Abstract
The physiological complex of yeast cytochrome c peroxidase and iso-1-cytochrome c is a paradigm for biological electron transfer. Using paramagnetic NMR spectroscopy, we have determined the conformation of the protein complex in solution, which is shown to be very similar to that observed in the crystal structure [Pelletier H, Kraut J (1992) Science 258:1748–1755]. Our results support the view that this transient electron transfer complex is dynamic. The solution structure represents the dominant protein–protein orientation, which, according to our estimates, is occupied for >70% of the lifetime of the complex, with the rest of the time spent in the dynamic encounter state. Based on the observed paramagnetic effects, we have delineated the conformational space sampled by the protein molecules during the dynamic part of the interaction, providing experimental support for the theoretical predictions of the classical Brownian dynamics study [Northrup SH, Boles JO, Reynolds JCL (1988) Science 241:67–70]. Our findings corroborate the dynamic behavior of this complex and offer an insight into the mechanism of the protein complex formation in solution.
Keywords: electron transfer, encounter state, transient complex, spin label, paramagnetic relaxation enhancement
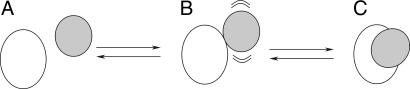
The process of protein complex formation can be described by a two-step model, in which a short-lived, dynamic encounter complex precedes a dominant, well defined state (Fig. 1). The former enables proteins to undergo reduced-dimensionality search of the optimal binding geometry, thereby accelerating molecular association as compared with 3D diffusion (1). Fast molecular association is essential for protein–protein complexes that require high turnover rates, like those involved in electron transfer (ET) in photosynthesis, respiration, and other metabolic processes (2). The physiological complex of yeast iso-1-cytochrome c (Cc) and yeast cytochrome c peroxidase (CcP) is a paradigm for the intermolecular ET (3) and is one of the few transient ET complexes for which a crystal structure has been solved (4). A recent study has confirmed that the protein–protein orientation observed in the crystal is ET-active (5). However, it has remained a matter of debate whether this structure represents the only form in solution (6–8). According to several studies, the complex is dynamic, and the crystal structure might represent only a subpopulation of protein orientations (9–15). Recent studies show that the photoinduced ET between Zn-substituted CcP and Cc, both in the crystal (8, 16) and in solution (17), occurs with faster backward than forward rates, indicating that the complex is present in multiple forms, only a few of which are ET-active (17).
Fig. 1.
Model for the formation of a protein complex. Free proteins (A) associate to form an encounter complex (B) consisting of an ensemble of protein orientations, which is in equilibrium with a single-orientation complex (C).
Characterization of the binding interface in the dynamic encounter state (Fig. 1B) has so far proven to be elusive. Investigation of protein complexes by using x-ray crystallography or conventional NMR spectroscopy addresses only the single-orientation species (Fig. 1C), and the only way to visualize the dynamic state is offered by theoretical modeling studies (11, 18). In the recent, elegant work of Clore and coworkers (19, 20), it was shown how paramagnetic relaxation can be applied to study the dynamic state of protein–DNA complexes. We report on the application of an analogous experimental approach that allows us to define both the dominant protein–protein orientation (Fig. 1C) and the conformational space sampled by the proteins in the dynamic encounter complex (Fig. 1B).
Results and Discussion
Solution Structure of the Complex.
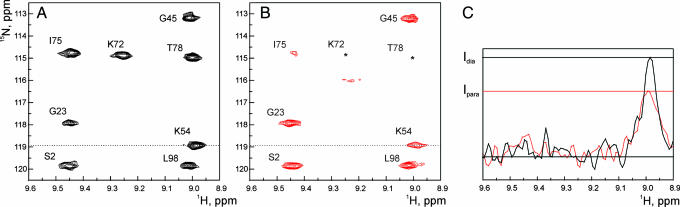
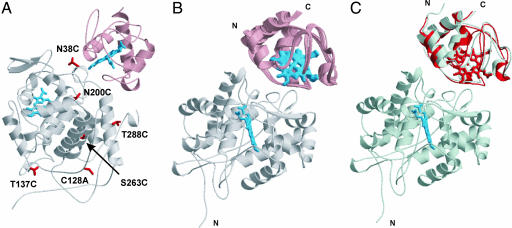
The concept of the approach is that NMR resonance intensities of one of the proteins in the complex are affected by a paramagnetic spin label covalently attached to the other protein (Fig. 2). The paramagnetic effects are converted into distance restraints (21–23), which can be used to calculate protein–protein orientations within the complex (24). Five single-cysteine CcP variants were prepared (Fig. 3A) and labeled with a paramagnetic nitroxide spin label, (1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanethiosulfonate (MTSL). For three of them (N38C, N200C, and T288C), this results in a broadening of backbone amide resonances of Cc when bound to CcP, whereas for the other two (S263C and T137C), no effects are observed [see supporting information (SI) Fig. 6]. A set of experimental restraints from the spin-labeled complexes was used for the subsequent docking calculations (SI Table 1). Reproducibly, a single set of orientations was found (Fig. 3B). Comparison of the best solution structure with the crystal structure of the complex shows that the two are very similar, with a root mean square deviation (rmsd) of the Cc backbone atoms of 2.2 Å after superposition of CcP (Fig. 3C). This finding provides direct evidence that the crystal structure represents the dominant form in solution.
