Abstract
The process of invasion and metastasis during tumor progression is often reminiscent of cell migration events occurring during embryonic development. We hypothesized that genes controlling cellular changes in the Spemann organizer at gastrulation might be reactivated in tumors. The Goosecoid homeobox transcription factor is a known executer of cell migration from the Spemann organizer. We found that indeed Goosecoid is overexpressed in a majority of human breast tumors. Ectopic expression of Goosecoid in human breast cells generated invasion-associated cellular changes, including an epithelial–mesenchymal transition. TGF-β signaling, known to promote metastasis, induced Goosecoid expression in human breast cells. Moreover, Goosecoid significantly enhanced the ability of breast cancer cells to form pulmonary metastases in mice. These results demonstrate that Goosecoid promotes tumor cell malignancy and suggest that other conserved organizer genes may function similarly in human cancer.
Keywords: epithelial–mesenchymal transition
Tumor cells acquire the ability to metastasize by evolving traits that allow them to overcome multiple barriers to dissemination. Such cells undergo changes in cell–cell adhesion, acquire anchorage independence, gain motility, invade into and out of the circulation, and colonize distant organs (1). The genetic bases of these highly complex steps are largely unknown. However, some analogies exist between metastasizing cells and migrating subpopulations of cells that mediate tissue reorganization during embryonic development (2). These analogies suggest that signaling pathways controlling such embryonic processes may be reactivated in tumor cells with significant consequences.
Gastrulation is an embryonic developmental process that displays some striking similarities to tumor invasion and metastasis. This critical process establishes the basic body plan by way of highly coordinated cell movements and is initiated by a conserved group of cells originally characterized in Xenopus laevis as the Spemann organizer (3). In higher vertebrates, the organizer equivalents (e.g., the anterior primitive streak in mouse and Hensen's node in birds) are regions in which epithelial cells break their cell–cell junctions and ingress into the interior of the embryo, migrating as individual mesenchymal cells (4). This shift of cell phenotype is defined as an epithelial–mesenchymal transition (EMT).
An EMT is marked by the loss of epithelial properties through down-regulation of epithelial components (e.g., E-cadherin and cytokeratins), and the acquisition of mesenchymal proteins (e.g., N-cadherin and vimentin) in their stead (5). This transition can impart additional mesenchymal properties to embryonic epithelial cells, such as motility and invasiveness, which enable various cell movements during gastrulation and other subsequent developmental processes requiring tissue remodeling (6). During cancer pathogenesis, EMTs are similarly thought to confer these phenotypes upon carcinoma cells, enabling them to complete some of the steps required for successful invasion and metastasis (2). Indeed, multiple experimental studies provide support for such a functional link between EMT and tumor metastasis. For example, certain proteins that can induce an EMT in mammalian mammary epithelial cells, such as Twist and TGF-β, were found to be necessary for the metastatic behavior of tumor cells in vivo (7, 8).
The parallels between gastrula organizer biology and tumor malignancy suggest that common signals may drive gastrulation and metastasis. We therefore directed our attention to the Goosecoid (Gsc) gene, which encodes a well conserved transcription factor that was first identified as the most highly expressed homeobox gene in the Spemann organizer (9–11). Gsc can recapitulate many of the properties of the organizer when ectopically expressed in the amphibian embryo (12) and is known to promote cell migration in X. laevis (13). Moreover, elements of the TGF-β superfamily and Wnt/β-catenin signaling pathways, which are known to be involved in tumor invasion and metastasis (2), can induce Gsc expression in embryonic cells and are required for Spemann organizer formation (14, 15). For these reasons we sought to ascertain whether Gsc also plays a role in neoplastic disease. Goosecoid and its encoded protein have not been previously studied in the context of human cancer pathogenesis. The results described here strongly support the notion that this embryonic transcription factor can indeed be appropriated opportunistically by human cancer cells, allowing such cells to acquire certain characteristics needed to overcome key barriers to tumor metastasis.
Results
Elevated Goosecoid Expression in Human Breast Tumors.
