Abstract
Sexual differentiation in Drosophila is regulated through alternative splicing of doublesex. Female-specific splicing is activated through the activity of splicing enhancer complexes assembled on multiple repeat elements. Each of these repeats serves as a binding platform for the cooperative assembly of a heterotrimeric complex consisting of the SR proteins Tra, Tra2 and 9G8. Using quantitative kinetic analyses, we demonstrate that each component of the enhancer complex is capable of recruiting the spliceosome. Surprisingly, Tra, Tra2 and 9G8 are much stronger splicing activators than other SR protein family members and their activation potential is significantly higher than expected from their serine/arginine content. 9G8 activates splicing not only through its RS domains but also through its RNA-binding domain. The RS domains of Tra and Tra2 are required but not sufficient for efficient complex assembly. Thus, the regulated assembly of the dsx enhancer complexes leads to the generation of an extended activation domain to guarantee the ‘all or none’ splicing switch that is required during Drosophila sexual differentiation.
INTRODUCTION
In higher eukaryotes, alternative pre-mRNA splicing is used as a common pathway to regulate the expression of genes (1–3). For classical cases of alternative splicing, it was shown that cis-acting RNA sequence elements increased exon inclusion by serving as binding sites for the assembly of multi-component splicing enhancer complexes. Because these sequence elements were usually located within the regulated exon, they were defined as exonic splicing enhancers (ESEs) (2,4). It has been demonstrated that the recognition of weak splice-site signals can be augmented significantly by the presence of ESEs, and, in contrast, that ESEs have little or no detectable effect on the activation of strong splice sites that conform to the consensus sequence (5,6). ESEs are usually recognized by at least one member of the essential serine/arginine (SR)-rich protein family and recruit the splicing machinery to the adjacent intron (2,4,5,7). SR proteins act at several steps during the splicing reaction (8–14) and require phosphorylation and dephosphorylation for spliceosomal assembly (15,16). Interestingly, SR protein-binding sites are present not only within alternatively spliced exons but also within the exons of constitutively spliced pre-mRNAs (17,18). It is therefore likely that SR proteins bind to sequences found in most, if not all, exons.
Although arginine-serine (RS) domains clearly function as activation domains, the mechanism by which they do so remains unclear. One model for the mechanism of SR protein action proposes that the RS domain of an enhancer-bound SR protein interacts directly with other splicing factors containing an RS domain, thus facilitating the recruitment of the spliceosome (5). An alternative model was suggested by experiments demonstrating that RS domains of SR proteins within the functional spliceosome contact the pre-mRNA (19,20). The sequential character of these contacts suggested that RS domain interactions with RNA promote spliceosome assembly. Irrespective of the mechanism of action, ESEs facilitate the recruitment of spliceosomal components to the regulated splice site (2,21,22). More recently, it was demonstrated that ESEs promote the recognition of both exon/intron junctions within the same step during exon definition. These results suggested that ESEs recruit a multi-component complex that minimally contains components of the splicing machinery required for 5′ and 3′ splice-site selection (23,24). Thus, ESEs function as general activators of exon recognition.
The relative activities of SR proteins depend on their affinity for splicing enhancers and the strength of their activation domains. Using an MS2 tethering approach it was possible to uncouple the spliceosomal recruitment and the RNA-binding activities of SR proteins (25). This system allowed a careful quantitative comparison of the activity of several RS domains (26) to demonstrate that the strength of ESEs is determined by the relative activities of the bound SR proteins, by the number of SR proteins assembled on ESEs, and by the distance between the enhancer and the intron. A mechanistically important observation was that the splicing activity of the bound SR proteins was proportional to the number of SR repeats contained within the RS domain of the proteins tested. Thus, the quantity of SR repeats apparently dictates the activation potential of SR proteins.
Genes involved in Drosophila sex determination are prototypes for the study of positive and negative control of alternative splicing (27). The best characterized are the doublesex (dsx) and fruitless (fru) genes, in which the use of two alternative 3′ (dsx) or 5′ (fru) splice sites leads to the expression of male- or female-specific gene products that initiate gender-specific sexual differentiation and orientation (28,29). Female-specific splicing of dsx requires the production of two proteins, Transformer (Tra) and Transformer2 (Tra2), and an ESE (dsx ESE) consisting of six repeat elements (dsxRE) (30–32). Similarly, female-specific splicing of fru depends on Tra, Tra2 and an ESE (fru ESE) consisting of three repeat elements identical to dsxREs (24,33). Both Tra and Tra2 contain RS-rich domains. Previous in vitro studies in HeLa extracts demonstrated that Tra, Tra2 and the SR protein 9G8 (the human homologue of Drosophila RBP1) assemble cooperatively on each dsxRE (34,35). While 9G8 and Tra2 bind to the 5′ and 3′ half, respectively, Tra appears to contact RNA only very weakly, if at all. Although the identity of the factors that make up the dsx enhancer complex has been revealed, it is currently unknown whether more than one molecule of each component is required for assembly. It is also unclear which one of the three proteins that make up a dsxRE complex is critical in communicating splice-site recognition to the splicing machinery. Each protein contains at least one SR domain that may be involved in crucial protein–protein interactions. Therefore, it is possible that the three proteins combined or one in particular function to recruit the splicing machinery.
