Abstract
In this study, we adapted the well known uracil DNA glycosylase (UNG) carry-over prevention system for PCR, and applied it to the analysis of DNA methylation based on sodium bisulfite conversion. As sodium bisulfite treatment converts unmethylated cytosine bases into uracil residues, bisulfite treated DNA is sensitive to UNG treatment. Therefore, UNG cannot be used for carry-over prevention of PCR using bisulfite treated template DNA, as not only contaminating products of previous PCR, but also the actual template will be degraded. We modified the bisulfite treatment procedure and generated DNA containing sulfonated uracil residues. Surprisingly, and in contrast to uracil, 6-sulfonyl uracil containing DNA (SafeBis DNA) is resistant to UNG. We showed that the new procedure removes up to 10 000 copies of contaminating PCR product in a closed PCR vessel without significant loss of analytical or clinical sensitivity of the DNA methylation analysis.
INTRODUCTION
DNA methylation is the most well characterized epigenetic DNA modification. DNA methylation is known to play a role in the regulation of gene expression during cell and cancer development. CpG island hypermethylation is one mechanism repressing tumour suppressor genes in many cancers (1–3). As genomic methylation patterns in somatic cells are generally stable, a change in these patterns, such as aberrant methylation of CpG island promoter regions, is a target for cancer diagnosis and treatment (1,2). Recent results of early cancer detection by identifying free-floating methylated cancer DNA in body fluids, such as blood and urine demonstrate the power of DNA methylation analysis (4–15).
Several PCR based assay formats have been developed to identify differentially methylated tumour DNA (5,16,17). The detection of a few methylated copies of a gene within a large excess of unmethylated copies requires optimized assays with the highest analytical sensitivity. The power of these PCR assays is due to the bisulfite conversion of the DNA prior to the analysis. As 5-methylcytosine and unmethylated cytosine show the same base pairing properties, bisulfite treatment of DNA is essential to transform epigenetic information into nucleic acid sequence alterations. The treatment converts unmethylated cytosines into uracil residues, but leaves 5-methylcytosines unconverted. It is thereby possible to analyse the methylation information trough PCR based assays (18,19). Although methylation sensitive restriction enzymes combined with PCR have successfully been used for the identification of methylated DNA without bisulfite conversion of DNA, the clinical sensitivity and specificity of this approach depends on the complete hydrolysis of unmethylated DNA, generally being below 99%, and is limited to the single CpG comprised by the restriction site (20,21).
Every PCR based system is susceptible to false positive results caused by contamination with products from previous reactions, and this is a major obstacle when used in diagnostic laboratories for sensitive tests (22). Several approaches have been reported to eliminate contamination by amplicons from previous reactions in the PCR mix of a subsequent amplification (23–28). In the widely used uracil DNA glycosylase (UNG) method, all PCRs in a laboratory are performed with dUTP instead of dTTP. In contrast to the genomic template DNA containing no uracil residues, PCR products can be selectively destroyed by the enzyme UNG in the closed vessel prior to PCR (29–32). However, this method is not applicable if bisulfite converted DNA, which contains uracil bases, is used as the template in PCR assays.
We have developed a modified bisulfite treatment method to enable the utilization of the UNG carry-over prevention method in methylation analysis. In this new procedure, the DNA is not desulfonated after bisulfite treatment. The bisulfite treatment itself generates sulfonated template DNA (SafeBis DNA), which is resistant to UNG cleavage. After treatment of the reaction mix with UNG, the PCR is carried out with an elongated initial denaturation step. During this step, the bisulfite DNA is desulfonated, the hot start Taq DNA polymerase is activated, and the UNG is inactivated. In this paper, we present the application of this novel procedure to the sensitive detection of methylated DNA, in which we applied the dUTP/UNG carry-over prevention method to bisulfite treated template DNA, which was subsequently analysed using the HeavyMethyl real-time PCR method.
MATERIAL AND METHODS
DNA preparation
The genomic DNA of 12 colon tumor (stage TN1-TN4) samples and normal adjacent tissues (BioCat, Heidelberg, Germany) was extracted using the QIAamp DNA mini kit protocol (Qiagen, Hilden, Germany). The tumor samples were obtained from five male and seven female patients. Universal methylated human DNA (Chemicon International, Inc., Temecula, CA) was used as reference DNA for quantitative real-time PCR.
Preparation of Bisulfite DNA
For standard bisulfite treatment (Bisulfite DNA), 0.5 μg DNA diluted in water was mixed with 354 μl of sodium bisulfite solution (5.89 mol/l) and 146 μl of dioxane containing a radical scavenger (6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid, 40 g/l). The reaction mixture was incubated using the following conditions: 99°C for 3 min, 50°C for 30 min, 99°C for 3 min, 50°C for 1.5 h, 99°C for 3 min and finally 50°C for 3 h. The DNA was subsequently purified by ultrafiltration using Millipore Microcon columns YM30 (Millipore Corporation, Bedford, MA). The purification was conducted essentially according to the manufacturer's instructions. For this purpose, the reaction mixture was mixed with 200 μl of water, loaded onto the ultrafiltration membrane, centrifuged for 15 min at 14 000 g, and subsequently washed with water. For desulfonation 100 μl of a 0.2 mol/l NaOH solution was added and incubated for 10 min. A centrifugation for 10 min at 14 000 g was followed by a final washing step with water. The DNA was eluted from the membrane with 75 μl of prewarmed TE buffer (50°C, pH 8.5). Then the column was turned over and centrifuged according to the manufacturer's instructions to retrieve the DNA from the membrane.