Fig. 2.
Intermolecular paramagnetic effects. (A and B) Parts of the 15N-1H HSQC spectra of Cc bound to CcP N200C modified with a diamagnetic (black, A) or paramagnetic (red, B) label. The asterisks indicate resonances broadened beyond detection. (C) A slice through the amide resonance of K54 in the 1H dimension, marked with a dotted line in A and B. The lines marked Ipara and Idia indicate the intensities of the resonance in the paramagnetic and diamagnetic complexes, respectively.
Fig. 3.
Solution structure of the Cc–CcP complex. (A) The residues of CcP used for mutagenesis are labeled and shown as red sticks. Ribbon representation of the complex was drawn from the crystal structure [PDB entry 2PCC (4)]. (B) Ensemble of best 20 solution structures. The average rmsd from the best structure for Cc backbone atoms is 0.7 Å. (C) Comparison of the solution and crystal structures. In A–C, CcP is at the bottom, and heme groups for both proteins are shown in sticks. In B and C, the CcP molecules are superimposed; the labels indicate protein termini. In C, the Cc backbones for the best solution and crystal structures are shown in red and gray, respectively. The backbone rmsd between the two Cc molecules is 2.2 Å. All images of the proteins used in this report were made with MOLSCRIPT (25) and rendered in Raster3D (26), except for Fig. 5, which was made with Swiss-PDB (27) and POV-Ray 3.6 (www.povray.org).
The approach used here provides information about the conformation of the proteins within the complex but gives no details on the orientation of the side chains. To establish whether the starting coordinate set influences the final structure, we compared the docking solution obtained with the coordinates of free Cc (28) and CcP (29) with those taken from the crystal structure (4). The two complexes exhibit an rmsd of Cc backbone atoms of 4.2 Å. With the coordinates of free CcP and those of Cc taken from the crystal structure, the difference disappears (rmsd 0.70 Å), indicating that the coordinates of free Cc affect the outcome of the docking. This effect may be attributable to the Cc side chain of Q16. In free Cc, Q16 protrudes from the surface, preventing a close approach of the proteins and causing a lateral translation within the docked complex as compared with the crystal structure. In the latter, the Q16 side chain is folded back to form a hydrogen bond with its own backbone (4), which, rather than being an artifact of crystallization, is induced by the binding (30). Apparently, this rearrangement is crucial for the formation of the correct complex.
Additional Paramagnetic Effects.
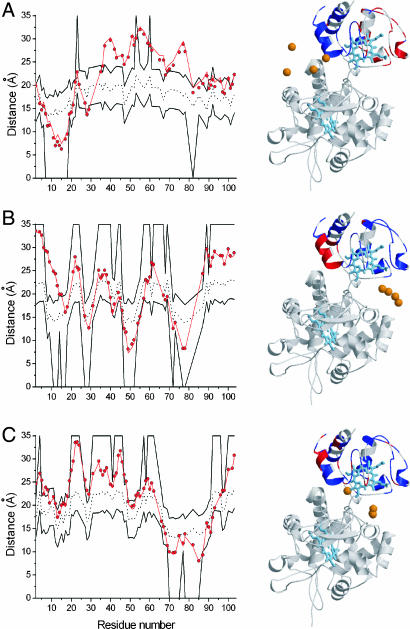
Analysis of the solution structure shows that most of the restraints are satisfied (Fig. 4). The violated Cc residues are nonrandomly distributed. For instance, the spin label attached to N38C CcP exerts paramagnetic effects on Cc residues that map on the back of the protein (red in Fig. 4A), whereas the spin label at N200C CcP strongly affects the N-terminal helix (red in Fig. 4B). In the solution structure, these residues are located too far away from the spin label to be affected. The “additional” effects, shown in Fig. 4, are reproducible and suggest that the calculated orientation is not sufficient to explain all observed paramagnetic effects. These additional effects could have several causes: (i) nonspecific interactions caused by random collisions; (ii) binding of more than one Cc to CcP (31–33); or (iii) dynamics within the complex. It should be noted that a 2:1 complex of Cc and CcP (possibility ii) is not expected to be observed under the present experimental conditions (0.1 M NaCl) (33). The first two possibilities imply the presence of distinct complexes in which Cc is bound with different affinities. If this is the case, lowering Cc concentration in a sample with a constant concentration of CcP would result in a decrease of the fraction of the weaker complexes. This decrease would lead to a reduction of the contributions from the weaker complexes to the paramagnetic relaxation enhancements (PREs), resulting in fewer violations and a nonuniform concentration-dependence of PREs. On the contrary, in the case of dynamics within the complex, lowering the Cc concentration should not influence the contribution of the dynamic encounter state to the PREs.