Here we evaluated in detail the potency of the dsxRE. By taking advantage of the MS2-fusion approach, we were able to demonstrate that each of its components has an activation potential that is significantly higher than expected from SR dinucleotide content alone. Our quantitative analysis shows that the SR domains of each component are necessary but not sufficient for maximal activation. For 9G8, the RNA-binding domain contributes considerably to splice-site activation. Furthermore, each dsxRE enhancer complex is significantly more potent than canonical SR proteins. These experiments demonstrate that the splicing enhancer complex assembled on dsxREs has evolved to possess exceptional activation potency as is required to induce a complete switch in sex-specific alternative pre-mRNA splicing for the dsx and fru genes.
MATERIALS AND METHODS
RNA
The plasmids, pdsx70(MS2)2, pdsx200-MS2, pdsx200(MS2)2, pdsx300(MS2)2, pdsx300-1RE, pdsx300-2RE, pdsx300-4RE and pdsx300-5RE, were previously described as transcription templates (25,35). Capped, 32P-labeled RNAs were transcribed with T7 RNA polymerase and gel-purified before use as splicing substrates (36).
Proteins
The expression and purification of MS2-fusion proteins was as described previously (25). The coding sequence of tra, tra-2 or 9G8 was amplified by PCR such that a BamHI site was inserted at the 5′ end and a HindIII site at the 3′ end. The purified fragment was inserted into pHIS-BIVT-MS2 vector (25), and digested to generate the corresponding baculovirus transfer vector.
All recombinant baculoviruses were generated in serum-free culture as described by the manufacturer (Gibco-BRL). Each protein was expressed in infected Sf9 cells for 3 days and batch purified under modified denaturing conditions. Briefly, the lysed cell protein pellet was extracted in a 6 M guanidine-HCl, pH 8, solution containing followed by the addition of Ni-NTA agarose slurry for binding to the 6×His-MS2-fusion protein. The slurry was funneled into a column and washed with an 8 M urea, pH 8, solution containing 20 mM imidizole. The MS2-fusion protein was eluted with stepwise increasing concentrations of imidizole. The protein was dialyzed against BC850 splicing buffer described previously (37).
In vitro splicing assays
Conditions for in vitro splicing reactions were 30% HeLa nuclear extract, 1 mM ATP, 20 mM creatine phosphate, 3.2 mM MgCl2, 5 U RNasin (Promega), 1 mM DTT, 72.5 mM KCl, 3% polyvinyl alcohol (MW 30–70 KDa) and 12 mM HEPES, pH 7.9. Reactions for Tra/Tra2-dependent or MS2-fusion protein-dependent substrates were performed in the absence or presence of saturating amounts of recombinant protein (∼400 nM), unless otherwise stated. Splicing reactions were performed for the indicated times at 30°C. Following incubation, reactions were proteinase K digested, phenol/chloroform extracted and ethanol precipitated before PAGE separation. Bands were visualized and quantified using PhosphorImager analysis (Bio-Rad). Percent spliced is defined as spliced products/(unspliced product + spliced products). The background was determined individually for each lane since the total number of substrate and product counts varied throughout a time course. Rate constants for each time course were determined as previously described (36). Experiments were performed at least three times. Variations between experiments were within 2-fold.
RESULTS
The role of Tra, Tra2 and 9G8 in splice-site activation and dsxRE complex formation are unknown. It is possible that only one protein factor of each dsxRE complex may be required for interactions with the splicing machinery while the others may be essential for complex assembly and/or to stabilize the protein network. We have made use of an MS2 hybrid approach to evaluate this possibility. Tra, Tra2 and 9G8 were fused with the MS2 bacteriophage coat protein and overexpressed in baculovirus infected Sf9 cells. A dsx pre-mRNA containing the cognate MS2-binding site in place of the dsx splicing enhancer was then used as a splicing substrate, thus placing the artificial enhancer site at various distances from the regulated intron, ranging from 70 to 300 nt. The rationale for this approach is to generate an extremely stable and specific interaction site for each of the enhancer complex components in isolation from each other.
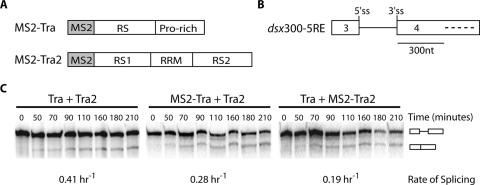
Using in vitro splicing assays, we first tested whether the fusion of MS2 to the N-terminus of Tra or Tra2 interfered with the protein's ability to activate dsx splicing from within the dsx ESE. Time course experiments demonstrated that both fusion proteins were able to activate female-specific splicing of the reporter substrate dsx300-5RE to extents similar to those measured in the presence of wt proteins (Figure 1 and Table 1). Observed rates of splicing were measured to be within 2-fold. We conclude that the MS2-fusion of Tra and Tra2 does not significantly inhibit dsx enhancer complex function. Additional experiments were carried out to determine the activation potential of one of the multiple dsxREs that make up the dsx ESE. In agreement with previous observations, the observed rate of intron removal increased as the number of repeat elements present within the dsx ESE increased (35). The average rate of splicing for one dsxRE was measured to be 0.11 h−1.