Preparation of SafeBis DNA
The preparation of SafeBis DNA was performed as described above with the following exceptions: The incubation with NaOH was omitted and replaced by a washing step with 100 μl water and the DNA was finally eluted from the membrane with 75 μl prewarmed water (50°C).
Quantification of Bisulfite and SafeBis DNA by a reference PCR
The total amount of bisulfite DNA was quantified using a real-time reference PCR amplifying a 130 bp fragment within the GSTP1 gene (GenBank accession no. X08058; nucleotide 2273–2402). The DNA amount was calculated against an external standard curve prepared with known concentrations of bisulfite converted universal methylated DNA. The concentration of the standard DNA was determined by UV at 260 nm after the bisulfite reaction. The PCR was performed on a LightCycler instrument 2.0 in a total volume of 20 μl containing 2 μl template DNA, 2 μl FastStart LightCycler Mix for hybridization probes (Roche Diagnostics, Mannheim, Germany), 3.5 mM MgCl2, 0.60 μM forward primer: GGAGTGGAGGAAATTGAGAT, 0.60 μM reverse primer: CCACACAACAAATACTCAAAAC and 0.2 μM Taqman probe: FAM-TGGGTGTTTGTAATTTTTGTTTTGTGTTAGGTT-BHQ1 (where FAM = carboxyfluorescein and BHQ1 = black whole quencher 1). The cycling conditions of the reference PCR were as follows: 95°C for 10 min, followed by 50 cycles at 95°C for 10 s, 56°C for 30 s and 72°C for 10 s. The detection was carried out during the annealing step at 56°C in channel F1 at 530 nm. For the quantification of SafeBis DNA, the cycling conditions were changed as follows: 37°C for 10 min, 95°C for 30 min, followed by 50 cycles at 95°C for 10 s, 56°C for 30 s and 72°C for 10 s.
Methylation specific real-time PCR
The TMEFF2-HeavyMethyl (HM) PCR selectively amplifies a 113 bp fragment of methylated bisulfite converted DNA in the promoter region of TMEFF2 (GenBank accession no. AF242221, nucleotide 1102–1214). The PCR was performed on the LightCycler device using FastStart LightCycler Mix for hybridization probes (Roche Diagnostics, Mannheim), using 3.5 mM MgCl2, 0.30 μM forward primer: AAAAAAAAAAAACTCCTCTACATAC, 0.30 μM reverse primer: GGTTATTGTTTGGGTTAATAAATG, 4.0 μM blocker: ACATACACCACAAATAAATTACCAAAAACATCAACCAA-PH (where PH = 3′-phosphate modification), 0.15 μM hybridization probe: TTTTTTTTTTCGGACGTCGTT-FL (where FL = fluorescein), and 0.15 μM hybridization probe: red640-TCGGTCGATGTTTTCGGTAA-PH (where red640 = LightCycler fluorescence label for channel 640 nm). The GSTP1-HM PCR (GenBank accession no. X08058, nucleotide 1183–1304) was performed with 0.30 μM forward primer: GGGATTATTTTTATAAGGTT, 0.30 μM reverse primer: TACTAAAAACTCTAAACCCCATC, 4.0 μM blocker: CCCATCCCCAAAAACACAAACCACACAT-PH, 0.15 μM hybridization probe: TTCGTCGTCGTAGTTTTCGTT-FL, 0.15 μM hybridization probe: red640- TAGTGAGTACGCGCGGTT-PH amplifying a 123 bp fragment. The cycling conditions were as follows: 95°C for 10 min, then 50 cycles at 95°C for 10 s, 56°C for 30 s and 72°C for 10 s. The detection was carried out during the annealing step monitoring the ratio of fluorescence at 640 and 530 nm. The cycle thresholds (Cts) were calculated according to the second derivative maximum method of the LightCycler software.
For the carry-over prevention procedure, the real-time PCRs were performed with the SafeBis DNA. In addition, the PCR contained 0.2 U UNG (Roche Diagnostics, Mannheim, Germany). The PCR cycling program was changed to: 37°C for 10 min, 95°C for 30 min, followed by 50 cycles at 95°C for 10 s, 56°C for 30 s and 72°C for 10 s.
Comparison of detection limit of real time PCR using Bisulfite and SafeBis template DNA
The positives signals of the GSTP1-HM, TMEEF2-HM and reference PCR of 16 replicates were determined. In the PCRs 0, 6, 12, 25 and 50 pg methylated Bisulfite or SafeBis template DNA were used. Probit regression analysis of percent positives versus log10 of pg template DNA/PCR was performed with SPSS for Windows (version 13.0) software. In addition, the 95% confidence intervals (CI) were determined at which 95% of the results were expected to be positive. The amount of template DNA, which was detected with a probability of 90% (LOD90), was defined as detection limit of the real time PCR. As the 95% CI becomes narrow at the detection probability of 50%, the 95% CI of the LOD50 values were determined. PCRs, showing no overlap of the 95% CI of LOD50, have statistically significant different LOD50 values.