The expression patterns of the Goosecoid gene have not been well characterized in human or murine adult tissues. To determine whether a role for the GSC developmental gene in cancer was plausible, we undertook to examine human tumor specimens for evidence of GSC mRNA. Because probes for this gene were not included in published microarray expression studies to the best of our knowledge, we were unable to assess GSC expression patterns through database mining. We therefore measured GSC levels in a cohort of microdissected human breast tumors of three prevalent pathological subtypes: atypical ductal hyperplasia (ADH), ductal carcinoma in situ (DCIS), and invasive ductal carcinoma (IDC) (16). The 72 tumor samples examined were each accompanied by a patient-matched sample of normal breast epithelium. The normal samples were presumably proliferative per published studies of normal human breast tissue (17). Because all samples in this cohort were obtained by laser capture microdissection, the samples do not contain significant numbers of stromal cells.
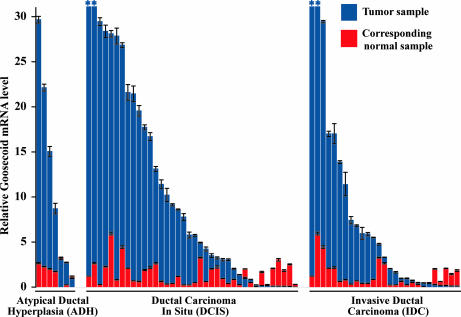
Quantitative real-time RT-PCR was used to compare levels of GSC mRNA represented in individual samples. The abundance of GSC mRNA in the normal tissue samples was found to be low, because signals were not detected until a high cycle number during PCR amplification. Strikingly, GSC expression was elevated in 56 of 72 tumors (78%) compared with corresponding patient-matched normal tissue samples (Fig. 1). By subtype, 71% of ADH samples, 79% of DCIS samples, and 78% of IDC samples contained a level of GSC mRNA above that of patient-matched normal tissue, and this pattern of GSC up-regulation was found to be significant in each case (P = 0.02 for ADH, P < 0.01 for DCIS, and P = 0.01 for IDC samples). The average extent of elevation of GSC mRNA across all samples per subtype was 5.9-, 9.6-, and 6.9-fold in the ADH, DCIS, and IDC samples, respectively, compared with corresponding normal samples. A more detailed view of the GSC expression data set can be found in Table 1, which is published as supporting information on the PNAS web site. Together, these results show that, in a majority of human ductal-type breast tumors, GSC expression is significantly elevated above normal levels, consistent with a role for this developmental gene in human cancer, as hypothesized.
Fig. 1.
Quantification of Goosecoid expression in human tumors. The relative level of GSC mRNA in each tumor (blue) and corresponding normal (red) tissue sample is shown with the lowest value of each pair in foreground. Pairs are grouped by tumor pathological subtype and sorted within groups according to the level of GSC mRNA in the tumor samples. All values displayed were normalized to the average of the GSC mRNA levels in the normal samples, which is set as the y value 1 in the graph. Values outside the scale of the y axis are marked by an asterisk.
Goosecoid Elicits an EMT and Enhances Cell Motility.
