The 1999 outbreak of mosquito-borne West Nile virus (WNV) in New York City (1) and its subsequent hemispheric spread by infected migratory birds have been met by urgency for new knowledge about this flavivirus. WNV is closely related to a number of other medically important flaviviruses that include the family's prototype, the storied yellow fever virus (YFV; flavi means yellow), dengue virus (DV), and the Japanese encephalitis and tick-borne encephalitis (TBE) viruses. Much of what we know about flavivirus epidemiology, biology, and pathogenesis derives from collective observations made about these particular flaviviruses. They exhibit both distinctive and shared clinical expression, and their biology appears to be more similar than dissimilar, so that lessons learned from one are likely to apply to the others. The flavivirus single-strand, positive-sense RNA genome encodes three structural (capsid, matrix, and envelope) and seven nonstructural (NS) proteins (2). Considerable attention has naturally been directed at the biology of the flavivirus envelope E protein because it subserves virus attachment and neutralization. E protein atomic structure has been solved for TBE, DV, and WNV, elegantly informing us about flaviviral entry mechanisms and how neutralizing antibodies protect (3–5). In the course of screening YFV monoclonal antibodies for protective activity in mice a number of years ago, investigators were quite surprised to find that, in addition to protective anti-YFV E monoclonal antibodies, passive transfer of some monoclonal antibodies against YFV NS1 glycoprotein (then known as “gp48”) protected mice against YFV encephalitis (6, 7). Remarkably, active immunization with YFV NS1 also protected monkeys against classic yellow fever (8). The work of Chung et al. (9) in this issue of PNAS brings us yet another surprise about NS1 that may enhance our knowledge of flavivirus pathogenesis and immunity: WNV NS1 is reported to exhibit an immunomodulatory function by regulating complement activity.
NS1 is a highly conserved, ≈48-kDa glycoprotein that is essential for flavi-virus RNA replication, although its precise function remains poorly defined (10, 11). It exists in the cell as a heat-labile homodimer that associates with cellular organelle membranes and is transported to the mammalian cell surface (12, 13) where it is vulnerable to immunological recognition. NS1 is also secreted by flavivirus-infected mammalian cells as a soluble hexamer (14, 15). DV NS1 is efficiently endocytosed by liver cells after i.v. injection of normal mice, and it associates with the surface of cultured normal human liver cells by an as-yet-undefined mechanism (16). It accumulates in late endosomes of cultured liver cells where it is quite resistant to degradation; remarkably, DV NS1 pretreatment also appears to enhance DV replication (16). Copious amounts of NS1 circulate in DV-infected patients (17, 18) in whom NS1 blood levels have been shown to correlate with disease severity (19). Microvascular leakage in such patients has been linked to complement activation by NS1–antibody complexes (20). Similarly, NS1 and WNV cocirculate early in the course of experimental infection in hamsters where NS1 abundance also correlates with disease severity (21). It seems highly probable that the same holds true for other flavivirus infections as well.
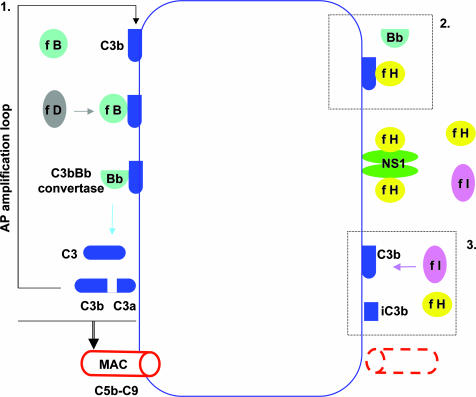
In the normal host, potentially neuroinvasive WNV faces the interactive mechanisms of the innate and adaptive immune systems. In mice, WNV appears to be susceptible to all three complement pathways, i.e., classical, lectin, and alternative, through the influence of individual complement activation components on adaptive immune responses and possibly by direct virolysis (22). Importantly, there is evidence that complement deficiencies, especially in two critical elements of alternative pathway activation, factor B (fB) and factor D (fD), lead to earlier WNV invasion of the mouse central nervous system (23). A key event in the alternative pathway (24) is the interaction between covalently surface-bound C3b (the convergence subunit molecule of the three complement pathways), fB, and fD that together generate the alternative pathway C3 convertase, C3bBb. Uninterrupted cleavage of serum C3 by C3bBb amplifies the amount of C3b available for deposition on cell membranes, leading, through subsequent steps, to assembly on the target cell membrane of the C5b-9 membrane attack complex (Fig. 1). Set into motion, and if unchecked, the activated alternative pathway consumes its components and threatens host tissues. Glycoprotein factor H (fH) is the predominant circulating regulator of the alternative pathway of complement activation (25). Its chief function is to protect innocent bystander host cells from collateral damage in the course of complement activation, and a number of progressive human microangiopathies have been linked to specific fH mutations and sequence polymorphisms. fH, having no intrinsic enzymatic activity, acts in concert with the circulating serine protease factor I (fI) to irreversibly inactivate newly formed C3b, the pivotal initiator of the terminal complement cascade that leads to formation of the membrane attack complex (C5b-9).