Calculation of the methylation rate
The relative level of gene methylation of the promoter of TMEFF2 was calculated according to the PMR value method (33), wherein the methylation level equals the percentage of methylated copies measured in a sample by the TMEFF2-HM assay in relation to the total DNA measured in the same sample by the reference PCR.
Generation of PCR contaminants
PCR amplificate was generated on methylated bisulfite converted DNA as described above. Subsequently, the PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) according to the manufacturers' recommendation. Finally, the concentration of the amplificate was determined by real-time PCR using bisulfite converted universal methylated DNA as the quantification standard. For calculation, 33 ng standard bisulfite converted DNA was assumed to represent 10 000 copies of gene product.
RESULTS
SafeBis DNA preparation and UNG carry-over prevention
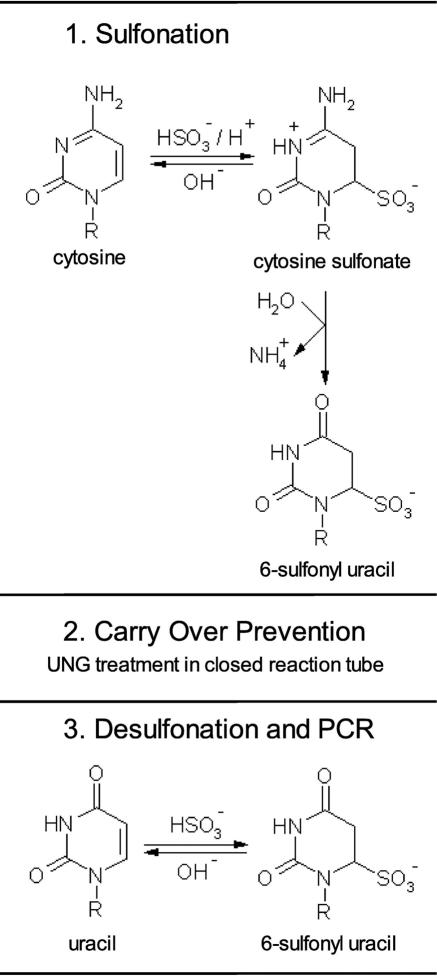
In order to use UNG for PCR product carry-over prevention in PCR based methylation analysis using bisulfite converted DNA, we developed the procedure shown in Figure 1. The new method, generating SafeBis DNA, comprises thermal denaturation of the DNA and subsequent sulfonation with sodium bisulfite. In contrast to published procedures (18), the sulfonated DNA was purified with a filter device using water without desulfonation. The SafeBis DNA was stored in water at −20°C until use. The desulfonation of the SafeBis DNA was performed in the PCR vessel. For quantitative desulfonation of the SafeBis DNA, the initial step activating the polymerase at 95°C was prolonged from 10 to 30 min. For carry-over prevention, the PCR contained 0.2 U UNG and was incubated for 10 min at 37°C prior to the heat activation step.
Figure 1.
Carry-over prevention procedure for PCR using bisulfite treated template DNA. Sulfonated uracils are a stable intermediate product of the bisulfite treatment process. The SafeBis DNA is purified and used as a template DNA for the PCR. For carry-over prevention the reaction is incubated with UNG in closed reaction tubes prior to PCR. The SafeBis DNA is not a substrate for UNG, but PCR amplificates containing uracil are destroyed by UNG. As SafeBis DNA is not amplifiable by the Taq polymerase, it needs to be desulfonated within the initial denaturation step of the PCR. The UNG is inactivated in the same step.
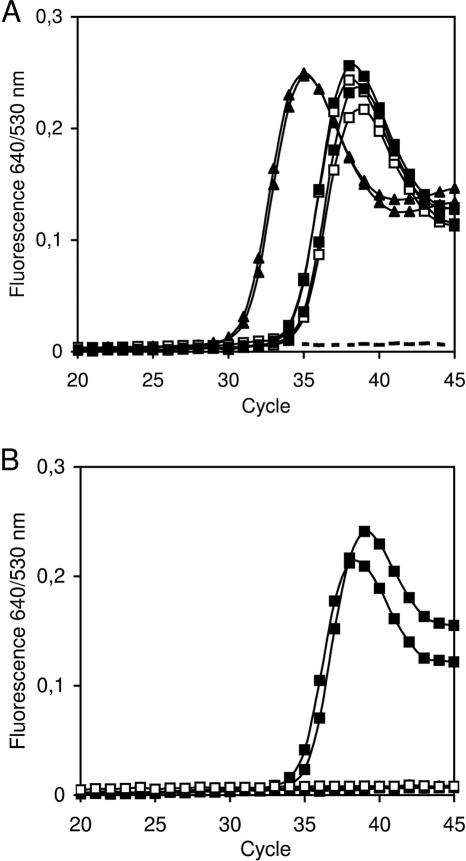
Standard bisulfite DNA (Bisulfite DNA), as prepared according to published procedures, is a single stranded uracil containing nucleic acid. As expected, 1 ng Bisulfite DNA was not amplifiable after UNG treatment (Figure 2B), whereas a Ct of 29.5 was determined for TMEFF2-HM PCR without prior UNG treatment (Figure 2A). Under the described conditions the SafeBis DNA was resistant to UNG (Figure 2B). The UNG activity was tested with a dilution series of products of the TMEFF2-HM PCR. Using up to 50 PCR cycles, 10 000 copies of the TMEFF2 PCR product contaminants were reproducibly not amplified, if the PCR was incubated with UNG prior to amplification (Supplementary Figure 1).