Figure 1.
Recombinant MS2-fusion proteins activate native enhancer elements for female-specific dsx splicing. (A) Schematic representation of recombinant fusion proteins. The RNA-binding domain of the bacteriophage MS2 coat protein was fused in-frame with the entire coding sequence of either Transformer (Tra) or Transformer2 (Tra2). (B) Schematic representation of the dsx mini-gene RNA substrate used for in vitro splicing assays. Open boxes represent exons 3 and 4 of the dsx gene. 5′ and 3′ splice sites (ss) are labeled. Dashes represent the 13 nt repeat elements (RE) of the native dsx enhancer located ∼300 nt downstream of the 3′ splice site. (C) In vitro splicing assays were performed in the presence of recombinant Tra and Tra2 proteins or MS2–Tra and MS2–Tra2 proteins to evaluate the potency of each recombinant fusion protein. Diagrams to the right indicate unspliced or spliced RNA.
Table 1.
Splicing rates promoted by recombinant Tra and Tra2
| RNA substrate | ||||
|---|---|---|---|---|
| dsx300-1RE | dsx300-2RE | dsx300-4RE | dsx300-5RE | |
| Tra + Tra2a | 0.11 | 0.31 | 0.33 | 0.39 |
| MS2–Tra + Tra2 | nd | nd | nd | 0.38 |
| Tra + MS2–Tra2 | nd | nd | nd | 0.19 |
All rate constants are expressed in units of h−1. Each value is the average from at least three independent splicing reactions consisting of 7–8 time points over a 4 h time period. To derive rate constants, the data at time points following the lag were fit to a first-order rate equation (43). Experimental error for the rate constants is within 50%. nd, not determined.
aAverage splicing rate per dsxRE with Tra + Tra2 = 0.11 ± 0.05 h−1.
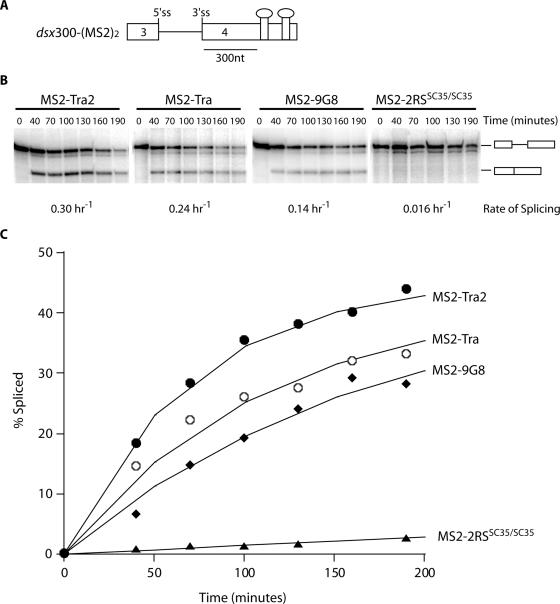
The activation potential of individual components of the dsxRE
To determine how efficient each fusion protein is in activating female-specific intron removal, the rate of splicing was measured for the dsx300-(MS2)2 substrate at saturating concentrations of fusion protein. Four hybrid proteins interact with dsx300-(MS2)2 because MS2 binds the MS2-hairpin as a dimer. The rate analysis shows that each recombinant enhancer protein is capable of independently activating splicing at the native distance when tethered to the RNA (Figure 2). Control experiments showed that, at the reaction conditions used, intron removal was only observed if the substrate contained a functional MS2-hairpin motif. Deletion or mutation of the MS2-hairpin motif did not activate intron removal, indicating that the recombinant fusion proteins used did not activate splicing through non-specific RNA interactions mediated through either their native or the MS2 RNA recognition motif (RRM) (data not shown). Interestingly, when evaluated at 300 nt downstream from the regulated splice site, the splicing activity observed for RS domain fusion protein MS2–RS(SC35/SC35), previously identified as a strong activator in the context of this MS2-hairpin system (26), was ∼10-fold lower than the activity measured for each of the full-length fusion proteins tested (Figure 2B and C). We conclude that each dsxRE complex component (Tra, Tra2 and 9G8) has the potential to contribute to the overall splicing potential of the multi-component dsx enhancer complex.
Figure 2.