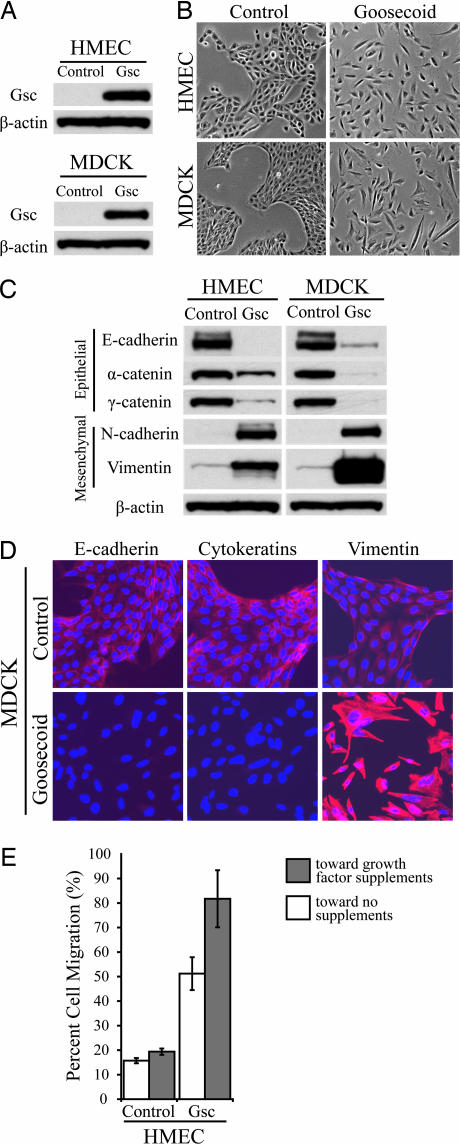
To identify the functional consequences of Gsc expression in adult epithelial cells, we stably expressed this protein in immortalized human mammary epithelial cells (HMECs) and in Madin–Darby canine kidney (MDCK) epithelial cells using retroviral transduction (Fig. 2A). Neither of these parental cell lines expressed substantial levels of Gsc protein by Western blotting (Fig. 2A). In both cell types, we observed that the population of cells expressing ectopic Gsc lost cell–cell contacts and displayed a scattered distribution in culture (Fig. 2B), whereas control cells retained their typical epithelial morphology, continuing to grow as groups of cobblestone-like cells. The morphological changes evident in the Gsc-expressing cells were suggestive of an EMT. We therefore examined the status of known EMT markers in these cells. The Gsc-expressing cells demonstrated marked down-regulation of E-cadherin, α-catenin and γ-catenin proteins, concordant with the apparent loss of adherens junctions (Fig. 2 C and D). These cells had replaced their cytokeratin-based intermediate filament network with one based on vimentin and stained positively for the mesenchymal protein N-cadherin (Fig. 2 C and D). Moreover, the Gsc-expressing HMECs were found to be substantially more migratory in transwell migration assays than control cells (Fig. 2E). Our results demonstrate that Gsc induces the central hallmarks of an EMT and cell motility in adult mammalian epithelial cells, recapitulating cellular changes driving gastrulation in higher vertebrates.
Fig. 2.
Effects of Goosecoid expression in immortalized human breast and canine kidney epithelial cells. (A) Ectopic expression of Gsc in HMECs and in MDCK epithelial cells by Western blotting. (B) Phase-contrast micrographs of HMECs and MDCK cells expressing either Gsc or GFP control. (C) Expression levels of epithelial proteins E-cadherin, α-catenin, and γ-catenin and mesenchymal proteins N-cadherin and vimentin in HMECs and MDCK cells expressing either Gsc or GFP control by Western blotting. β-Actin protein is shown as a loading control. (D) Immunofluorescence staining for epithelial proteins E-cadherin and cytokeratins and mesenchymal protein vimentin in MDCK cells expressing either Gsc or GFP control. Antibody staining is shown in red, and Hoechst nuclear staining is shown in blue. (E) Quantification of the migratory abilities of HMECs expressing Gsc or GFP control by transwell migration assay. Movement toward medium with or without growth factor supplements (EGF, insulin, and hydrocortisone) is graphed as the percentage of total cells assayed that migrated after 48 h. Assays were done in triplicate, and the averages with SEM are shown.
TGF-β Signaling Induces Goosecoid Expression in Adult Breast Epithelial Cells.