Figure 2.
Susceptibility of Bisulfite DNA and SafeBis DNA to UNG treatment. Detection of methylated DNA with the TMEFF2-HM real-time PCR using 1 ng Bisulfite DNA (open squares), SafeBis DNA (black squares) and 10 000 copies of TMEFF2 PCR product (black triangles). Broken lines indicate no template controls. The PCR products were previously generated with dUTP. Whereas all three different template DNAs are amplified without prior UNG treatment (A), only the SafeBis DNA shows amplification after UNG treatment (B). The treatment was performed with 0.2 U UNG in closed PCR vessels for 10 min at 37°C.
The conditions for the desulfonation step in the PCR were evaluated. The Ct differences of the GSTP1-HM PCR using 1 ng SafeBis and 1 ng Bisulfite DNA were measured for different times of initial incubation at 95°C. If SafeBis DNA was desulfonated for only 10 min at 95°C, the amplification of 1 ng SafeBis-DNA was delayed for 2.7 cycles compared to 1 ng Bisulfite DNA. However, there was only a delay of 0.2 Ct, if the desulfonation time was prolonged to 25 or 30 min. When TMEFF2-HM PCR was applied, the amplification of 1 ng SafeBis DNA was delayed for 0.5 Ct using the described conditions (Table 1). In addition, different PCR buffers were tested for their desulfonation ability. Using the reference PCR, the FastStart reaction buffer (Roche Diagnostics, Mannheim Germany) and the QuantiTect Probe reaction buffer (Qiagen, Hilden Germany) showed similar performance indicated by a 0.4 Ct delay of the amplification of 1 ng SafeBis DNA compared to standard bisulfite treated DNA. In contrast, the amplification of 1 ng SafeBis DNA was delayed for 5.2 Ct, when qPCR Core Kit (Eurogentec, Seraing, Belgium) was applied (Table 1 and Supplementary Figure 2A–F).
Table 1.
Desulfonation of SafeBis-DNA in PCR buffers
| Δ Cta | ||||
|---|---|---|---|---|
| PCR | Desulfonation time (min) | FastStart LightCycler mix | QuantiTect kit | qPCR core kit |
| GSTp1-HM | 10 | 2.7 | Nd | Nd |
| 15 | 1.9 | Nd | Nd | |
| 20 | 0.6 | Nd | Nd | |
| 25 | 0.2 | Nd | Nd | |
| 30 | 0.2 | Nd | Nd | |
| TMEFF2-HM | 30 | 0.5 | Nd | Nd |
| Reference-PCR | 30 | 0.4 | 0.4 | 5.2 |
adetermined as (Ct of 1 ng SafeBis DNA)—(Ct of 1 ng Bisulfite DNA).
Performance of the methylation specific real-time PCR applying the carry over prevention work flow
In order to evaluate, if the sensitivity of detection is limited by the application of the carry over prevention workflow, the performances of different real-time PCRs were comparatively analysed. The 50% limit of detection (LOD50) of 3 real-time PCR on SafeBis and Bisulfite DNA were determined. As these values showed overlapping confidence intervals for the individual PCRs, there was no statistically significant difference in the sensitivity of the assays, when using either SafeBis or Bisulfite DNA as template (Table 2). The 90% detection limits (LOD90) determined for the TMEFF2-HM, GSTP1-HM and the reference PCR on SafeBis DNA were 16.1, 16.1 and 16.3 pg, respectively. Supplementary Figure 3 shows the detailed results of the LOD determination.
Table 2.
Determination of the detection limits of three real-time PCRs on SafeBis and Bisulfite template DNA
| PCR | SafeBis DNA | Bisulfite DNA | ||
|---|---|---|---|---|
| LOD50 (pg)a | 95% CI (pg)b | LOD50 (pg)a | 95% CI (pg)b | |
| GSTP1-HM | 4.9 | 2.3–6.9 | 4.8 | 0.5–8.1 |
| TMEFF2-HM | 7.0 | 3.7–9.3 | 4.1 | 0.0–6.8 |
| Reference-PCR | 5.7 | 1.3–8.3 | 6.9 | 2.7–9.9 |
a50% detection limits were calculated by a Probit regression analysis based on the results using 50, 25, 12, 6 and 0 pg methylated DNA analysed in 16 replicates each.
b95% confidence interval of LOD50 value.