Individual enhancer complex proteins activate the female-specific dsx 3′ splice site. (A) Schematic representation of a dsx mini-gene RNA substrate with two synthetic MS2-hairpin enhancer elements positioned ∼300 nt downstream of the female-specific 3′ splice site. Four RS fusion proteins are expected to interact with dsx300-(MS2)2 because MS2 binds the MS2-hairpin RNA as a dimer. (B) In vitro splicing assays were performed in the presence of saturating amounts of recombinant MS2-fusion proteins. Diagrams to the right indicate unspliced and spliced products. Average splicing rates for each MS2-fusion protein are listed below the corresponding gel. (C) The percent spliced is plotted as a function of time to determine a rate of splicing for each fusion protein tested. Filled circles indicate MS2–Tra2, open circles indicate MS2–Tra, filled diamonds indicate MS2–9G8 and filled triangles indicate MS2–2RS(SC35/SC35).
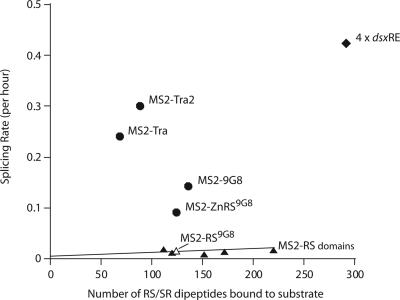
To further evaluate the activation potential of Tra, Tra2 and 9G8, we carried out comparative splicing reactions with a variety of MS2-fusion proteins. In agreement with previous observations, fusing the RS domains of the SR proteins SRp55, SC35, ASF/SF2 and 9G8 to MS2 resulted in strong activation when tethered close to the regulated splice site (25) (Table 2). However, the activation potential of these SR fusion proteins decreased dramatically when located where the dsx ESE usually resides, ∼300 nt downstream of the female-specific splice site. At this distance, the activation potential of the MS–RS fusion proteins increased proportional to the number of SR dipeptide repeats (Figure 3). Strikingly, tethered Tra, Tra2 or 9G8 are significantly more active than one would predict from their SR content. Furthermore, the splicing activity of the dsxRE complex is close to the sum of the activation potential measured for each individual component. These observations are consistent with the idea that each component of the dsxRE complex contributes functionally to the recruitment of the spliceosome to the regulated splice site. We conclude that the components of the dsxRE splicing complex are significantly more active than the RS content would predict, suggesting contributions beyond the RS domains.
Table 2.
Splicing rates promoted by MS2–RS fusion proteins
| RNA substrate | |||||
|---|---|---|---|---|---|
| Fusion protein | No. of RS/SR | dsx70-(MS2)2 | dsx200-MS2 | dsx200-(MS2)2 | dsx300-(MS2)2 |
| MS2–2RS(SC35/SC35) | 55 | 1.7 | 0.026 | nd | 0.016 |
| MS2–2RS(SC35/SF2) | 43 | 1.5 | 0.027 | nd | 0.013 |
| MS2–2RS(SF2/SF2) | 30 | 1.9 | 0.020 | nd | 0.011 |
| MS2–RS(p55) | 38 | Nd | 0.016 | nd | 0.009 |
| MS2–RS(SC35) | 28 | Nd | 0.016 | nd | 0.019 |
| MS2–9G8 | 34 | 1.4 | 0.41 | 0.72 | 0.14 |
| MS2–ZnRS(9G8) | 31 | Nd | 0.28 | 0.52 | 0.091 |
| MS2–RS(9G8) | 31 | 0.92 | 0.016 | nd | 0.014 |
| MS2–Tra | 17 | 1.0 | 0.18 | 0.60 | 0.24 |
| MS2–Tra(ΔPro) | 17 | Nd | nd | 0.61 | 0.24 |
| MS2–Tra(ΔRS) | 4 | nd | nd | 0.0041a | 0.015 |
| MS2–Tra2 | 22 | 0.59 | 0.29 | 0.74 | 0.30 |
| MS2–Tra2(ΔRS1) | 9 | 0.067 | nd | 0.014 | 0.010 |
| MS2–Tra2(ΔRS2) | 14 | 0.46 | nd | 0.17 | 0.068 |
All rate constants are expressed in units of h−1. Each value was determined from at least three independent splicing reactions consisting of 7–8 time points over a 4 h time period. To derive rate constants, the data at time points following the lag were fit to a first-order rate equation (43). Experimental error for the rate constants is within 50%. nd, not determined.
aThe splicing of the dsx200-(MS2)2 substrate was not significantly above background.
Figure 3.
Individual dsx enhancer complex proteins are exceptionally potent activators relative to their RS content. The average splicing rate for the dsx300-(MS2)2 RNA substrate is plotted as a function of the RS/SR dipeptide content for each MS2-fusion protein (data from Tables 1 and 2). The line indicates a linear fit for five MS2–RS domain proteins previously evaluated for splicing efficiency (25). Filled triangles indicate MS2–RS domain proteins utilized in these experiments. The open triangle denotes MS2–RS(9G8). Filled circles indicate the dsx enhancer MS2-fusion proteins. The RS content for complexes formed on the dsx300-(MS2)2 substrate containing two MS2-binding sites is four times that of each hybrid protein because MS2 binds the MS2-hairpin RNA as a dimer. The filled diamond represents the average splicing rate of four dsxRE complexes and assumes one molecule of each enhancer protein (Tra, Tra2 and 9G8) is bound to a single 13 nt repeat element.