Fig. 4.
Violation analysis of the best solution structure for the Cc–CcP complex. (Left) Graphs illustrate the distances from the Cc backbone amide protons in the best solution structure (red circles) and crystal structure (red line) to the averaged position of the oxygen atom of the spin label attached to CcP at N38C (A), N200C (B), and T288C (C). The black dotted and solid lines indicate the PRE-derived distances and error margins, respectively, used in the structure calculations. (Right) Cartoon representations of the best solution structure, indicating the residues with satisfied (blue) and violated (red) restraints. Heme groups for both proteins are in cyan. For each of the spin-label positions, four oxygen atoms, representing the conformational freedom of the spin label, are shown as orange spheres. These were used for ensemble averaging in the structure calculations (see Materials and Methods).
Control experiments in which Cc concentration was five times lower than in the experiments described above, although the concentration of CcP remained the same, were carried out for N200C CcP. This variant was selected because the spin label attached at this position produces the largest violations (Fig. 4B). Under the conditions used, the fraction of Cc available for the weaker interactions decreases from 0.10 to 0.015 (see Materials and Methods). However, the results show the same pattern for the observed paramagnetic effect as those for the 1:1 protein ratio, with the largest deviation of the restraint target distance of ±2 Å for several residues (SI Fig. 6F). Therefore, we conclude that additional paramagnetic effects are independent of the Cc concentration in the range used. This finding is in agreement with multiple protein–protein orientations within the complex.
Dynamics Within the Complex.
The presence of multiple orientations within Cc–CcP complex previously has been suggested by several studies (9–15, 17). A complex comprising an equilibrium between a well defined form and a dynamic state (Fig. 1) adequately could explain the additional paramagnetic effects. Because of the sixth-power distance-dependence of the paramagnetic relaxation, a dynamic state will contribute to the PREs for the residues that get close to the spin label, even if this state is populated for only a few percent of the time. Thus, the observed paramagnetic effect represents a sum of PRE contributions from all protein orientations within the complex, corresponding to the combination of the dominant complex and the multiple forms in the dynamic ensemble (Fig. 1 B and C; also see Materials and Methods).
We have made an estimate for the lower limit of the fraction of time spent by the proteins in the dominant orientation (fdom), based on the assumption that the effects for all residues with satisfied restraints arise mostly from this orientation. To obtain an actual PRE (R2para) for these residues, the observed PRE (R2, obspara) must be multiplied by 1/fdom (see Eq. 3 in Materials and Methods). Protein docking performed with the R2para values corresponding to fdom ≤ 0.7 results in an energy increase for all solutions attributable to both van der Waals and restraint violations. To satisfy the shorter distance restraints resulting from low fdom values, Cc should come unrealistically close to MTSL and, accordingly, CcP atoms, resulting in solutions that are sterically forbidden. From these calculations, we conclude that the proteins spend >70% of the lifetime of the complex in the dominant orientation. This finding is consistent with other studies that have proposed the existence of a single-orientation complex in solution (10, 34, 35). In particular, large NMR chemical-shift perturbations of Cc resonances upon complex formation suggest the presence of a dominant orientation (30) because for more dynamic protein complexes, the perturbations tend to be smaller (36, 37).
Conformational Space of the Encounter Complex.
It is impossible to identify the orientations constituting the dynamic encounter state without knowing how many of these comprise the ensemble and what fraction of the lifetime of the complex is spent in each of them. However, we can define the conformational space occupied by Cc in the encounter complex (Fig. 5). This definition is achieved by mapping out the areas around those spin labels that give rise to additional PREs (N38C, N200C, and T288C) and those that do not affect Cc atoms at all (S263C and T137C). In the dynamic state, Cc must come close to the spin labels that cause additional PREs (the extent of approach indicated by the red area in Fig. 5) and be far from those showing no effects (the areas to be avoided are colored blue in Fig. 5). The remaining space around CcP also could harbor Cc molecules; however, those would not contribute to the observed paramagnetic effects.
