Abstract
A method is introduced for subdiffraction imaging that accumulates points by collisional flux. It is based on targeting the surface of objects by fluorescent probes diffusing in the solution. Because the flux of probes at the object is essentially constant over long time periods, the examination of an almost unlimited number of individual probe molecules becomes possible. Each probe that hits the object and that becomes immobilized is located with high precision by replacing its point-spread function by a point at its centroid. Images of lipid bilayers, contours of these bilayers, and large unilamellar vesicles are shown. A spatial resolution of ≈25 nm is readily achieved. The ability of the method to effect rapid nanoscale imaging and spatial resolution below Rayleigh criterion and without the necessity for labeling with fluorescent probes is proven.
Keywords: diffusion controlled, fluorescence, single molecule
For the purpose of visualizing biological processes and structures, it is essential to develop new methods that can determine the spatial locations of molecules with the highest possible precision. The submolecular methods of spatial resolution such as x-ray and electron topography, atomic force microscopy, and near-field optical microscopy have contributed enormously toward this goal. However, new approaches that are not invasive to biological objects and are accessible to bulk, as well as surface structures, could have a significant impact and could address many new questions in the life sciences (1). Optical methods based on fluorescence detection do have single-molecule sensitivity and can be arranged to be nondestructive even when performed in natural environments, incorporating such complex and sensitive structures as living cells. In principle, optical responses can probe the entire 3D space. Although diffraction limits the spatial resolution of optical methods, many approaches have been developed recently that allow distance measurements on scales that are much shorter than the wavelength of the light.
It is possible to specify the location of a single molecule with very high precision from measurements of its fluorescence by fitting the emitted intensity distribution to the 2D spatial parameters of the point-spread function (PSF). This approach has been shown to locate emitters with ≈1-nm precision (2). The distance between two optically different molecules can be estimated by FRET methods that can serve as molecular rulers at nanometer accuracy (3). Two molecules emitting light at different frequencies have also been resolved by means of PSF measurements. Even for molecules having the same spectra, the process of photobleaching causes the fluorescence source to be switched between molecules. When combined with a PSF measurement, this approach has been able to distinguish pairs (4) and quartets (5) of molecules with nanometer precision. Another approach has been to analyze the blinking trajectory of the emission of quantum dots to distinguish between a single dot and groups of them clustered on the nanometer scale (6). The intermittency in the fluorescence signal caused by diffusion-controlled collisions of a fluorescent probe with objects of interest was recently introduced as a method of subdiffraction imaging (7). It permitted the identification of two closely localized 50-nm-radius vesicles. Most of these methods of subdiffraction visualization of the structure of groups of particles are based on the principle of time separation of detection; only one molecule at a time is in a bright state.
Objects having different shapes can be distinguished from the distribution of point positions of fluorescent probes that are bound to them. If the probes are localized at nanometer precision by PSF methods, the boundaries of the object may also be visualized on the nanometer scale from a high-resolution image of the distribution of points. Nanometer-localized multiple single-molecule fluorescence microscopy (8) has been used to create images of short DNA fragments by combining centroid localization and photobleaching of a small number of single chromophores. Localization of a much larger set of probes, sufficient to provide an outline of a complete structure, was recently demonstrated for intracellular proteins in a method termed photoactivated localization spectroscopy (9). DNA fragments were visualized at a resolution of 20 nm with photoswitchable fluorophores by stochastic optical reconstruction (10). These approaches have much-improved statistics, yet they rely on the photophysical characteristics of photoactivatable fluorescent proteins or photoswitchable dyes.
In the present paper, a method of high-resolution imaging is introduced that combines the concepts of intermittency caused by bimolecular collisions (7), photobleaching, and PSF measurements. In this approach, the collision followed by binding generates a fluorescence signal spike, so an almost unlimited number of probe molecules can be used to form entire images that could readily be acquired with nanometer resolution.
Principles of Point Accumulation for Imaging in Nanoscale Topography.