The domains responsible for exceptional potency of dsx enhancer proteins
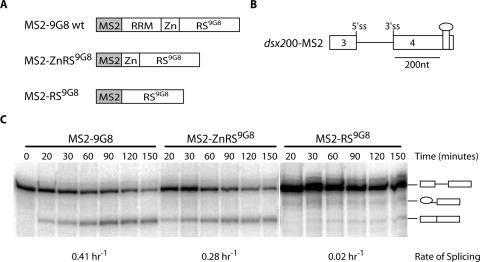
To determine which domains of Tra, Tra2 and 9G8 are responsible for the strong splicing activation, we performed a series of truncation and deletion experiments. In the case of 9G8 we compared full-length 9G8 with an RRM deletion and an RRM/Zinc-knuckle deletion. Splicing analysis of the substrate dsx200-(MS2)2 indicated that a significant drop in activity was observed when the Zinc-knuckle domain was deleted from the fusion protein (Figure 4). Although the presence of the RS domains is required for splicing activity (25), our results suggest that the Zinc-knuckle and the RS domain of 9G8 cooperate in promoting splicing. However, the mechanisms for this cooperation are currently unclear.
Figure 4.
The Zinc-knuckle and RRM domains add significant potency to 9G8 splicing activation. (A) Schematic representation of MS2–9G8 recombinant fusion proteins. MS–9G8 is full-length 9G8 fused in-frame to the bacteriophage MS2 coat protein. Likewise, MS2–ZnRS(9G8) contains only the Zinc-knuckle and RS domain of 9G8, and MS2–RS(9G8) contains only the RS domain of 9G8, fused in-frame to the MS2 protein. (B) Schematic representation of the dsx mini-gene RNA substrate used (dsx200-MS2) with one synthetic MS2-hairpin enhancer element positioned ∼200 nt downstream of the female-specific 3′ splice site. (C) In vitro splicing assays were performed with a dsx mini-gene RNA substrate containing a single MS2-hairpin ∼200 nt from the 3′ splice site (dsx200-MS2) in the presence of recombinant MS2-fusion proteins. Diagrams to the right indicate unspliced, spliced or intermediate products. Average splicing rates for each MS2-fusion protein are listed below the corresponding gel (data from Table 2).
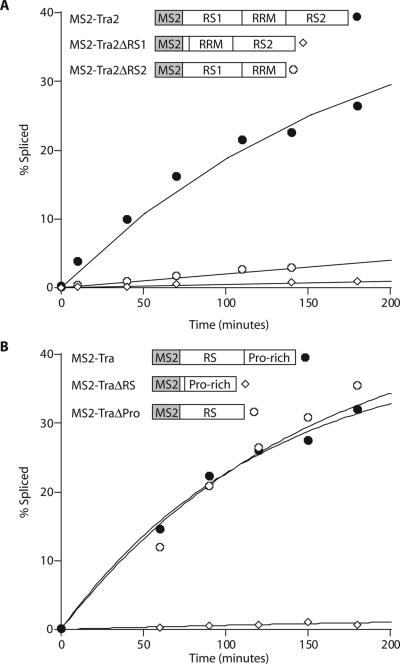
Similar experiments were carried out with wt and mutant versions of Tra and Tra2 (Figure 5 and Table 2). The results show that both RS domains of Tra2 contribute significantly to splicing activation, especially at a distance of 300 nt from the regulated splice site. However, the loss of the N-terminal RS domain (30-fold reduction) is much more detrimental to the splicing activity than the loss of the C-terminal RS domain (4-fold reduction). As was observed for 9G8, the RS domains of Tra2 cooperate in mediating splicing activation. This is because the sum of the RS1 and RS2 activation potential is considerably less than what was observed for full-length Tra2 (Figure 5A and Table 2). We conclude that both RS domains of Tra2 are required for maximal splicing activation.
Figure 5.
The RS domains of Tra and Tra2 are essential for splicing activation. In vitro splicing assays were performed with the dsx300-(MS2)2 RNA substrate in the presence of the recombinant MS2-fusion proteins represented by schematic diagrams. (A) Splicing activities for full-length Tra2 (MS2–Tra2) indicated by filled circles, RS1-deletion mutant (MS2-Tra2ΔRS1) indicated by open diamonds and RS2-deletion mutant (MS2-Tra2ΔRS2) indicated by open circles are plotted as a function of time. (B) Splicing activities for full-length Tra (MS2–Tra) indicated by filled circles, RS-deletion mutant (MS2-TraΔRS) indicated by open diamonds and proline-rich region-deletion mutant (MS2-TraΔPro) indicated by open circles are plotted as a function of time. Average splicing rates for these recombinant proteins are listed in Table 2.