Also, the PCR efficiency of real-time PCRs was not significantly changed using SafeBis DNA as compared to Bisulfite DNA. Using dilution series of 20, 5, 2, 0.8 and 0.2 ng methylated SafeBis DNA, the TMEFF2-HM assay showed an efficency of 1.85 compared to an efficiency of 1.82 on Bisulfite DNA. The efficencies of the reference PCR were determined with 1.99 (SafeBis DNA) and 1.98 (Bisulfite DNA), respectively. The data points showed a good alignment with R2 > 0.99, when the log10 DNA amount was plotted as a function of Ct (Supplementary Figure 2A, B, C and D). These data indicate that SafeBis DNA (i) is not a substrate for UNG, and (ii) is sufficiently desulfonated by the PCR during the initial incubation at 95°C for 30 min.
Stability of SafeBis DNA under different storage conditions
Due to the low stability of the sulfonyl group at the C6-position of the uracil at elevated pH values, the storage conditions of SafeBis DNA at pH 7, pH 8 and pH 9 were investigated with the GSTP1-HM PCR. SafeBis DNA was stable for 144 h when stored at 4–8°C and pH 7 (Table 3). Applying the UNG treatment, which reliably destroyed 3000 copies of GSTP1 PCR product, 10, 1 and 0.1 ng SafeBis template DNA stored at 4–8°C and pH 7 for up to 144 h showed comparable performance to the standard bisulfite DNA without UNG treatment. The cycle thresholds were increased by a maximum of 0.3 Ct when compared to the same amount of standard bisulfite converted DNA. Storage at 40°C, particularly at pH 9, caused significant desulfonation of the SafeBis template DNA. No amplification was detected on 0.1 ng SafeBis DNA stored under these conditions. Using 1 and 10 ng SafeBis DNA, the Cts were increased by 6.4 and 4.9, respectively. At −20°C and pH 7 no significant desulfonation of SafeBis DNA was detected up to a storage time of 2 weeks (data not shown).
Table 3.
Stability of SafeBis DNA under different storage conditions
| Ct of GSTP1 HM real-time PCR assaya | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Standard Bis pH 8 | Safe Bis pH 7 | Safe Bis pH 8 | Safe Bis pH 9 | |||||||||||||
| Storage | 12 h 4°C | 12 h 4°C | 144 h 4°C | 22 h 40°C | 12 h 4°C | 12 h 4°C | 144 h 4°C | 22 h 40°C | 12 h 4°C | 12 h 4°C | 144 h 4°C | 22 h 40°C | 12 h 4°C | 12 h 4°C | 144 h 4°C | 22h 40°C |
| UNGb | − | + | + | + | − | + | + | + | − | + | + | + | − | + | + | + |
| 10 ng template | 26.8 | − | − | − | 27.2 | 27.4 | 27.3 | 28.5 | 27.0 | 27.9 | 27.3 | 30.4 | 27.0 | 28.1 | 28.2 | 33.2 |
| 1 ng template | 30.4 | − | − | − | 31.0 | 31.3 | 31.6 | 32.1 | 31.6 | 31.4 | 31.3 | 34.0 | 31.7 | 31.9 | 31.8 | 35.3 |
| 0.1 ng template | 34.6 | − | − | − | 34.9 | 34.7 | 35.2 | 36.3 | 36.2 | 35.9 | 36.1 | − | 35.5 | 36.2 | 35.4 | − |
aThe mean of triplicate experiments is shown; − indicates no amplification curve.
b+: 0.2 U UNG were added and the reaction incubated for 10 min 37°C prior to PCR.
Methylation analysis of clinical samples using SafeBis DNA
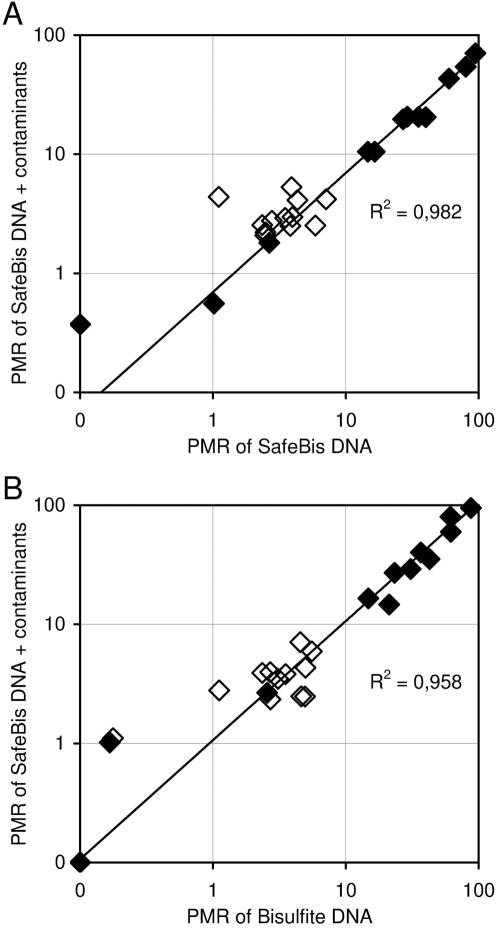
The new procedure was investigated on a subset of 12 matched sample pairs of normal and colon cancer tissue. After DNA extraction 500 ng of tissue DNA was bisulfite treated with the two procedures described in this study. The TMEFF2-HM PCR was performed on bisulfite DNA, SafeBis DNA and SafeBis DNA spiked with 10 000 copies of TMEFF2 amplificate generated on bisulfite converted universally methylated DNA. The total amounts of bisulfite DNA and SafeBis DNA were determined by the reference assay without prior UNG treatment. Subsequently, the PMR value of the TMEFF2 promoter fragment was determined. The PMR values of TMEFF2, determined on SafeBis and SafeBis DNA spiked with 10 000 copies of TMEFF2 amplificates, were highly correlated with R2 = 0.982 (Figure 3A). The methylation signal was not influenced by the addition of up to 10 000 copies of PCR contaminants. This indicates that the re-amplification of contaminants was efficiently prevented by the presented technology. Furthermore, a high correlation (R2 = 0.958) between the PMR values of TMEFF2 determined on standard bisulfite DNA and SafeBis DNA spiked with 10 000 copies of TMEFF2 was observed (Figure 3B). These results demonstrate that TMEFF2-HM PCR showed the same analytical results using either SafeBis DNA or standard bisulfite DNA as template DNA.