In the points accumulation for imaging in nanoscale topography (PAINT) method, the object to be imaged is continuously targeted by fluorescent probes present in the solution. The flux of molecules incident on the object depends on the diffusion coefficient and concentration gradient of the probes. A fluorescent signal appears as a diffraction-limited spot on the object when a label binds to it and is immobilized; it is destroyed when that label dissociates from the object or is photobleached. In general, it is not necessary to specify the nature of the binding that may arise from specific or nonspecific interactions. For example, it might depend on an electrostatic coupling or a hydrophobic interaction, as in the examples we provide herein. The collision rate of probes with a single object is easily controlled, because it depends linearly on the concentration of the probes. The rate of appearance of fluorescent spikes is then approximated by the diffusion-controlled bimolecular reaction rate constant (11) times the probability that a collision will lead to a binding configuration that generates a signal from the probe (12). Further discrimination between locations in an image can be obtained if the probe fluorescence is sensitive to the chemical composition of the object.
In the PAINT procedure, the density of probes on the object surface should be kept below one molecule/μm2 during each observation period. For this condition, every fluorescent image can be treated as a PSF from a single molecule and can be fitted to a 2D Gaussian function to achieve its absolute position with high precision. To meet this requirement, the time between subsequent observation frames is set to be longer than the on- time of the fluorescence burst of a probe, which may be determined by either the natural residence time or the photobleaching time, but the frame length of an image is set to be shorter than the period between collisions. The signaling molecules are then guaranteed to be deactivated between successive image recordings. More quantitatively, the number of molecules targeting a circular area πR2 in unit time from a solution containing N molecules per unit volume is given by a Smoluchowski-type (13) relation, 4DRN (s−1), where D is the diffusion coefficient. Therefore, in the steady state, the number of probes on the object is 4DRN 〈τon〉, where 〈τon〉 is the mean time that the probe spends associated with the object before it undergoes dissociation or is photobleached.
In general, the deactivation rate, 〈τon〉−1, is the sum of the mean rates of photobleaching and dissociation of the probe from the object, if it is assumed that the probes all act independently of one another. The rate of bleaching depends on the excitation intensity and the photochemical and photophysical properties of the probe in its surrounding medium (14). Its contribution can be controlled in an imaging experiment by varying the power density of the excitation and the ratio between the observation time for a single frame and period between frames. The time spent by the probe on the object is an intrinsic property that depends on the interaction of the probe with the object being imaged. Thus, by varying the concentration of probes in the solution and the light intensity, the proper conditions of probe density can easily be found. Because the number of probe molecules in the surrounding solution is not significantly depleted by the photobleaching that occurs on only one relatively small illuminated object, the required image can be achieved by the accumulation of as many image frames as are needed. The image is formed by overlapping these frames. The total recording time is limited by the mechanical stability and possible movements of the objects relative to the light collection system.
Results and Discussion
Imaging of a Large Unilamellar Vesicles (LUVs).
To demonstrate the method we imaged LUVs, which have relatively well defined shapes when attached to a glass surface. These vesicles have been well characterized (15). The probe was chosen to be Nile red because the collisional kinetics of Nile red with LUVs has been studied in detail (12). Nile red does not fluoresce in water but exhibits strong fluorescence in a hydrophobic environment, such as the lipid core. The mean residence time that was reported previously for Nile Red associated with liquid-phase LUVs is ≈6 msec when the vesicle radius was chosen as 50 nm (12). With the present conditions of Nile red concentration (10−9 M or N = 6·1011 cm−3) and laser-power density (1 kW/cm2), we found that the probability of observing two fluorescent molecules on a single LUV is very low. The fluorescence image contains approximately one to two fluorescent bright spots per μm2, each one corresponding to a vesicle, reminiscent of a single-molecule image. We collected 4,095 frames of an area of 40 × 40 μm2 at a rate of 50 frames/sec. A short piece of fluorescence trajectory obtained by averaging of the signal in the diffraction-limited area of a single LUV is shown in Fig. 1; it exhibits the intermittency that has been observed in previous work, attributed to collisions of Nile red with the LUV. The large fluctuations in the peak intensity arise because of the broad distribution of residence times. The fragments (3 × 3 μm2) of the regular fluorescent image and a synthetic image obtained as described above are shown in Fig. 2a and b. The single vesicles are well resolved in a large area of observation, and the synthetic image contains details that could not be resolved by standard fluorescence imaging. A total of 6,494 spots were overlapped to form the synthetic image. On average, there was a 10% chance of a probe molecule appearing on a single LUV in each of the 20-msec frames. Thus the apparent collision rate was 5 Hz, somewhat lower than seen in the confocal experiments (12), which had better time resolution.
Fig. 1.
The total integrated intensity vs. time for a single vesicle probed by Nile red. The intermittency is caused by fluorescence bursts of Nile red molecules undergoing collisions with the lipid bilayer.