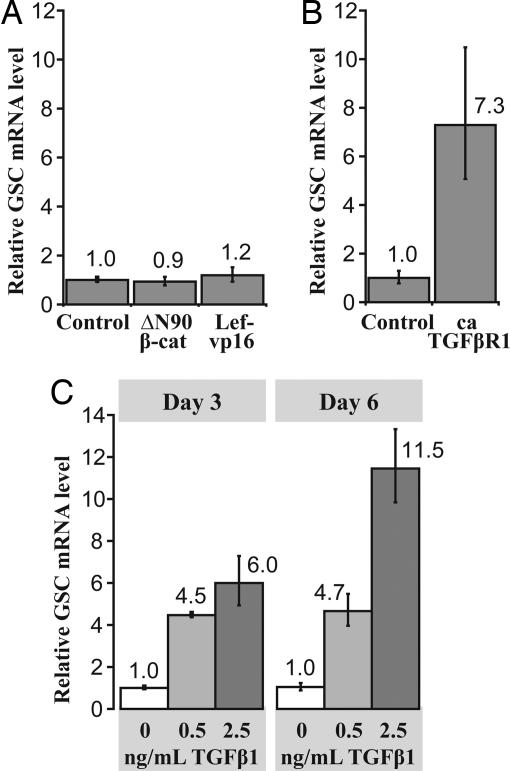
The Wnt/β-catenin and TGF-β superfamily signaling cascades are required for Spemann organizer formation and Gsc gene expression (18), and these same pathways have been implicated in tumor metastasis (2). Because Gsc recapitulated aspects of its embryonic organizer function in adult mammalian epithelial cells, we tested whether these two organizer-associated signaling cascades induce GSC expression in these cells. We found that the enhancement of Wnt/β-catenin signaling by two approaches failed to activate GSC expression. Specifically, GSC mRNA expression was not increased in HMECs either by expression of a nondegradable form of β-catenin (ΔN90 β-catenin) (19) or by a constitutively active form of Lef-1 (Lef-vp16) (20), a DNA-binding protein that associates with β-catenin to induce transcription of target genes (Fig. 3A). We confirmed these constructs were transcriptionally functional using the Topflash/Fopflash reporter system (data not shown) (21).
Fig. 3.
Induction of Goosecoid in HMECs. (A) Relative GSC mRNA expression levels in HMECs containing empty vector, nondegradable β-catenin (ΔN90 β-cat), or constitutively active Lef-1 (Lef-vp16). Each bar represents the average with SEM of triplicate assays. (B) Relative GSC mRNA expression levels in HMECs expressing either empty vector or constitutively active TGF-β type 1 receptor. Each bar represents the average with SEM of triplicate assays. (C) Relative GSC mRNA expression levels in HMECs treated with activated TGF-β1 ligand at various concentrations for 3 or 6 days. Each bar represents the average with SEM of triplicate assays.
In contrast, expression of constitutively active TGF-β type 1 receptor (22) in these cells using retroviral transduction did induce GSC mRNA expression (Fig. 3B). GSC mRNA was also induced in nontransduced HMECs in a dose-dependent manner by the addition of soluble, activated TGF-β1 to the cell culture medium (Fig. 3C). When TGF-β1 was applied to HMECs expressing nondegradable β-catenin or the constitutively active form of Lef-1 or GFP control, GSC expression was not induced to a level greater than that achieved without activation of the Wnt/β-catenin pathway (data not shown). Together, these experiments demonstrate that TGF-β signaling induces GSC in adult breast epithelial cells as do related mesoderm-inducing signals in gastrulating embryos and other cells (14, 23, 24). We did not observe β-catenin acting synergistically with these signals in our system, contrary to observations in Xenopus embryos (14). This may reflect distinct roles for Goosecoid in Xenopus and humans, as well as distinct mechanisms of regulation.
Goosecoid Enhances the Metastatic Ability of Cancer Cells.
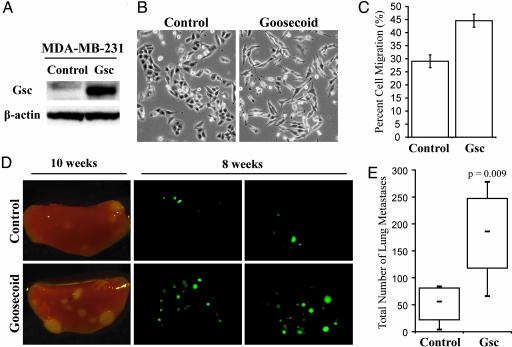
Because Goosecoid triggered an EMT and enhanced cell motility in adult epithelial cells (both known correlates of invasive and metastatic ability) we tested whether this gene could also promote tumor metastasis. Gsc was ectopically expressed in GFP-labeled MDA-MB-231 human breast cancer cells (Fig. 4A). The cells of this line are weakly metastatic and quasi-mesenchymal, in that they do not express E-cadherin and do express vimentin, yet they display an epithelial-like morphology in culture (25). We observed that upon the introduction of Gsc, the MDA-MB-231 cells acquired a spindle-like morphology more typical of mesenchymal cells (Fig. 4B) as well as an increased degree of motility (Fig. 4C).