Figure 3.
Methylation analysis of the promoter region of TMEFF2. 12 colon cancer (black) and 12 healthy adjacent tissues (white) were processed with and without carry-over prevention procedure. The PMR values were calculated from the ratio of DNA values obtained by TMEFF2-HM and a reference real-time PCR. (A) Results from two measurements of SafeBis DNA using UNG for carry-over prevention are plotted. The values on the y-axis were obtained from samples spiked with 10 000 copies of uracil containing TMEFF2 PCR products, representing a carry-over contamination. On the x-axis, the PMR values from the same SafeBis DNA samples, without spiked amplificates, were plotted. (B) A correlation plot of the results from the standard method without carry-over prevention (x-axis) and the results obtained from SafeBis DNA spiked with 10 000 copies contaminating PCR products and prior UNG treatment (y-axis).
DISCUSSION
Cross-contamination by previously generated PCR products is a serious problem for PCR based assays in clinical diagnostics, and also for the sensitive detection of tumour DNA based on its epigenetic properties. Although alternative methods that allow for the enzymatic sterilization of single closed PCR vessels have been developed (25,34), the dUTP/UNG method exhibits some major advantages. The dUTP/UNG method destroys double stranded and single stranded uracil containing DNA fragments, a single abasic site generated by UNG inactivates the contaminating PCR product as a template for the next PCR. Therefore, the dUTP/UNG is a highly effective carry-over prevention technique (31). Another method using type IIS restriction enzymes requires 5′-modified primers and is not applicable with established PCR assays (34). In addition, single stranded PCR amplificates might be either not, or only incompletely destroyed. The UNG technology is, to our knowledge, the only ‘closed vessel’ carry-over prevention method for PCR based diagnostic screening assays.
In this paper we present, a new procedure that enables the use of the dUTP/UNG system as a carry-over prevention system for DNA methylation detection with PCR using bisulfite converted DNA as the template. The workflow combines a modified bisulfite treatment procedure that generates 6-sulfonyl uracil containing DNA with an elongated initial activation step of hot start Taq polymerase in order to obtain also a complete desulfonation of the template DNA in the PCR. In contrast to uracil containing DNA molecules, such as standard bisulfite converted DNA, the sulfonated DNA is not a substrate for the UNG. Under the conditions used, no significant loss of HM real-time PCR performance was detected using SafeBis DNA in comparison to bisulfite treated DNA prepared with standard procedures.
As only SafeBis DNA is resistant to UNG, the storage conditions of SafeBis DNA are very important for avoiding inadvertent desulfonation. The sulfonyl residue of the uracil is easily hydrolyzed at alkaline pH and elevated temperatures. Accordingly, SafeBis becomes sensitive to UNG if stored at pH 8–9 and higher temperatures. However, SafeBis DNA stored at pH 7 is stable for several days at 4–8°C or up to 2 weeks at −20°C. However, a complete desulfonation of uracil is necessary for effective amplification of the SafeBis DNA. DNA containing 6-sulfonyl uracil is not a template for Taq polymerases. The desulfonation happens during the initial heat denaturation step of the PCR at 95°C, at the same time inactivation of the UNG occurs. An initial PCR activation of 25 or 30 min showed the best results; a shorter initial activation step reduced the PCR efficiency by incomplete desulfonation of the SafeBis DNA. A longer activation step is not recommended, because of partial deactivation of the Taq polymerase. Furthermore, we tested the SafeBis DNA desulfonation capabilities of two additional PCR systems. The QuantiTect Kit (Qiagen, Hilden, Germany) showed comparable results to the FastStart LightCycler Mix (Roche Diagnostics, Mannheim, Germany). However, desulfonation using the qPCR Core Kit (Eurogentec, Seraing) was less efficient compared to the other PCR buffers. As a higher desulfation efficiency is expected in a more alkaline solution, the pH of the individual buffers, in combination with the reaction time, is a critical parameter. Both, the FastStart LightCycler Mix and the QuantiTect Kit have a pH 8.3–8.5, in contrast to the qPCR Core Kit with pH 7.9. The results demonstrate that quantitative desulfonation within the PCR initial denaturation step is achievable, but might vary between buffers, which therefore need to be tested previously.