Fig. 2.
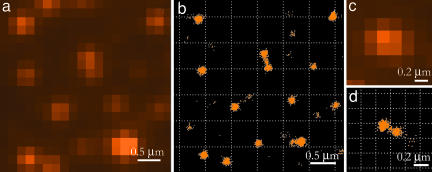
Imaging of vesicles. Fluorescence image (a) and synthetic high-resolution image (b) of vesicles attached to a glass surface. The synthetic image, obtained as described in the text, displays the average coordinates of single fluorophores. (c and d) Fluorescence and synthetic images of two vesicles with a center-to-center distance of ≈200 nm.
To minimize the effects of thermal and mechanical instability, the overall recording time was kept as short as possible. This constraint is met by making the concentration large. On the other hand, the concentration must be low enough that two probes do not arrive on a single vesicle within a single time frame. On average, we arranged to have one probe appearing on an LUV every 10 frames, so that there was a 1% chance of a probe appearing on each of two vesicles within one time frame. Some of the images in Fig. 2b show a connection between nearby vesicles. This connecting region is caused by the infrequent occurrence during one frame of one probe on each vesicle. Under the present conditions, a single collision leading to light emission occurs about every 10 frames, whereas the double collision occurs every 100 frames. When there is a double event, the fitting procedure gives the mean location, which is a point between the two vesicles. The dumbell shape near the center of Fig. 2b consists of 775 total points, with 48 of them located between the vesicles, close to these expectations. Vesicles that are further apart do not show these connecting regions because of the constraints on the eccentricity of the fitting function. In this experiment, the average probe density over the entire image was ≈10 probes/μm2, 10 times higher than required for the wide-field imaging of extended shapes; however, it was low enough for imaging of separately located vesicles on the glass surface.
An averaging effect is expected if the imaged molecule is undergoing significant motion during the frame time. One of many experimental distributions of the density (number per unit area) of fluorescent spots versus their distance from the centroid is shown in Fig. 3a. For a circular or hemispherical vesicle, the centroid is at the center of the circular distribution. The 1/e widths of these distributions (Fig. 3b) were not correlated with the number of points present in the synthetic images of the vesicles. The average width was 27 nm; the sizes of the LUVs in Fig. 2 b and d appear larger in the synthetic image, because the pileup of dots at the center is not easily visualized from the figure. According to the foregoing discussion, the collision rate and hence the number of fluorescent probes accumulated on a vesicle surface during a give time interval should be proportional to the vesicle radius. This has been proven experimentally in other types of experiments (12). However, a diminution of the correlation between the number of spots and the apparent vesicle radius is expected if the fluorescent probe moves significantly during the frame time. Indeed, one would expect to measure an average coordinate if sufficiently rapid diffusion were present. This situation prevails for Nile red in the vesicles. The lateral diffusion coefficient for the bilayer at room temperature is 2–5 μm2/sec (16), and the residence lifetime of the Nile red probe on the vesicle surface is in the millisecond range. Therefore, if the probe is diffusing, its average displacement is expected to be larger than the dimensions of the R = 50-nm vesicle.
Fig. 3.
Spatial distribution of fluorescent spots. (a) Example of a density distribution of fluorescent spots around the center of a vesicle. The solid line represents a Gaussian fit with σ = 28 nm. (b) Histogram of distribution widths obtained as in a for 185 vesicles. (c) Distribution of points density obtained by simulations for 50-nm-radius disks without any diffusive motion of the probes (dots) and with diffusion (D = 3 μm2/sec) for different residence times of 1 (dash), 10 (solid), and 100 (dash-dot) msec.
We performed simulations for a probe diffusing on a circular disk with radius 50 nm. One thousand initial positions of the probes were chosen from a uniform distribution on the circle. Each initial point was assumed to undergo Brownian diffusion with mean-square displacement 4Dt, where D is the diffusion coefficient, and t is the on time. All of the resulting coordinates of a probe, each assumed to have a 148-nm PSF, were overlapped to define the simulated image of that diffusing fluorophore. This image was then fitted to a 2D Gaussian as described in the processing section.