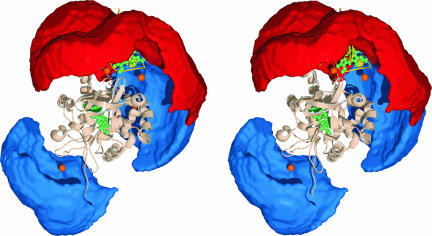
Fig. 5.
Conformational space occupied in the Cc–CcP encounter complex. The best solution structure of the complex in the same orientation as that in Fig. 3 B and C is shown in a stereoview. CcP and Cc are in light gray and yellow ribbons, respectively, with heme groups in green and spin-label positions indicated by orange spheres. The red surface defines the area that must contain the center of mass of Cc for at least 3% of the lifetime of the complex (outer perimeter of the red area) or less (moving from the periphery toward a spin label). The blue surface corresponds to the area that cannot be occupied by the Cc center of mass for longer than 3% of the lifetime of the complex (outer perimeter of the blue area) or less (moving to the center toward a spin label).
In the dynamic state, the binding interface on CcP is localized around the Cc position in the dominant orientation. This finding is in agreement with the view that an encounter complex facilitates formation of the dominant one via preorientation of the protein molecules and reduced-dimensionality search (1, 9). The CcP binding surface contains a part of the negatively charged region predicted to interact with Cc by a classical Brownian dynamics study (11), suggesting that electrostatic attraction plays a dominant role in determining the nature of the encounter complex. Clearly, the binding interface on CcP is larger than that observed in the crystal structure (4), with an earlier NMR chemical-shift perturbation analysis showing the same to be true for Cc (30). Several reports described CcP binding effects on the rear of Cc in solution (13, 14, 30), and a recent study of complexes between CcP and different variants of Cc has reported crystal structures in which Cc is positioned with its back toward CcP (16). Such binding modes, sampled in the dynamic state of the complex, can explain the paramagnetic effects at the back of Cc (Fig. 4A).
Implications for Intermolecular ET.
As we show in this work, the dominant form of the Cc–CcP complex in solution is the same as the protein–protein orientation observed in the crystal structure. If the dominant form of the complex also is the most ET-reactive, as indeed was suggested by Poulos and coworkers (5), this could explain similar ET rates observed in the crystal structure (8) and in solution (17). However, the forward ET between Cc and CcP is likely to be conformationally gated, implying that the most favorable binding geometry is not necessarily the most ET-active (8, 17). In fact, there might be a multitude of reactive ET configurations as suggested by the study of variant Cc–CcP complexes (16). In this case, the ET may be rationalized by the protein–protein orientations, some of which could be ET-active, sampled in the dynamic encounter state of the complex. The conformational space of the encounter state delineated in this work therefore could represent the extent of conformational fluctuations in Cc–CcP complex that modulate the ET in solution (17).
Summary.
By defining the conformational space of an ensemble of protein orientations and characterizing the dominant structure of the complex, we were able to address both sides of the equilibrium of the complex formation between Cc and CcP (Fig. 1). The dominant orientation of the protein complex in solution is the same as that in the crystal structure. This form is in equilibrium with a dynamic state that contributes to the observed intermolecular effects. The experimental approach outlined in the present report will be useful for study of other biological macromolecular complexes with a particular focus on the role of dynamics in biomolecular interactions.
Materials and Methods
Protein Preparation.
Isotopically enriched 15N Cc (T-5A/C102T) was expressed in Escherichia coli and purified as previously described (38, 39). To prepare CcP mutants, its single native cysteine (C128) was changed into alanine, and another cysteine residue was introduced at a desired position on the protein surface (Fig. 3A). Site-directed mutagenesis was carried out by using the Quik Change PCR protocol (Stratagene, La Jolla, CA) with the plasmid CCP(MKT) as a template (40). All constructs were verified by DNA sequencing. Mutant proteins were expressed and purified analogously to the wild type (40), except for the gel-filtration step, which was carried out in the presence of 1 mM DTT.