The application of the carry-over prevention on clinical samples was also successful. Using the new procedure, the measured methylation values of colon samples in the presence of 10 000 copies of contaminating PCR product showed high correlation with those determined with the standard method lacking any contamination. The data also suggest, that putative PCR artefacts due to incomplete desulfonation did not interfere in a major way with the analysis. Up to now, four different HM assays showing detection limits of 10–25 pg methylated DNA, were successfully tested with the present carry-over prevention procedure. In this work, up to 10 000 copies of TMEFF2-HM PCR products were reproducibly neutralized by the work flow presented. At least 3000 copies of products of the GSTP1-HM and the reference PCR were not amplified after UNG treatment (data not shown). Based on the LODs of the individual PCRs (LOD90 = max. 16.3 pg; ∼5 copy equivalents), the efficiency of degradation of PCR products was found to be between 99.8 and 99.95%. The variability of sterilization rates might be dependent on the fragment length and the uracil content of the PCR product. Nevertheless, the determined degradation rates were in the range described for other quantitative real-time PCRs (35). This paper describes, for the first time, a PCR carry-over prevention procedure that can be used with bisulfite converted DNA—a prerequisite for high throughput epigenetic analysis in diagnostic laboratories.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to Matthias Schuster and Kurt Berlin, for critically reviewing the manuscript. This work was supported by a grant from the Bundesministerium für Bildung und Forschung (BioChancePlus AkZ 0313166). Funding to pay the Open Access publication charges for this article was provided by the Epigenomics AG.
Conflict of interest statement. The authors are employees of the Epigenomics AG. RT and JD are shareholders of the Epigenomics AG.
REFERENCES
- 1.Das P.M., Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 2.Dulaimi E., Hillinck J., Ibanez de Caceres I., Al-Saleem T., Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin. Cancer Res. 2004;10:6189–6193. doi: 10.1158/1078-0432.CCR-04-0597. [DOI] [PubMed] [Google Scholar]
- 3.Verma M., Srivastava S. Epigenetics in cancer: implications for early detection and prevention. Lancet Oncol. 2002;3:55–63. doi: 10.1016/s1470-2045(02)00932-4. [DOI] [PubMed] [Google Scholar]
- 4.Lofton-Day C., Lesche R. DNA methylation markers in patients with gastrointestinal cancers. Current understanding, potential applications for disease management and development of diagnostic tools. Dig. Dis. 2003;21:299–308. doi: 10.1159/000075352. [DOI] [PubMed] [Google Scholar]
- 5.Cottrell S.E., Laird P.W. Sensitive detection of DNA methylation. Ann. NY Acad. Sci. 2003;983:120–130. doi: 10.1111/j.1749-6632.2003.tb05967.x. [DOI] [PubMed] [Google Scholar]
- 6.Cottrell S.E. Molecular diagnostic applications of DNA methylation technology. Clin. Biochem. 2004;37:595–604. doi: 10.1016/j.clinbiochem.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Herman J.G. Circulating methylated DNA. Ann. NY. Acad. Sci. 2004;1022:33–39. doi: 10.1196/annals.1318.006. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z.J., Maekawa M. Polymerase chain reaction-based methods of DNA methylation analysis. Anal. Biochem. 2003;317:259–265. doi: 10.1016/s0003-2697(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 9.Hoque M.O., Topaloglu O., Begum S., Henrique R., Rosenbaum E., Van Criekinge W., Westra W.H., Sidransky D. Quantitative methylation—specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J. Clin. Oncol. 2005;23:6569–6575. doi: 10.1200/JCO.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Belinsky S.A., Klinge D.M., Dekker J.D., Smith M.W., Bocklage T.J., Gilliland F.D., Crowell R.E., Karp D.D., Stidley C.A., Picchi M.A. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin. Cancer Res. 2005;11:6505–6511. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara K., Fujimoto N., Tabata M., Nishii K., Matsuo K., Hotta K., Kozuki T., Aoe M., Kiura K., Ueoka H., et al. Identification of epigenetic aberrant promoter methylation in serum DNA is useful for early detection of lung cancer. Clin. Cancer Res. 2005;11:1219–1225. [PubMed] [Google Scholar]
- 12.Ichikawa D., Koike H., Ikoma H., Ikoma D., Tani N., Otsuji E., Kitamura K., Yamagishi H. Detection of aberrant methylation as a tumor marker in serum of patients with gastric cancer. Anticancer Res. 2004;24:2477–2481. [PubMed] [Google Scholar]
- 13.Sabbioni S., Miotto E., Veronese A., Sattin E., Gramantieri L., Bolondi L., Calin G.A., Gafa R., Lanza G., Carli G., et al. Multigene methylation analysis of gastrointestinal tumors: TPEF emerges as a frequent tumor-specific aberrantly methylated marker that can be detected inperipheral blood. Mol. Diagn. 2003;7:201–207. doi: 10.1007/BF03260039. [DOI] [PubMed] [Google Scholar]