The distributions of the density of points are shown in Fig. 3c for different residence times of 1-msec (dash), 10-msec (solid), and 100- msec (dash-dot) lines. The dotted line represents the distribution in the absence of diffusion (D = 0), and in this case the distribution width is equal to the disk radius. For diffusing probes, the width is significantly smaller; at our experimental conditions (t = 1–10 ms), the expected width is only several nanometers if the probe is undergoing diffusion. Thus, the diffusion of the probes on the small vesicles allows the location of the vesicle center to be determined with high subdiffraction accuracy, but it influences the apparent shape. The width of the experimental distribution is 27 nm, which originates from the uncertainty in the probe localization. However, the absolute location of the centroid of the area over which the probe is moving is, in principle, determined to subnanometer accuracy; the standard deviation is , where σm refers to a single measurement, and N is the number of probes forming the image. In the present experiments, where N was typically 500, the precision was 0.85 nm.
It has been demonstrated that two vesicles in close contact can be resolved by the PAINT method. The diffraction-limited spot in the regular image of Fig. 2c shows a single peak, whereas the synthetic image (see Materials and Methods) of Fig. 2d clearly shows two separate distributions of spots whose center-to-center distance is 202.4 nm. We conclude these are two vesicles. The resolution is limited by the width of the points density distributions, which in the present case is 27 nm.
Imaging of a Supported Bilayer.
To demonstrate the ability of the PAINT method in imaging of more extended shapes, we carried out experiments on a lipid bilayer that is a relatively large object. A supported bilayer was prepared as described in Materials and Methods. The experiment was performed similarly to that described in the LUV imaging section. The image size was 188 μm2, whereas the area occupied by membrane was ≈124 μm2. Nile red was added to the sample chamber to maintain a density on the membrane that was on average 0.23 probes/μm2 in every 20-msec time frame. The density is low enough that the probability of two molecules being colocalized within the diffraction limit is sufficiently low to consider with high accuracy that each fluorescent spot on the image originates from a single probe. A total of 118,163 fluorescent spots were collected in 4,095 frames at 50 frames/sec. A small fragment of the conventional fluorescent image and the synthetic image are shown in Fig. 4, where such structural details as the sharply defined contour of the bilayer and separately located membrane pieces are well resolved by the PAINT method.
Fig. 4.
Image of a supported bilayer on glass, probed by Nile red. (a) Conventional fluorescence image. (b) High-resolution synthetic image obtained by locating 2,778 single Nile red probes collected in 4,095 frames.
As mentioned earlier, it is not essential that there are changes in the spectroscopic properties of the probe when it binds to the surface of the object. Almost any fluorescent probe that binds can be used for imaging by the PAINT method. To demonstrate this, we present an example using the fluorescently labeled protein (transferrin) adsorbed on a clean glass surface. It is presumed that transferrin adsorbs to the glass by an electrostatic interaction with the negatively charged groups on the glass surface. Not all collisions lead to adsorption, which depends on the flux of proteins onto the surface and the ionic strength of the solution. Transferrin protein was dissolved in 100 mM NaCl/10 mM Hepes buffer (pH 7.4) at a concentration of 1.2·10−8 M. The protein does not bind to the neutral lipid bilayer for a long enough time to be observed in the experiments, so with the bilayer on the surface, only the uncovered clean glass area immobilizes the transferrin. The deactivation of the adsorbed probe fluorescence between successive frames was achieved by photobleaching. The sample was continuously excited by the laser at 1 kW/cm2 power, and the recording was performed with 20-msec frame periods with the time interval between frames set at 150 msec, an interval that was long enough to photobleach the fluorophore before the next frame in the sequence. At these conditions, the observed surface density of the fluorophores in a frame never exceeded 0.05 μm−2, and the probability of finding two molecules in the diffraction limited area during one frame was negligible. The result of overlapping of the 20,553 coordinates in 4,095 frames is shown in Fig. 5. In this figure, the bright areas originate from molecules adsorbed to the glass surface, whereas the dark ones correspond to the membrane. The sharp contours of the bilayer are readily visualized at subdiffraction limited resolution. In this example, the diffusion of the probes is sufficiently slow that the image is not affected by probe motion. It should be emphasized that the distribution of binding sites can be obtained at subdiffraction spatial resolution for any labeled protein interacting with any surface using the proposed method.
Fig. 5.
High-resolution synthetic image of the contour of a supported bilayer. Transferrin conjugated with Alexa Fluor 568 creates the image by targeting areas on the glass surface that are not covered by the bilayer (bright regions), whereas the bilayer appears to be unaffected by the flux of labeled proteins (dark area).