The architecture of Tra is different from Tra2 as Tra does not contain a recognizable RNA-binding domain. The N-terminal RS domain is followed by a C-terminal proline-rich domain (Figure 5B). To determine whether the proline-rich domain contributes to splice-site activation, we compared the splicing efficiency of the dsx300-(MS2)2 substrate in the presence of Tra fused to MS2, in the presence of a proline-rich domain deletion of Tra fused to MS2, or in the presence of an RS domain deletion of Tra fused to MS2. Splicing experiments indicated that the deletion of the RS domain reduced splicing efficiency to background levels. In contrast, deletion of the proline-rich domain did not affect the activation potential of Tra (Figure 5B). We conclude that the RS domain of Tra is essential and sufficient to fully activate splicing at the female-specific 3′ splice site of the dsx300-(MS2)2 substrate.
The components of the dsxRE are functionally unique relative to other RS domain proteins
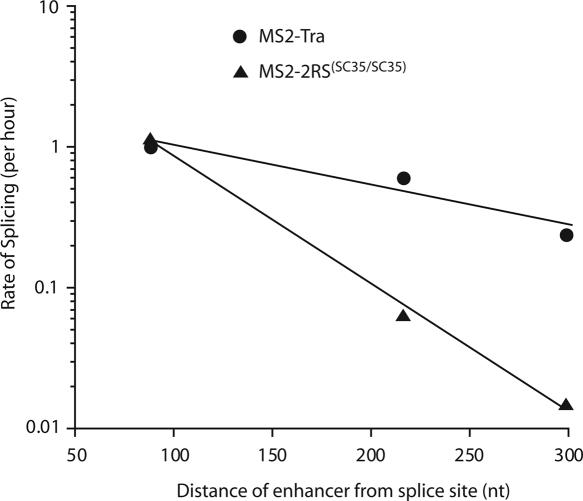
Our kinetic analysis indicated that the components of each dsxRE are more potent splicing activators compared with canonical SR proteins. To evaluate the activation potential of Tra, Tra2 and 9G8 at various distances from the regulated splice site, we carried out in vitro splicing reactions with substrates that harbor MS2-hairpin domains at various distances from the regulated splice site. In agreement with previous observations (26), a large decrease in splice-site activation was observed for the test substrates that were activated by MS2–RS fusion proteins (Figure 6 and Table 2). Moving the MS2-hairpin from 70 to 300 nt downstream from the female-specific splice site resulted in a >50-fold reduction in splicing efficiency. However, substituting MS2–Tra for MS2–2RS(SC35/SC35) in the same experiment resulted in only a 4-fold reduction in the observed rate of splicing. Similar trends were also determined for MS2–9G8 and MS2–Tra2 (Table 2). These results show that the components of the dsxRE complex are highly potent activators, even at a distance of 300 nt from the regulated splice site.
Figure 6.
Tra, Tra2 and 9G8 are more potent female-specific dsx splicing activators at greater distances than other RS domain-containing proteins. Representative splicing rates for MS2–Tra, represented by filled circles, and MS2–2RS(SC35/SC35) represented by filled triangles. Observed rates of splicing are plotted as a function of the distance between the enhancer elements and the female-specific dsx 3′ splice site.
DISCUSSION
The activation potential of the dsxRE
The formation of splicing enhancer complexes is essential for the activation of most constitutive and differential splicing patterns. To gain insights into the mechanisms of enhancer complex assembly and function, we investigated the role of Tra, Tra2 and 9G8 in the recruitment of the splicing machinery. Each dsxRE serves as a binding platform for the cooperative assembly of multiple multi-component splicing enhancer complexes consisting of the SR proteins 9G8, Tra and Tra2 (34,35). To determine the activation potential of the SR proteins that comprise the dsxRE, we generated a series of fusion proteins between the MS2 coat protein and Tra, Tra2, 9G8 or other SR proteins. Because MS2 interacts specifically with a hairpin loop, we were able to form stable interactions of individual components of the dsxRE complex in isolation. Splice-site activation was tested in vitro with each of the fusion proteins present at concentrations that saturate binding to the MS2-hairpin but that did not support non-specific RNA binding. Using this tethering approach, we demonstrated that each fusion protein in isolation was able to activate splicing. However, they differed drastically in the extent of activation. A previous work indicated that enhancer-dependent splice-site activation correlates with the number of RS dipeptide repeats present in each protein (25). Tra, Tra2 and 9G8 do not follow this trend, as they activate splicing much more efficiently than other SR proteins (Figure 3). Our analysis demonstrated that in the context of dsx, Tra, Tra2 and 9G8 are highly active splicing activators that may use mechanisms of activation distinct or in addition to those described for canonical SR proteins.
The roles of 9G8 in dsx splicing
Tra, Tra2 and 9G8 mutants were generated to determine which domains are responsible for their high activation potential. In all cases, the presence of their RS domains was required for maximal splice-site activation. However, the overall contributions of the RS domains tested differed significantly. In the case of the SR protein 9G8, the most significant drop in activity was observed when the Zn-knuckle was deleted from the fusion protein. These observations suggest that the Zn-knuckle domain of 9G8 contains a stretch of amino acids that promotes splicing activity, however, it is unknown how the effect is mediated. Perhaps, the Zn-knuckle domain could be involved in direct interactions with components of the splicing machinery. Alternatively, the presence of the Zn-knuckle could induce a specific structural fold within the RS domain that renders it more potent or accessible for phosphorylation and dephosphorylation, thus overcoming the intrinsically disordered structure of RS domains (38).
