 2006;14(Suppl. 1):365. [Google Scholar]
- 10.Bobadilla J.L., Macek M., Jr, Fine J.P., Farrell P.M. Cystic fibrosis: a worldwide analysis of CFTR mutations—correlation with incidence data and application to screening. Hum. Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 11.Hardison R.C., Chui D.H., Giardine B., Riemer C., Patrinos G.P., Anagnou N., Miller W., Wajcman H. HbVar: A relational database of human hemoglobin variants and thalassemia mutations at the globin gene server. Hum. Mutat. 2002;19:225–233. doi: 10.1002/humu.10044. [DOI] [PubMed] [Google Scholar]
- 12.Patrinos G.P., Giardine B., Riemer C., Miller W., Chui D.H., Anagnou N.P., Wajcman H., Hardison R.C. Improvements in the HbVar database of human hemoglobin variants and thalassemia mutations for population and sequence variation studies. Nucleic Acids Res. 2004;32:D537–D541. doi: 10.1093/nar/gkh006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patrinos G.P., Wajcman H. Recording human globin gene variation. Hemoglobin. 2004;28:v–vii. [PubMed] [Google Scholar]
- 14.Giardine B., van Baal S., Kaimakis P., Riemer C., Miller W., Samara M., Kollia P., Anagnou N.P., Chui D.H., Wajcman H., et al. HbVar database of human hemoglobin variants and thalassemia mutations: 2007 update. Hum Mutat. 2007 doi: 10.1002/humu.9479. in press. [DOI] [PubMed] [Google Scholar]
- 15.Scriver C.R., Waters P.J., Sarkissian C., Ryan S., Prevost L., Cote D., Novak J., Teebi S., Nowacki P.M. PAHdb: a locus-specific knowledgebase. Hum. Mutat. 2000;15:99–104. doi: 10.1002/(SICI)1098-1004(200001)15:1<99::AID-HUMU18>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.Scriver C.R., Hurtubise M., Konecki D., Phommarinh M., Prevost L., Erlandsen H., Stevens R., Waters P.J., Ryan S., McDonald D., et al. PAHdb 2003: What a locus-specific knowledgebase can do. Hum. Mutat. 2003;21:333–344. doi: 10.1002/humu.10200. [DOI] [PubMed] [Google Scholar]
- 17.Scriver C.R., Nowacki P.M., Lehväslaiho H. Guidelines and recommendations for content, structure and deployment of mutation databases. Hum. Mutat. 1999;13:344–350. doi: 10.1002/(SICI)1098-1004(1999)13:5<344::AID-HUMU2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Patrinos G.P., Brookes A.J. DNA, diseases and databases: Disastrously deficient. Trends Genet. 2005;21:333–338. doi: 10.1016/j.tig.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Horaitis O., Cotton R.G. The challenge of documenting mutation across the genome: the human genome variation society approach. Hum. Mutat. 2004;23:447–452. doi: 10.1002/humu.20038. [DOI] [PubMed] [Google Scholar]
- 20.Cotton R.G., Kazazian H.H. Toward a Human Variome Project. Hum. Mutat. 2005;26:499. [Google Scholar]
- 21.Pagon R.A., Tarczy-Hornoch P., Baskin P.K., Edwards J.E., Covington M.L., Espeseth M., Beahler C., Bird T.D., Popovich B., Nesbitt C., et al. GeneTests-GeneClinics: Genetic testing information for a growing audience. Hum. Mutat. 2002;19:501–509. doi: 10.1002/humu.10069. [DOI] [PubMed] [Google Scholar]