Summary and Prospects.
Optical wide-field imaging with subdiffraction resolution can fill the gap between nonoptical methods (<1 nm) and far-field conventional microscopy (>200 nm). Recently, several methods based on single-molecule localization were presented (5, 6, 8) that have subdiffraction resolution along with photon statistics limitations (8) or the requirement of very special probes (9, 10). In this paper, we describe a method of superresolution imaging that is also based on the principle of a precise determination of the coordinates of individual fluorophores for which the accuracy could be as high as 1 nm (2). In our method, we subject the surface of the studied object to a flux of fluorescent probes that attach transiently to the surface. The surface density of probes is easily adjusted by varying their concentration in the surrounding solution and/or the incident power to levels that ensure that the fluorescent bursts are optimally separated in time and space. The sequential localization of large numbers of molecules (usually >104) then defines the shape of the object with nanometer accuracy. In the presence of rapid diffusion of the probe, the centroid of the region explored by the probe is measured very accurately.
We have demonstrated the PAINT technique by imaging a supported bilayer and large lipid vesicles 100 nm in diameter. A fluorescently labeled transferrin protein and a small environment-sensitive dye (Nile red) were used to demonstrate the possible wide selection of probes that might be used for imaging with this method. With the PAINT method, micrometer-sized images were recorded in seconds. The number of points forming the image can be as large as required to achieve the appropriate image sharpness.
The PAINT method can be used without any modifications for imaging objects such as cell organelles, cell membranes, and other lipid objects. By varying the probe molecule, it will be possible to achieve selectivity in imaging specific objects. For example, many fluorescently labeled proteins having high specificity to particular cell compartments are commercially available and can serve as probes for imaging. The essential requirement of the proposed imaging method is that the probe molecules interact with and are temporarily immobilized by the object. Probe molecules could also be delivered to the interior of objects by various techniques. PAINT microscopy will have applications in imaging large molecular ensembles in vivo. The spatial resolution of slow dynamical processes is also feasible by means of PAINT.
Materials and Methods
Materials.
Lipids (POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) were purchased from Avanti Polar Lipids (Alabaster, AL); Alexa Fluor 568-conjugated transferrin and Nile red were purchased from Invitrogen (Carlsbad, CA) and used without purification as received. Hepes buffer and sodium chloride were from Fisher Scientific (Hampton, NH), and chloroform was from Acros Organics (Geel, Belgium).
Glass coverslips and cylinders (Fisher Scientific) were cleaned by sonication for 1 h in piranha solution (3:1 mixture of sulfuric acid and 30% hydrogen peroxide) with sequential extensive washing in UltraPure water (Millipore, Billerica, MA).
LUVs were prepared by the standard extrusion technique (15). Briefly, 0.1 ml of 10 mg/ml POPC lipids in chloroform was evaporated by nitrogen flow in a small glass vial to yield a dry phospholipid film. After drying for 1 h in vacuum, the film was hydrated in 1 ml of 10 mM Hepes buffer, pH 7.4, with continuous stirring at room temperature for 1.5 h. After five freeze/thaw cycles, the resulting multilamellar vesicles were extruded 21 times through 100-nm polycarbonate membranes (Whatman, Florham Park, NJ) by means of a MiniExtruder (Avanti Polar Lipids).
Supported bilayers were prepared on a glass surface by rupturing of giant unilamellar vesicles (GUVs). The GUVs were grown by the electroformation method (17). A solution of lipids in chloroform (10 μl of 0.2 mg/ml) were deposited on indium tin oxide (ITO)-coated glass and subsequently dried in a nitrogen stream and in vacuum for 1 h. The preparation chamber consisted of two ITO-coated glass plates with a 2-mm rubber spacer between them. A 10-Hz sinusoidal voltage was applied to the electrodes at 0.5 V/mm while the chamber was filled with 0.2 M sucrose solution. The voltage was slowly increased to 1.5 V/mm in steps of 0.1 V every 5 min by means of a software-controlled functional generator, constructed around a NI-DAC board (National Instruments, Austin, TX). Vesicle formation was continued for 1 h at 1.5 V/mm. The growth of vesicles was visually controlled by observation using differential interference contrast microscopy. The GUVs were then gently removed by a syringe and stored at 4°C. For preparation of a supported bilayer, 1 μl of GUV suspension was added to a 100-μl chamber (cell separation glass cylinder) containing 0.1 M NaCl in 10 ml of Hepes solution having similar osmolarity. Because of differences in the densities of the interior and exterior solutions, the GUVs sank to the glass bottom and either stayed attached to surface or ruptured. The ruptured vesicles formed the uniform bilayers used in our experiments.