Previous studies suggested that the RNA-binding domain of the SR protein ASF/SF2 is directly involved in mediating spliceosomal assembly (39). It is possible that the RRM of ASF/SF2 makes up a binding platform involved in specific protein–protein interactions. Alternatively, the RRM-mediated interaction of ASF/SF2 with the pre-mRNA may modulate splicing by competing with the binding of other regulatory factors, thus suggesting that antagonism between positive and negative splicing factors may be derived from competition for overlapping pre-mRNA binding sites (40). Our observation that the Zn-knuckle of 9G8 drastically increases 9G8's activation potential is more consistent with the formation of a binding platform involved in specific protein–protein interactions. This is because all fusion proteins tested interacted with the same MS2-hairpin structure on the pre-mRNA. Thus, the potent splice-site activation of 9G8 is mediated not only by the RS domain but also by portions of its RRM.
The roles of Tra and Tra2 in dsx splicing
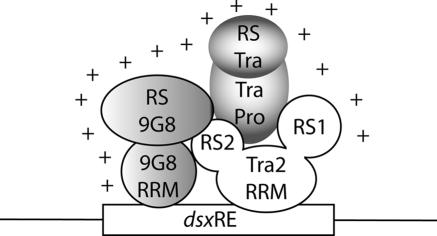
In the case of Tra2, both RS domains were shown to contribute to splicing activation, however, the N-terminal RS1 domain was at least 5-fold more potent than the C-terminal RS2 domain. These observations appear to be in disagreement with results obtained from mutational analyses carried out in transgenic flies (41). It was demonstrated that the deletion of RS2 abolishes sex-specific dsx splicing in flies while the deletion of RS1 had only marginal effects. Interestingly, the same study also showed that RS2 deletions of Tra2 significantly reduced protein interactions with other SR proteins. However, the results from both studies can be reconciled by proposing that RS2 of Tra2 is essential for the formation of the heterotrimeric complex on each of the dsxREs. This model suggests that in the absence of RS2 no stable heterotrimeric complex forms and female-specific splicing of dsx is severely compromised. In the absence of RS1, heterotrimeric complex formation is still possible and the stable recruitment of 9G8 and Tra is sufficient to mediate female-specific splicing of dsx. Thus, RS1 and RS2 of Tra2 may have distinct roles in activating female-specific splicing of dsx. RS1 may be involved in recruiting spliceosomal components, whereas RS2 may be engaged in protein–protein interactions that ensure high stability of the heterotrimeric complex assembled on each dsxRE (Figure 7).
Figure 7.
Extended activation domain of the dsxRE. The heterotrimeric complex that binds each dsxRE generates a potent splicing activation domain that is composed of contributions from each complex member. The RS domains of 9G8, Tra and the RS1 domain of Tra2 may be involved in mediating spliceosomal recruitment to the regulated splice site. RS2 may be involved in securing complex stability through essential protein–protein interactions.
In the case of Tra, the mutational analysis clearly indicates that the RS domain is sufficient to promote high levels of splice-site activation. No splicing activity was observed for the proline-rich domain of Tra, suggesting that this domain is not involved in direct interactions with the spliceosome.
Spliceosome activation by the dsx ESE
Because none of the fusion proteins alone, but the sum of all, can account for the activation potential of the fully assembled dsx enhancer complex (Figure 3), we propose that each component of the heterotrimeric enhancer complex participates in interactions with the spliceosome, generating an extended activation domain that is stabilized through the cooperative interactions between the components of the dsxRE (34,35) (Figure 7). Perhaps, this hypothesis could have been tested more directly by generating MS2-fusion proteins linking the SR domains of Tra, Tra2 and 9G8 or by creating a fusion between MS2 and all three full-length proteins. However, such fusion experiments did not guarantee to fully reproduce the cooperative interactions between RS1 and RS2 of Tra2 nor the contributions of the Zn-knuckle domain of 9G8.
Tra and Tra2 regulate sexual differentiation and behavior of Drosophila through alternative splicing of the dsx and fru genes (42). To avoid mixed expression of male- and female-specific transcription factors, the regulation of dsx and fru alternative splicing requires tight control and an efficient switch between male and female splicing. The composition and location of the dsx ESE 300 nt downstream of the regulated splice site is conserved among different Drosophila species, suggesting evolutionary influences in generating a unique exon architecture (43). Indeed, it was shown that the regulation of dsx splicing could only be achieved if the dsx ESE is 300 nt downstream of the regulated splice site. Constitutive activation was observed when the dsx ESE was within 100 nt of the splice site (44). Thus, the dsx ESE must have evolved to communicate female-specific splice-site recognition with high efficiency from a distance that usually does not support splicing regulation. We previously demonstrated that the cooperative assembly and the multiple presence of the dsxRE are contributing factors in achieving the elevated potency required for female-specific splicing (35). The analysis presented here reveals an additional mechanism to ensure efficient splice-site selection. Each repeat element within the dsx ESE recruits the unusually effective splicing activators Tra, Tra2 and 9G8 to form a splicing enhancer complex of great potency. Thus, multiple enhancer complexes with high stability and elevated activity ensure practically exclusive alternative splicing of dsx and fru.