Fig. 4.
Goosecoid expression changes the behavior of MDA-MB-231 human breast cancer cells in vitro and in mice. (A) Gsc expression in MDA-MB-231 cells expressing either Gsc or GFP control by Western blotting. (B) Phase-contrast micrographs of MDA-MB-231 cells expressing either Gsc or GFP control. (C) Quantification of the migratory abilities of MDA-MB-231 cells expressing Gsc or GFP control by transwell assay, graphed as the percentage of total cells assayed that migrated after 16 h. Assays were done in triplicate, and the averages with SEM are shown. (D) Representative brightfield and fluorescence images of mouse lung lobes 10 or 8 weeks after tail vein injection of MDA-MB-231 cells expressing either Gsc or GFP control. (E) Quantification of the number of metastatic foci in the lungs of mice 8 weeks after tail vein injection of MDA-MB-231 cells expressing either Gsc or GFP control (n ≥ 6; trend was confirmed by four independent experiments). Quartiles, medians, and the P value of the mean are shown.
Control or Gsc-expressing MDA-MB-231 cells were injected into the tail veins of mice and lungs were examined for metastases 8–10 weeks after injection (Fig. 4D). At both time points, a greater number of pulmonary metastases were visible in the mice injected with Gsc-expressing cells. Quantification of the observed lung nodules at 8 weeks using image analysis indicated a 4-fold increase in the average number of metastases in the mice injected with Gsc-expressing cells compared with control animals (Fig. 4E).
This demonstrated enhancement of metastasis might have arisen as a consequence of a Gsc-induced stimulation of proliferation in vivo. To address this possibility, we directly compared the in vivo proliferation rates of these two cell populations by injecting them either into the s.c. space or into the mammary glands of mice. In fact, the resulting primary tumors generated by the Gsc-expressing cells grew more slowly than did control tumors at both sites (data not shown). The demonstration that Gsc-expressing breast cancer cells formed significantly greater numbers of metastases in murine lungs despite proliferating more slowly in vivo provide strong indication that Goosecoid expression enhances the metastatic ability of MDA-MB-231 human breast cancer cells.
Discussion
Our results demonstrate that the Goosecoid homeobox transcription factor, a major orchestrator of Spemann organizer biology during gastrulation, plays an important role in activating cell properties associated with tumor progression to malignancy. To date, a limited number of developmental transcription factors, including SNAI1 (Snail), SNAI2 (Slug), and Twist, have also been linked to metastasis (7, 26–28). During embryogenesis these transcription factors are required for mesoderm formation in Drosophila and neural crest development in vertebrates (5), two processes in which an EMT and cell migration are critical (6). Although the concept of the EMT as a driving force behind human cancer metastasis is well described, there are still very limited in vivo data demonstrating that genes inducing the mesenchymal state contribute functionally to tumor metastasis (5). Here we have found that Gsc is sufficient to enhance metastatic behavior in an in vivo model of experimental metastasis. Gsc may augment metastatic colonization by promoting extravasation, cell survival in the environment of the lung, or migration to hospitable microenvironments within the lung.
Our finding that GSC expression is up-regulated in the vast majority of clinical ductal-type tumors supports a role for this embryonic transcription factor in human breast cancer. The up-regulation of GSC occurs quite early in multistep cancer progression rather than concurrently with the overt display of the invasive phenotype. This result is not unusual for breast carcinoma progression; for example, the HER2/neu gene, known to promote invasive cell behavior (29) and routinely used to inform both patient treatment and prognosis, is likewise already overexpressed in human tumors before the overt onset of invasiveness (30). Our observations are in concordance with other gene expression studies examining different stages of ductal-type breast cancer progression, which have shown that most expression changes associated with invasiveness are already present in preinvasive tissue (16, 31). Moreover, our observations are consistent with other published results demonstrating that several genes shown to promote the metastatic behaviors and poor prognosis of aggressive cancers, such as Slug and HOXB13, are expressed in clinical specimens before the appearance of the malignant tumor phenotype (32, 33). Thus, it possible that, in human ductal-type breast tumors, Goosecoid primes cells for the expression of aggressive phenotypes, which manifest themselves in the context of subsequent alterations.