Purified single-cysteine CcP mutants were incubated with 10 mM DTT in 0.1 M Tris·HCl (pH 8.0)/0.1 M NaCl for 2 h at room temperature to reduce intermolecular disulfide bonds. DTT was removed by passing the CcP solution through a PD-10 column (Amersham Pharmacia, Uppsala, Sweden). The resulting monomeric protein was reacted with a 7- to 10-fold excess of paramagnetic (MTSL) or diamagnetic control (MTS) label (SI Fig. 7) and incubated overnight at room temperature. MTSL and MTS [(1-acetyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanethiosulfonate] were purchased from Toronto Research Chemicals (North York, ON, Canada) and used without further purification. Upon completion of the reaction, protein solution was passed through a PD-10 column to remove any unreacted label, exchanged into 20 mM NaPi/100 mM NaCl buffer (pH 6.0), and concentrated by using Amicon Ultra concentrators (Millipore, Billerica, MA). In each case, the yield of labeling was close to 100% as estimated from double-integrated EPR spectra of mutant CcP-MTSL and a control sample containing a known amount of free MTSL.
Throughout the study, we used ferric Cc and five-coordinated high-spin ferric CcP with previously reported purity criteria (30). Concentrations of Cc and CcP were determined according to the optical absorbance peaks at 410 nm (ε = 106.1 mM−1 cm−1) and 408 nm (ε = 98 mM−1 cm−1), respectively (30).
NMR Samples and Experiments.
NMR samples contained 0.3–0.5 mM 1:1 complex of 15N Cc with wild-type CcP, mutant CcP-MTSL, or CcP-MTS in 20 mM NaPi/100 mM NaCl (pH 6.0)/6% D2O for lock and 0.1 mM CH3CO15NH2 as an internal reference. For the control experiment with low Cc concentration, described in Results and Discussion, samples of 0.3 mM CcP-MTS or CcP-MTSL with 0.06 mM 15N Cc were used. The pH of the samples was adjusted to 6.00 ± 0.05 with small aliquots of 0.1 M HCl or 0.1 M NaOH. Measurements were performed at 301 K on a Bruker DMX600 spectrometer equipped with a triple-resonance gradient probe or, in case of the samples containing 0.06 mM 15N Cc, a TCI-Z-GRAD CryoProbe (Bruker, Karlsruhe, Germany). 2D 15N-1H heteronuclear single-quantum coherence (HSQC) spectra were obtained with 2,048 and 512 points in the direct and indirect dimensions, respectively, and spectral widths of 32 ppm (15N) and 16 ppm (1H). All data were processed with the Azara suite of programs (provided by Wayne Boucher and the Department of Biochemistry, University of Cambridge, Cambridge, U.K.) and analyzed in Ansig for Windows (41, 42). Assignments of the 15N and 1H nuclei of Cc were taken from a previous report (30). In the present work, a number of Cc amides (A3, F10, H26, L32-H33, H39, N56, N63, M64, E66, Y74, K79-A81, G83-G84, K86, E88, and L94) were either not observed or not analyzed because of spectral overlap.
Determination of Distance Restraints from PREs.
For each observed amide proton of Cc, an MTSL-induced PRE was calculated from Eq. 1 (23):
where Ipara and Idia are measured intensities of HSQC peaks of Cc in the complex with CcP-MTSL and CcP-MTS, respectively (SI Fig. 6); R2, dia is the transverse relaxation rate of Cc amide protons in the complex with CcP-MTS; R2, para is the paramagnetic contribution to the relaxation rate (PRE); and t is the total polarization transfer time of the HSQC. For each amide peak, R2, dia was estimated from the width at half-height (Δν1/2) of its Lorentzian fit in proton dimension by using R2, dia = πΔν1/2. For the residues whose resonances disappear in the paramagnetic spectrum, Ipara was estimated from the noise level of the spectrum. Calculated PRE rates were converted into distances by using Eq. 2 (23):
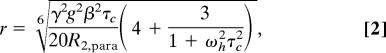 |
where r is the distance between the unpaired electron of the MTSL and a given amide proton of Cc; τc is the rotational correlation time of the electron-nucleus vector; ωh and γ are proton Larmor frequency and gyromagnetic ratio, respectively; g is the electronic g factor; and β is the Bohr magneton.
Three classes of intermolecular distance restraints, used in subsequent docking calculations, were defined (SI Table 1) (23). Residues that are strongly affected by MTSL and whose resonances disappear in the paramagnetic spectrum are restrained with only an upper bound. Those not affected by MTSL have only a lower limit. Finally, the residues affected by the spin label and whose resonances are observed in the paramagnetic spectra are restrained with both upper and lower bounds. The margins of ±4 Å for the calculated restraints accommodate the experimental error and enhance the convergence of docking solutions as compared with tighter bounds (23).
Protein Docking.