Fig. 1.
Possible effects of cell surface NS1-complexed fH on alternative complement pathway (AP) activation. (Box 1) The normal AP amplification loop. Interactions between cell surface-bound C3b, fB, and protease factor D generate the C3bBb convertase that amplifies C3b production, which finally leads to C5b-9 membrane attack complex (MAC) formation and cell membrane damage. (Box 2) Recruitment of fH by NS1 disrupts C3bBb convertase assembly and function. (Box 3) In addition, fH, acting with protease fI, cleaves cell surface-bound C3b to produce the functionally inactivated form of C3b, iC3b. The net result of events in boxes 2 and 3 is inhibition of MAC formation. (A comprehensive summary of alternative complement pathway activation is found in ref. 24.)
The novel finding of Chung et al. (9) originated in the course of preparing WNV recombinant NS1 for monoclonal antibody development (26). A ≈150-kDa protein was copurified with rNS1 if FBS was present in the system. Suspicion that the protein might be a ligand for NS1 led to its identification as bovine complement fH, and further tests revealed the same propensity of NS1 to bind to human fH. After confirming that fH bound to NS1, Chung et al. (9) established that the NS1–fH complex accelerated C3b digestion by fI in solution and that NS1 had no intrinsic cofactor activity. They then demonstrated the very same effect on Chinese hamster ovary (CHO) cells that displayed recombinant NS1 in amounts equivalent to those found on WNV-infected cells. Here, fH engaged by cell surface NS1 accelerated the breakdown of C3bBb convertase so that C3b deposition was preferentially reduced in the NS1 transfectants, and consequently, so was the formation of the terminal C5b-9 membrane attack complex (Fig. 1). The alternative pathway complement amplification loop mechanism was thereby significantly retarded by NS1 expression, and, very likely, WNV-infected cells also would be protected from alternate complement pathway-mediated damage in the same way.
A number of bacterial and fungal pathogens evade complement-dependent phagocytosis by binding fH, but viruses usually evade immunological defenses either by escape mutations in the case of RNA viruses whose replication is unedited or by deceptions that involve hijacking and altering host genes of the humoral and cellular immune system in the case of some large DNA viruses (27). The finding by Chung et al. (9) that fH binding by a viral protein (WNV NS1) interferes with C3b function has not been previously recognized as a mechanism of immune evasion by any viruses. It will be immediately important to find out whether their discovery applies to other flavivirus family members, especially in cell types known to support replication of specific flaviviruses in vivo, such as cells of nervous or liver tissue, and cells of monocyte/macrophage lineage that are likely to be the earliest site of flavivirus replication after insect bite. In view of the large amounts of circulating NS1 early in flavivirus infection, it also would be interesting to see whether soluble NS1 deposited on uninfected cells will behave in the same fashion with respect to fH as NS1 displayed on transfected (or naturally infected) cells. If so, NS1 would then appear to perform double duty by enhancing infection of flavi-virus-susceptible cells (16) and then, as the results of Chung et al. (9) might suggest, by defending them against the action of activated complement. A key question that arises from their findings is how they might relate to NS1 as the target of protective antibodies. Earlier work from the same laboratory linked the activity of some strongly protective anti-WNV NS1 monoclonal antibodies to host Fcγ receptor (FcγR) function, although at least one antibody conferred solid protection in FcγR and C1q (classical complement pathway)-deficient mice (26). In light of Chung et al.'s (9) current findings, it seems reasonable to speculate that protective anti-NS1 antibodies might also operate by blocking fH attachment to NS1. More complete understanding of NS1–fH interactions and their consequences will require knowledge of their respective topographies at the atomic level. This knowledge will further inform us of how protective anti-NS1 antibodies work and may well offer insight into the NS1–fH complex as a potential target for antiviral drug treatment, which currently does not exist for any insect-borne flavivirus disease.