Acknowledgments
We are grateful to Tom Maniatis for providing Tra and Tra2, and members of the Hertel laboratory for helpful comments on the manuscript. HeLa cells were obtained from the National Cell Culture Center (Minneapolis, MN). This work was supported by NIH grant GM 62287 (K.J.H.) Funding to pay the Open Access publication charges for this article was provided by NIH grant GM 62287.
Conflict of interest statement. None declared.
REFERENCES
- 1.Smith C.W., Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 2.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Pagani F., Baralle F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nature Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 4.Blencowe B.J. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 5.Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveley B.R., Hertel K.J., Maniatis T. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA. 2001;7:806–818. doi: 10.1017/s1355838201010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 8.Fu X.D., Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 9.Krainer A.R., Conway G.C., Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 10.Ge H., Manley J.L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 11.Zahler A.M., Lane W.S., Stolk J.A., Roth M.B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 12.Fu X.-D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 13.Roscigno R.F., Garcia-Blanco M.A. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 14.Chew S.L., Liu H.X., Mayeda A., Krainer A.R. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc. Natl Acad. Sci. USA. 1999;96:10655–10660. doi: 10.1073/pnas.96.19.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mermoud J.E., Cohen P., Lamond A.I. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 1992;20:5263–5269. doi: 10.1093/nar/20.20.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mermoud J.E., Cohen P.T., Lamond A.I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaal T.D., Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 19.Shen H., Kan J.L., Green M.R. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 20.Shen H., Green M.R. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 21.Hertel K.J., Lynch K.W., Maniatis T. Common themes in the function of transcription and splicing enhancers. Curr. Opin. Cell Biol. 1997;9:350–357. doi: 10.1016/s0955-0674(97)80007-5. [DOI] [PubMed] [Google Scholar]
- 22.Graveley B.R., Hertel K.J., Maniatis T. SR proteins are ‘locators’ of the RNA splicing machinery [letter; comment] Curr. Biol. 1999;9:R6–R7. doi: 10.1016/s0960-9822(99)80032-3. [DOI] [PubMed] [Google Scholar]
- 23.Lam B.J., Hertel K.J. A general role for splicing enhancers in exon definition. RNA. 2002;8:1233–1241. doi: 10.1017/s1355838202028030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam B.J., Bakshi A., Ekinci F.Y., Webb J., Graveley B.R., Hertel K.J. Enhancer-dependent 5′-splice site control of fruitless pre-mRNA splicing. J. Biol. Chem. 2003;278:22740–22747. doi: 10.1074/jbc.M301036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graveley B.R., Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 26.Graveley B.R., Hertel K.J., Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker B.S. Sex in flies: the splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 28.Ryner L.C., Baker B.S. Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes Dev. 1991;5:2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- 29.Ryner L.C., Goodwin S.F., Castrillon D.H., Anand A., Villella A., Baker B.S., Hall J.C., Taylor B.J., Wasserman S.A. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 30.McKeown M., Belote J.M., Boggs R.T. Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male Drosophila. Cell. 1988;53:887–895. doi: 10.1016/s0092-8674(88)90369-8. [DOI] [PubMed] [Google Scholar]
- 31.Nagoshi R.N., McKeown M., Burtis K.C., Belote J.M., Baker B.S. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- 32.Nagoshi R.N., Baker B.S. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- 33.Heinrichs V., Ryner L.C., Baker B.S. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell. Biol. 1998;18:450–458. doi: 10.1128/mcb.18.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch K.W., Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 35.Hertel K.J., Maniatis T. The function of multisite splicing enhancers. Mol. Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 36.Hicks M.J., Lam B.J., Hertel K.J. Analyzing mechanisms of alternative pre-mRNA splicing using in vitro splicing assays. Methods. 2005;37:306–313. doi: 10.1016/j.ymeth.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 37.Tian M., Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992;256:237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- 38.Haynes C., Iakoucheva L.M. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34:305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J., Krainer A.R. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eperon I.C., Makarova O.V., Mayeda A., Munroe S.H., Caceres J.F., Hayward D.G., Krainer A.R. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amrein H., Hedley M.L., Maniatis T. The role of specific protein–RNA and protein–protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 42.Forch P., Valcarcel J. Splicing regulation in Drosophila sex determination. Prog. Mol. Subcell. Biol. 2003;31:127–151. doi: 10.1007/978-3-662-09728-1_5. [DOI] [PubMed] [Google Scholar]
- 43.Hertel K.J., Lynch K.W., Hsiao E.C., Liu E.H., Maniatis T. Structural and functional conservation of the Drosophila doublesex splicing enhancer repeat elements. RNA. 1996;2:969–981. [PMC free article] [PubMed] [Google Scholar]
- 44.Tian M., Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]