Coordinates of both proteins were taken from the Protein Data Bank [PDB ID code 2PCC (4)]. For the docking of individual proteins, the molecular coordinates from PDB ID codes 2YCC (28) for Cc and 1ZBY (29) for CcP were used. Surface cysteine mutations of CcP were introduced in silico, followed by addition of the MTSL atoms and energy minimization of the labeled protein. To take into account mobility of the MTSL attached to the surface of CcP, we performed r−6 ensemble averaging of the intermolecular distance restraints with four different MTSL orientations. To generate these orientations, we systematically rotated the attached MTSL around five single bonds that join its ring to the Cα atom of the cysteine and retained only the sterically allowed conformers for each mutant. From these, four representative orientations were selected. Using 10 MTSL orientations produced similar results, consistent with the finding that averaging over a larger set of atoms does not alter the outcome (43).
A set of distance restraints from four MTSL positions (SI Table 1) was used to drive docking of the protein complex in Xplor-NIH version 2.13 (44). First, a rigid-body docking of the protein molecules was carried out. Only two energy terms were specified during this calculation, corresponding to restraints and van der Waals forces. The latter were defined as repel forces for the protein atoms, whereas for MTSL atoms they were set to zero. For each run performed, a single cluster of low-energy solutions was consistently produced. During the second step, 30 to 40 best structures were subjected to energy minimization and side-chain dynamics with fixed position of backbone atoms for both proteins and active van der Waals parameters for MTSL. For the refined structures, the entire docking procedure was repeated until no further reduction in energy was observed. The 20 best structures of the final solution showed an average rmsd from the lowest energy structure of 0.7 ± 0.2 Å for the backbone atoms of Cc after superposition of CcP molecules (Fig. 3B).
Additional Caveats for Intermolecular Distance Calculations from PRE.
The presence of a spin label on the surface of CcP can potentially interfere with Cc binding. For all labeled CcP mutants reported in this study, chemical-shift perturbations in the complex with 15N Cc are equal to those in the wild-type complex at the same protein ratio, indicating that an attached spin label does not alter the binding (data not shown).
The complex is in fast exchange on the NMR time scale; therefore, only one set of Cc resonances is observed in the spectrum. Given an association constant Ka = (1.9 ± 0.3)·105 M−1 (obtained by isothermal titration calorimetry at 30°C in 20 mM NaPi/0.1 M NaCl, pH 6.0; A.N.V., J.A.R.W., and M.U., unpublished data), 88–91% of Cc is bound to CcP in a 1:1 complex for the protein concentrations used (0.3−0.5 mM). Thus, the observed PRE (R2, obspara) represents a fraction (F) of the actual PRE (R2para), so that R2, obspara = FR2para, with F = 0.88−0.91. The experimentally determined PREs were corrected by 1/F, to obtain the R2para values that were used in further calculations.
Although τc is different for each amide proton, the use of a single value for all nuclei has been justified (21, 22). For the complex of Cc and CcP, the upper limit of τc of 16 ns was estimated from the Einstein–Stokes equation (45); it is in good agreement with the previously reported value for a protein system of a similar size (23). Considering a wide range of τc values used in the literature for conversion of PRE into distances (21–23), two control sets of distance restraints were generated with τc of 12 and 4 ns for each mutant. In each case, docking calculations resulted in the same best solution as that for the restraints based on τc of 16 ns (data not shown).
To check for possible nonspecific binding effects, a spin label was placed at two CcP positions farther away from the Cc binding site (Fig. 3A). Cc experiences no paramagnetic effect in the complex with spin-labeled T137C CcP, whereas with S263C CcP, only two Cc residues are affected (SI Fig. 6), consistent with the best solution structure. This finding suggests that a nonspecific protein binding does not take place in solution.
Conformational Space of the Encounter Complex.
For each Cc residue, the observed paramagnetic effect (R2, obspara) represents a sum of PRE contributions (R2para) from n protein orientations within the complex, weighted by fraction of time (f) spent in each of them:
 |
To delineate the conformational space explored by Cc in the complex with CcP as shown in Fig. 5, the largest distance between a backbone nucleus of Cc and a spin label that would result in a significant (≥7 Hz) R2, obspara with f = 0.03 was calculated to be 12 Å. Thus, nuclei that experience additional PREs (Fig. 4) must spend at least 3% of the time at this distance from the spin label or less time at a shorter distance. Cc nuclei spend at most 3% of the time at 12 Å from those spin labels that exert no paramagnetic effects and even less time at shorter distances. Use of a somewhat arbitrary value of f = 0.03 is justified because, due to r−6 dependence, very similar results are obtained for other values for the encounter complex as can be appreciated from SI Fig. 8. See SI Data Sets 1–3 for the coordinates of the best solution structure and the conformational space generated at f = 0.01, f = 0.03, and f = 0.10, respectively.