Footnotes
The author declares no conflict of interest.
See companion article on page 19111.
References
- 1.Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, Crise B, Volpe KE, Crabtree MB, Scherret JH, et al. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- 2.Chambers TJ, Hahn CS, Galler R, Rice CM. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 3.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 4.Modis Y, Ogata S, Clements D, Harrison SC. Proc Natl Acad Sci USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y. J Virol. 2006;80:11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlesinger JJ, Brandriss MW, Walsh EE. J Immunol. 1985;135:2805–2809. [PubMed] [Google Scholar]
- 7.Gould EA, Buckley A, Barrett AD, Cammack N. J Gen Virol. 1986;67:591–595. doi: 10.1099/0022-1317-67-3-591. [DOI] [PubMed] [Google Scholar]
- 8.Schlesinger JJ, Brandriss MW, Cropp CB, Monath TP. J Virol. 1986;60:1153–1155. doi: 10.1128/jvi.60.3.1153-1155.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, Atkinson JP, Diamond MS. Proc Natl Acad Sci USA. 2006;103:19111–19116. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackenzie JM, Jones MK, Young PR. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 11.Muylaert IR, Galler R, Rice CM. J Virol. 1997;71:291–298. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkler G, Randolph VB, Cleaves GR, Ryan TE, Stollar V. Virology. 1988;162:187–196. doi: 10.1016/0042-6822(88)90408-4. [DOI] [PubMed] [Google Scholar]
- 13.Winkler G, Maxwell SE, Ruemmler C, Stollar V. Virology. 1989;171:302–305. doi: 10.1016/0042-6822(89)90544-8. [DOI] [PubMed] [Google Scholar]
- 14.Crooks AJ, Lee JM, Easterbrook LM, Timofeev AV, Stephenson JR. J Gen Virol. 1994;75:3453–3460. doi: 10.1099/0022-1317-75-12-3453. [DOI] [PubMed] [Google Scholar]
- 15.Flamand M, Megret F, Mathieu M, Lepault J, Rey FA, Deubel V. J Virol. 1999;73:6104–6110. doi: 10.1128/jvi.73.7.6104-6110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alcon-LePoder S, Drouet MT, Roux P, Frenkiel MP, Arborio M, Durand-Schneider AM, Maurice M, Le Blanc I, Gruenberg J, Flamand M. J Virol. 2005;79:11403–11411. doi: 10.1128/JVI.79.17.11403-11411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcon S, Talarmin A, Debruyne M, Falconar A, Deubel V, Flamand M. J Clin Microbiol. 2002;40:376–381. doi: 10.1128/JCM.40.2.376-381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young PR, Hilditch PA, Bletchly C, Halloran W. J Clin Microbiol. 2000;38:1053–1057. doi: 10.1128/jcm.38.3.1053-1057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libraty DH, Young PR, Pickering D, Endy TP, Kalayanarooj S, Green S, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. J Infect Dis. 2002;186:1165–1168. doi: 10.1086/343813. [DOI] [PubMed] [Google Scholar]
- 20.Avirutnan P, Punyadee N, Noisakran S, Komoltri C, Thiemmeca S, Auethavornanan K, Jairungsri A, Kanlaya R, Tangthawornchaikul N, Puttikhunt C, et al. J Infect Dis. 2006;193:1078–1088. doi: 10.1086/500949. [DOI] [PubMed] [Google Scholar]
- 21.Macdonald J, Tonry J, Hall RA, Williams B, Palacios G, Ashok MS, Jabado O, Clark D, Tesh RB, Briese T, Lipkin WI. J Virol. 2005;79:13924–13933. doi: 10.1128/JVI.79.22.13924-13933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samuel MA, Diamond MS. J Virol. 2006;80:9349–9360. doi: 10.1128/JVI.01122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mehlhop E, Diamond MS. J Exp Med. 2006;203:1371–1381. doi: 10.1084/jem.20052388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walport MJ. N Engl J Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez de Cordoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Chung KM, Nybakken GE, Thompson BS, Engle MJ, Marri A, Fremont DH, Diamond MS. J Virol. 2006;80:1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcami A, Koszinowski UH. Immunol Today. 2000;21:447–455. doi: 10.1016/S0167-5699(00)01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]