Taken together, the present results implicate the Spemann organizer gene, Goosecoid, in tumor metastasis. Moreover, they suggest that the reactivation of conserved organizer genes may be a recurrent theme in human cancer metastasis. Our findings therefore warrant a comprehensive examination of these genes in multiple types of human malignancies.
Materials and Methods
RNA Preparation and RT-PCR.
The clinical cohort examined was previously described (16). Seventy-two tumor samples were obtained from 40 patients, 28 of whom had two or more pathological subtypes of breast cancer detectable at diagnosis, and each was accompanied by a patient-matched normal breast tissue sample. The Massachusetts Institute of Technology Committee on the Use of Humans as Experimental Subjects and the Massachusetts General Hospital Human Research Committee approved this study of deidentified samples. cDNAs from the previous study were additionally analyzed for GSC by real-time quantitative PCR analysis by using the ABI 7900HT system as previously described (16). The sequences of the GSC-specific fluorogenic MGB probe (5′ to 3′) and the PCR primer pair, respectively, were as follows: VIC, CCCACCGTAGTATTTAT, GCCGCCCGCGACTAG, and CACTTTATTGTACTGTCACCCTTAATTTAAC. Statistical significance was calculated for this clinical data set by using the paired Student t test, and relative expression was calculated as described (7).
For cell line analyses, total RNA was purified by using RNA STAT-60 (Tel-Test, Friendswood, TX) and RNase-free DNase set (Qiagen, Valencia, CA) according to the manufacturer's instructions. Hexanucleotide mix (Roche, Indianapolis, IN) was used for reverse transcription. Quantitative real-time RT-PCR was performed in triplicate by using the iCycler apparatus (Bio-Rad, Cambridge, MA) and SYBR-Green detection reagent, either from stock (Molecular Probes, Eugene, OR) or in commercial master mix (PerkinElmer Applied Biosystems, Foster City, CA). The sequences of the GSC-specific primer pairs were (5′ to 3′) TCTCAACCAGCTGCACTGTC (left) and GGCGGTTCTTAAACCAGACC (right), and those of the GAPDH-specific pairs were AGCCACATCGCTCAGACAC (left) and AATGAAGGGGTCATTGATGG (right). Experimental data were normalized to GAPDH, and relative expression was calculated as described (7).
Expression Constructs and Virus Generation.
Full-length mouse Goosecoid cDNA (34) (provided by Martin Blum, Universitât Hohenheim, Stuttgart, Germany) was subcloned with or without an HA-antigen tag at the amino terminus into the pWZL-Blasticidin vector. A corresponding vector containing the GFP gene was used as control. ΔN90 β-catenin consisting of mouse β-catenin containing amino-terminal deletions of 90 aa (19) and Lef-vp16 consisting of mouse Lef-1 fused to the transactivation domain from the herpes simplex virus VP16 protein (20) (provided by Masahiro Aoki, The Scripps Research Institute, La Jolla, CA) were expressed by using the pBabe-Puromycin vector. Activated, myc-tagged, human TGFβ type I receptor cDNA (22) (provided by Joan Massagué, Sloan–Kettering Cancer Center, New York, NY) was expressed by using the pWZL-Blasticidin vector. pWZL and pBabe amphotropic viruses and lentiviruses were generated and used for target cell infection as previously described (35).
Cell Culture.
The MDCK cell line was obtained from American Type Culture Collection (Manassas, VA) and cultured in DMEM supplemented with 10% heat-inactivated FCS. The immortalized, nontransformed HMEC line, expressing the SV40 early region and hTERT, was previously described (36) and cultured in DMEM and F12 medium (1:1) containing the supplements EGF (10 ng/ml), insulin (10 μg/ml), and hydrocortisone (0.5 μg/ml), with noted exceptions. The Gsc-expressing HMEC cells were generated by using differential trypsinization of the polyclonal population of Gsc-transduced cells to separate out the scattered, less adherent cells from those not expressing substantial amounts of Gsc, as confirmed by Western blotting. Soluble, activated TGF-β1 ligand (R & D Systems, Minneapolis, MN) was used at a working concentration of 100 pM, or 2.5 ng/ml, in the presence of 5% calf serum. The MDA-MB-231 cell line was maintained in DMEM supplemented with 10% FBS.