To define the space that must be occupied or is avoided by the center of mass of Cc (Fig. 5 and SI Fig. 8), red and blue spheres, respectively, were constructed around the oxygen atom of MTSL with the radius of 12 Å plus the average distance from the Cc center of mass to its surface (14 Å). Only the segments of the red and blue spheres that are between 12 and 19 Å from the surface of CcP are retained. The former corresponds to the closest approach of the center of mass of Cc to the surface of CcP, whereas the latter denotes the farthest distance from CcP at which the proteins still form a complex. In case of an overlap, blue space is given priority over red. The segments around spin labels at N38C, N200C, and T288C are in red, whereas those around T137C and S263C are in blue.
Supplementary Material
Acknowledgments
We thank Prof. R. Boelens for valuable suggestions, Prof. Gerard W. Canters and Dr. Derek S. Bendall for critical reading of the manuscript, and an anonymous reviewer for useful advice. This work was supported by The Netherlands Organization for Scientific Research (NWO-CW Grants 700.50.514, 700.52.425, and 98S1010).
Abbreviations
- ET
electron transfer
- Cc
yeast iso-1-cytochrome c
- CcP
yeast cytochrome c peroxidase
- PRE
paramagnetic relaxation enhancement
- MTSL
(1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanethiosulfonate
- MTS
(1-acetyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl)-methanethiosulfonate
- HSQC
heteronuclear single-quantum coherence
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2GB8).
This article contains supporting information online at www.pnas.org/cgi/content/full/0603551103/DC1.
References
- 1.Adam G, Delbruck M. In: Structural Chemistry and Molecular Biology. Rich A, Davidson N, editors. San Francisco: Freeman; 1968. pp. 198–215. [Google Scholar]
- 2.Bendall DS. In: Protein Electron Transfer. Bendall DS, editor. Oxford: BIOS; 1996. pp. 43–68. [Google Scholar]
- 3.Mathews FS, Mauk AG, Moore GR. In: Protein–Protein Recognition. Kleanthous C, editor. New York: Oxford Univ Press; 2000. pp. 60–101. [Google Scholar]
- 4.Pelletier H, Kraut J. Science. 1992;258:1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 5.Guo M, Bhaskar B, Li H, Barrows TP, Poulos TL. Proc Natl Acad Sci USA. 2004;101:5940–5945. doi: 10.1073/pnas.0306708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nocek JM, Zhou JS, de Forest S, Priyadarshy S, Beratan DN, Onuchic JN, Hoffman BM. Chem Rev. 1996;96:2459–2490. doi: 10.1021/cr9500444. [DOI] [PubMed] [Google Scholar]
- 7.Erman JE, Vitello LB. Biochim Biophys Acta. 2002;1597:193–220. doi: 10.1016/s0167-4838(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 8.Kang SA, Marjavaara PJ, Crane BR. J Am Chem Soc. 2004;126:10836–10837. doi: 10.1021/ja049230u. [DOI] [PubMed] [Google Scholar]
- 9.McLendon G. Struct Bond. 1991;75:159–174. [Google Scholar]
- 10.Moench SJ, Chroni S, Lou BS, Erman JE, Satterlee JD. Biochemistry. 1992;31:3661–3670. doi: 10.1021/bi00129a015. [DOI] [PubMed] [Google Scholar]
- 11.Northrup SH, Boles JO, Reynolds JCL. Science. 1988;241:67–70. doi: 10.1126/science.2838904. [DOI] [PubMed] [Google Scholar]
- 12.McLendon G, Zhang Q, Wallin SA, Miller RM, Billstone V, Spears KG, Hoffman BM. J Am Chem Soc. 1993;115:3665–3669. [Google Scholar]
- 13.Jeng MF, Englander SW, Pardue K, Rogalskyj JS, McLendon G. Nat Struct Biol. 1994;1:234–238. doi: 10.1038/nsb0494-234. [DOI] [PubMed] [Google Scholar]
- 14.Yi Q, Erman JE, Satterlee JD. Biochemistry. 1994;33:12032–12041. doi: 10.1021/bi00206a004. [DOI] [PubMed] [Google Scholar]