- 14.Muller H.M., Widschwendter M. Methylated DNA as a possible screening marker for neoplastic disease in several body fluids. Expert Rev. Mol. Diagn. 2003;3:443–458. doi: 10.1586/14737159.3.4.443. [DOI] [PubMed] [Google Scholar]
- 15.Cairns P., Esteller M., Herman J.G., Schoenberg M., Jeronimo C., Sanchez-Cespedes M., Chow N.H., Grasso M., Wu L., Westra W.B., et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin. Cancer Res. 2001;7:2727–2730. [PubMed] [Google Scholar]
- 16.Herman J.G., Graff J.R., Myohanen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cottrell S.E., Distler J., Goodman N.S., Mooney S.H., Kluth A., Olek A., Schwope I., Tetzner R., Ziebarth H., Berlin K. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 2004;32:e10. doi: 10.1093/nar/gnh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark S.J., Harrison J., Paul C.L., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olek A., Oswald J., Walter J.A. Modified and improved method for bisulphite based cytosine methylationanalysis. Nucleic Acids Res. 1996;24:5064–5066.. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuzawa R., Breslow N.E., Morison I.M., Dwyer P., Kusafuka T., Kobayashi Y., Becroft D.M., Beckwith J.B., Perlman E.J., Reeve A.E. Epigenetic differences between Wilms' tumours in white and east-Asian children. Lancet. 2004;363:446–451. doi: 10.1016/S0140-6736(04)15491-3. [DOI] [PubMed] [Google Scholar]
- 21.Melnikov A.A., Gartenhaus R.B., Levenson A.S., Motchoulskaia N.A., Levenson Chernokhvostov V.V. MSRE-PCR for analysis of gene-specific DNA methylation. Nucleic Acids Res. 2005;33:e93. doi: 10.1093/nar/gni092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimino G.D., Metchette K., Isaacs S.T., Zhu Y.S. More false-positive problems. Nature. 1990;345:773–774. doi: 10.1038/345773b0. [DOI] [PubMed] [Google Scholar]
- 23.Borst A., Box A.T., Fluit A.C. False-positive results and contamination in nucleic acid amplification assays: suggestions for a prevent and destroy strategy. Eur. J. Clin. Microbiol. Infect Dis. 2004;23:289–299. doi: 10.1007/s10096-004-1100-1. [DOI] [PubMed] [Google Scholar]
- 24.Niederhauser C., Hofelein C., Wegmuller B., Luthy J., Candrian U. Reliability of PCR decontamination systems. PCR Methods Appl. 1994;4:117–123. doi: 10.1101/gr.4.2.117. [DOI] [PubMed] [Google Scholar]
- 25.Walder R.Y., Hayes J.R., Walder J.A. Use of PCR primers containing a 3′-terminal ribose residue to prevent cross-contamination of amplified sequences. Nucleic Acids Res. 1993;21:4339–4343. doi: 10.1093/nar/21.18.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rys P.N., Persing D.H. Preventing false positives: quantitative evaluation of three protocols for inactivation of polymerase chain reaction amplification products. J. Clin. Microbiol. 1993;31:2356–2360. doi: 10.1128/jcm.31.9.2356-2360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dwyer D.E., Saksena N. Failure of ultra-violet irradiation and autoclaving to eliminate PCR contamination. Mol. Cell. Probes. 1992;6:87–88. doi: 10.1016/0890-8508(92)90075-9. [DOI] [PubMed] [Google Scholar]
- 28.Cimino G.D., Metchette K.C., Tessman J.W., Hearst J.E., Isaacs S.T. Post-PCR sterilization: a method to control carryover contamination for the polymerase chain reaction. Nucleic Acids Res. 1991;19:99–107. doi: 10.1093/nar/19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longo M.C., Berninger M.S., Hartley J.L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 30.Pang J., Modlin J., Yolken R. Use of modified nucleotides and uracil-DNA glycosylase (UNG) for the control of contamination in the PCR-based amplification of RNA. Mol. Cell. Probes. 1992;6:251–256. doi: 10.1016/0890-8508(92)90024-r. [DOI] [PubMed] [Google Scholar]
- 31.Pruvost M., Grange T., Geigl E.M. Minimizing DNA contamination by using UNG-coupled quantitative real-time PCR on degraded DNA samples: application to ancient DNA studies. Biotechniques. 2005;38:569–575. doi: 10.2144/05384ST03. [DOI] [PubMed] [Google Scholar]
- 32.Udaykumar, Epstein J.S., Hewlett I.K. A novel method employing UNG to avoid carry-over contamination in RNA-PCR. Nucleic Acids Res. 1993;21:3917–3918. doi: 10.1093/nar/21.16.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eads C.A., Lord R.V., Wickramasinghe K., Long T.I., Kurumboor S.K., Bernstein L., Peters J.H., DeMeester S.R., DeMeester T.R., Skinner K.A., et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 34.Ashkenas J., Dennis J.W., Ho C.Y. Simple enzymatic means to neutralize DNA contamination in nucleic acid amplification, supplementary material. Biotechniques. 2005;39:69–73. doi: 10.2144/05391ST02. [DOI] [PubMed] [Google Scholar]
- 35.Pennings J.L., Van De Locht L.T., Jansen J.H., Van der Reijden B.A., De Witte T., Mensink E.J. Degradable dU-based DNA template as a standard in real-time PCR quantitation. Leukemia. 2001;15:1962–1965. doi: 10.1038/sj.leu.2402290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.