Microscope.
The setup, based on a commercial Olympus (Melville, NY) IX81 inverted microscope, has been described (18, 19). The laser beam from a Kr+ ion laser (568 nm) passes through a quarter-wave plate to generate circularly polarized light, which is then focused with appropriate lenses onto the back focal plane of an oil-immersion objective (Olympus ×60 NA = 1.45). A lens, coupled with a translation stage, was used to align the excitation beam across the objective's back aperture to achieve easy illumination, angle adjustment, and interconversion of the setup between through-the-objective TIR and epifluorescence microscopy. The excitation light was filtered by a laser band pass filter and a dichroic mirror (Chroma Technologies, Rockingham, VT). Fluorescence from the sample was collected by the objective and directed to a CCD camera with multiplication-on-chip capability (Roper Scientific, Tucson, AZ; Cascade 512F) by means of a beam splitter and an appropriate set of band pass filters (Omega Optical, Brattleboro, VT). With the ×1.6 lens, the total magnification of the microscope was ×96, which yields a pixel size in the image plane of the camera of ≈167 nm. Specially designed LabView (National Instruments)-based software was developed to record sequences of images with various exposure times, frames, and collection rates. All experiments were conducted in TIRFM (20) mode at room temperature.
Image Processing and Molecular Localization.
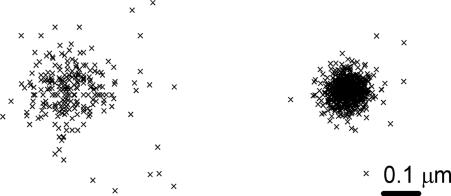
All algorithms for molecular localization were developed in house. Programs were written by using MATLAB (MathWorks, Natick, MA) The recorded sequences of frames were treated in two stages. The purpose of the first step is to find all of the fluorescent spots in a frame and determine the approximate locations of these spots. The coordinates were determined by finding the centroid of intensity of every fluorescent spot in each frame. This process includes several steps: 2D cutoff filtering to reduce high-frequency noise of the CCD detector, discrimination of fluorescent spots from background by means of a threshold value, and finding contours of equal intensity for each spot and finding the mean x and y coordinates of the contour. These coordinates represent the absolute position of the fluorescent peak in the image. The coordinate precision is subpixel and estimated to be ≈50 nm, well below the diffraction limit. The results of the first stage were then saved to file, which contains information about the frame number and all x,y absolute coordinates for each frame. This file can be used to create a synthetic image, where the coordinates of all spots from all frames are superimposed and represented graphically (Fig. 6Left). The synthetic image already contains details of the studied object that are not resolvable by regular florescence imaging. The data processing in this stage takes very little time because it does not include any nonlinear procedures of minimization: the average computation speed was ≈118 points/sec. The areas of interest on the synthetic image can then be chosen for more precise fitting in stage two. In this manner, the overall time for processing can be significantly reduced.
Fig. 6.
Stages in the determination of the locations of Nile red molecules on the surface of an LUV. (Left) Stage 1 (see text) determines the x,y coordinates to a precision of ≈50 nm. (Right) After fitting to 2D Gaussian (stage 2), the precision becomes ≈25 nm.
The second processing stage is a precise adjustment of the spot locations from step 1. The diffraction pattern of a single fluorophore has an Airy distribution that is well approximated by a Gaussian (21). The exact positions of spots were determined by fitting the PSF to a 2D Gaussian function (22, 23):
 |
where I is an intensity or PSF, xo and yo are coordinates of the centroid, A and Ao are the PSF amplitude and background level, and σx and σy are the width of the PSF in the x and y directions. After subtraction of the background Ao, which is flat within the area of a single PSF, five fitting parameters, x, y, σx, σy, and A, were determined by least-square algorithms. In principle, the PSF has cylindrical symmetry (σx = σy), but we treated σx and σy independently to verify the quality of the fit procedure. The original nonprocessed images were used as a source for fitting. The initial parameters xo and yo were predetermined in step 1 with subpixel precision, significantly reducing the overall time for data processing, because these parameters are close to the final ones. To reconstruct the image, the locations of >104–105 molecular probes were determined. The speed of the fitting for this stage was ≈14 spots/sec. The values of σx and σy in our experiment are 0.89 pixels, corresponding to 148 nm. All points with σx or σy outside the range 0.4–1.4 pixels were discarded; we also discarded points having | σx − σy | /(σx + σy) > 0.15. The final synthetic image (Fig. 6Right) is then formed as described above for step 1. The precision of fitting and hence the resolution of the image depend on the background, the photon noise, and the pixel size of the detector, but the photon statistics is the main contribution (24). The number of emitted photons before photobleaching for certain fluorophores can exceed ≈104, in which case the accuracy in positioning could be as high as ≈1 nm (2).