- 15.Moore GR, Cox MC, Crowe D, Osborne MJ, Rosell FI, Bujons J, Barker PD, Mauk MR, Mauk AG. Biochem J. 1998;332:439–449. doi: 10.1042/bj3320439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang SA, Crane BR. Proc Natl Acad Sci USA. 2005;102:15465–15470. doi: 10.1073/pnas.0505176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman BM, Celis LM, Cull DA, Patel AD, Seifert JL, Wheeler KE, Wang J, Yao J, Kurnikov IV, Nocek JM. Proc Natl Acad Sci USA. 2005;102:3564–3569. doi: 10.1073/pnas.0408767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabdoulline RR, Wade RC. J Mol Biol. 2001;306:1139–1155. doi: 10.1006/jmbi.2000.4404. [DOI] [PubMed] [Google Scholar]
- 19.Iwahara J, Schwieters CD, Clore GM. J Am Chem Soc. 2004;126:12800–12808. doi: 10.1021/ja046246b. [DOI] [PubMed] [Google Scholar]
- 20.Iwahara J, Clore GM. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 21.Gillespie JR, Shortle D. J Mol Biol. 1997;268:158–169. doi: 10.1006/jmbi.1997.0954. [DOI] [PubMed] [Google Scholar]
- 22.Gaponenko V, Howarth JW, Columbus L, Gasmi-Seabrook G, Yuan J, Hubbell WL, Rosevear PR. Prot Sci. 2000;9:302–309. doi: 10.1110/ps.9.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battiste JL, Wagner G. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 24.Ubbink M, Ejdebäck M, Karlsson BG, Bendall DS. Structure (London) 1998;6:323–335. doi: 10.1016/s0969-2126(98)00035-5. [DOI] [PubMed] [Google Scholar]
- 25.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 26.Merritt EA, Bacon DJ. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 27.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 28.Berghuis AM, Brayer GD. J Mol Biol. 1992;233:959–976. doi: 10.1016/0022-2836(92)90255-i. [DOI] [PubMed] [Google Scholar]
- 29.Bonagura CA, Bhaskar B, Shimizu H, Li H, Sundaramoorthy M, McRee DE, Goodin DB, Poulos TL. Biochemistry. 2003;42:5600–5608. doi: 10.1021/bi034058c. [DOI] [PubMed] [Google Scholar]
- 30.Worrall JAR, Kolczak U, Canters GW, Ubbink M. Biochemistry. 2001;40:7069–7076. doi: 10.1021/bi0025823. [DOI] [PubMed] [Google Scholar]
- 31.Zhou JS, Hoffman BM. Science. 1994;265:1693–1696. doi: 10.1126/science.8085152. [DOI] [PubMed] [Google Scholar]
- 32.Zhou JS, Nocek JM, DeVan ML, Hoffman BM. Science. 1995;269:204–207. doi: 10.1126/science.7618081. [DOI] [PubMed] [Google Scholar]
- 33.Mauk MR, Ferrer JC, Mauk AG. Biochemistry. 1994;33:12609–12614. doi: 10.1021/bi00208a011. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Pielak GJ. Biochemistry. 1999;38:16876–16881. doi: 10.1021/bi992005i. [DOI] [PubMed] [Google Scholar]
- 35.Pielak GJ, Wang X. Biochemistry. 2001;40:422–428. doi: 10.1021/bi002124u. [DOI] [PubMed] [Google Scholar]
- 36.Worrall JAR, Liu Y, Crowley PB, Nocek JM, Hoffman BM, Ubbink M. Biochemistry. 2002;41:11721–11730. doi: 10.1021/bi026296y. [DOI] [PubMed] [Google Scholar]
- 37.Volkov AN, Ferrari D, Worrall JAR, Bonvin AMJJ, Ubbink M. Prot Sci. 2005;14:799–811. doi: 10.1110/ps.041150205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollock WBR, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Biochemistry. 1998;37:6124–6131. doi: 10.1021/bi972188d. [DOI] [PubMed] [Google Scholar]
- 39.Morar AS, Kakouras D, Young GB, Boyd J, Pielak GJ. J Biol Inorg Chem. 1999;4:220–222. doi: 10.1007/s007750050307. [DOI] [PubMed] [Google Scholar]
- 40.Goodin DB, Davidson MG, Roe JA, Mauk AG, Smith M. Biochemistry. 1991;30:4953–4962. doi: 10.1021/bi00234a017. [DOI] [PubMed] [Google Scholar]
- 41.Kraulis PJ. J Magn Reson. 1989;84:627–633. [Google Scholar]
- 42.Helgstrand M, Kraulis PJ, Allard P, Hard T. J Biol NMR. 2000;18:329–336. doi: 10.1023/a:1026729404698. [DOI] [PubMed] [Google Scholar]
- 43.Iwahara J, Schwieters CD, Clore GM. J Am Chem Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 44.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J Magn Res. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 45.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. London: Academic; 1995. pp. 17–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.