Acknowledgments
We thank Prof. David Talaga and Dr. Erwen Mei for critical reading of the manuscript. This research was supported by the National Institutes of Health Research Resource (Grant RR 001348) and in part by Grant RFA-RM-04–001.
Abbreviations
- LUV
large unilamellar vesicle
- GUV
giant unilamellar vesicle
- PSF
point-spread function
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- PAINT
points accumulation for imaging in nanoscale topography.
Footnotes
The authors declare no conflict of interest.
References
- 1.Tinnefeld P, Sauer M. Angew Chem Int Ed. 2005;44:2642–2671. doi: 10.1002/anie.200300647. [DOI] [PubMed] [Google Scholar]
- 2.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 3.Forster T. Ann Phys. 1948;2:55–75. [Google Scholar]
- 4.Gordon MP, Ha T, Selvin PR. Proc Natl Acad Sci USA. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muls B, Uji-i H, Melnikov S, Moussa A, Verheijen W, Soumillion J-P, Josemon J, Mullen K, Hofkens J. ChemPhysChem. 2005;6:2286–2294. doi: 10.1002/cphc.200500235. [DOI] [PubMed] [Google Scholar]
- 6.Lidke KA, Rieger B, Jovin TM, Heintzmann R. Opt Express. 2005;13:7052. doi: 10.1364/opex.13.007052. [DOI] [PubMed] [Google Scholar]
- 7.Mei E, Gao F, Hochstrasser RM. Phys Chem Chem Phys. 2006;8:2077–2082. doi: 10.1039/b601670g. [DOI] [PubMed] [Google Scholar]
- 8.Qu X, Wu D, Mets L, Scherer NF. Proc Natl Acad Sci USA. 2004;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betzig E, Patterson GH, O R S, Lindwasser W, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 10.Rust MJ, Bates M, Zhuang X. Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smoluchowski M. Phys Z. 1916;17:557. [Google Scholar]
- 12.Gao F, Mei E, Lim M, Hochstrasser RM. J Am Chem Soc. 2006;128:4814–4822. doi: 10.1021/ja058098a. [DOI] [PubMed] [Google Scholar]
- 13.Hill TL. Proc Natl Acad Sci USA. 1975;72:4918–4922. doi: 10.1073/pnas.72.12.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggeling C, Widengren J, Rigler R, Seidel CAM. Anal Chem. 1998;70:2651–2659. doi: 10.1021/ac980027p. [DOI] [PubMed] [Google Scholar]
- 15.Johnson JM, Ha T, Chu S, Boxer SG. Biophys J. 2002;83:3371–3379. doi: 10.1016/S0006-3495(02)75337-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaz WLC, Stümpel J, Hallmann D, Gambacorta A, Rosa MD. Eur Biophys J. 1987;15:111–115. [Google Scholar]
- 17.Angelova MI, Dimitrov Prog Colloid Polym Sci. 1988;76:59–67. [Google Scholar]
- 18.Mei E, Sharonov A, Ferris JH, Hochstrasser RM. Appl Phys Lett. 2005;86:043102. [Google Scholar]
- 19.Mei E, Sharonov A, Gao F, Ferris JH, Hochstrasser RM. J Phys Chem A. 2004;108:7339–7346. [Google Scholar]
- 20.Axelrod D. Methods Cell Biol. 1989;30:245–270. doi: 10.1016/s0091-679x(08)60982-6. [DOI] [PubMed] [Google Scholar]
- 21.Saxton MJ, Jacobson K. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 22.Cheezum MK, Walker WF, Guilford WH. Biophys J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bobroff N. Rev Sci Inst. 1986;57:1152. [Google Scholar]
- 24.Thompson RE, Larson DR, Webb WW. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]