Antibodies, Immunoblotting, and Immunofluorescence.
A rabbit polyclonal antibody against Gsc was generated by using a KLH-conjugated peptide of the sequence CSENAEKWNKTSSSKA, and resulting antisera were affinity-purified (Covance, Philadelphia, PA). The specificity of the antibody was confirmed by Western immunoblotting using whole-cell lysates expressing either tagged or untagged ectopic Gsc. Other primary antibodies used were vimentin (V9, catalog no. MS129P from Neomarkers, Fremont, CA), N-cadherin [catalog no. 180224 from Zymed (San Francisco, CA) and catalog no. 610920 from BD Transduction Labs, San Jose, CA], β-actin (catalog no. 8226 from Abcam, Cambridge, MA), pan-cytokeratin (catalog no. 071M from Biogenex, San Ramon, CA), α-catenin, γ-catenin, and E-cadherin (catalog nos. C21620, 610254, and 610182 from BD Transduction Labs). Standard procedures were used for immunoblotting and immunofluorescence.
Transwell Migration Assays.
Cells were plated on cell culture inserts (Falcon, West Chester, PA) containing a filter with 8.0-μm pores. Total cells and migrated cells were quantified by using crystal violet staining after time indicated and compared with control for differences in cell number as described (37).
Mice and Injection of Tumor Cells.
Female NOD-SCID mice (propagated on site) and nude mice (NCR nude; Taconic, Hudson, NY) were used in these studies, and all protocols were approved by the Massachusetts Institute of Technology Committee on Animal Care. Nude mice received 400 rad of γ-radiation using a dual 137Cesium source 1 day before tumor cell injection. Mice were anesthetized with either avertin (i.p.) or isoflurane (inhalation). For orthotopic injections, 1 million cells in 30 μl of Matrigel (Becton Dickinson, San Jose, CA) diluted 1:2 in medium were injected into each of two mammary glands per NOD-SCID mouse. For s.c. injections, 2 × 106 cells in 160 μl of Matrigel diluted 1:2 in medium were injected at each of three sites per nude mouse. For tail vein injections, 2 × 106 cells in 200 μl PBS were injected per mouse. Tumor diameters were measured multiple times per week by using precision calipers.
Visualization and Quantification of GFP-Labeled Lung Metastases.
Upon necropsy, lungs of injected mice were removed, separated into individual lobes, and examined under a Leica MZ 12 fluorescence dissection microscope. Images of both faces of all lobes were captured at identical settings, and the fluorescent metastatic nodules in each image were analyzed by using Cell-Profiler image analysis software developed in the laboratory of David Sabatini (www.cellprofiler.org) (38). The unpaired Student t test was used for statistical comparisons of these data.
Supplementary Material
Acknowledgments
We thank I. Ben-Porath, J. Yang, S. Mani, C. Kuperwasser, B. Elenbaas, R. Hynes, J. Lees, T. Ince, S. McCallister, L. Xu, R. Lee, A. Orimo, L. Spirio, C. Scheel, A. Karnoub, S. Stewart, and other members of the R.A.W. laboratory for invaluable input during the course of this work. We also thank M. Blum, M. Aoki, J. Massagué, M. van de Wetering, B. Elenbaas, C. Kuperwasser, and L. Spirio for reagents; M. Brooks and M. Rockas for technical assistance; and G. Bell for help with statistical analysis. We acknowledge the support of the W. M. Keck Biological Imaging Facility at Whitehead Institute. K.A.H. is the recipient of a U.S. Army Predoctoral Breast Cancer Fellowship. R.A.W. is a Daniel K. Ludwig Cancer Research Professor and an American Cancer Society Research professor. This research was supported by National Institutes of Health Grant R01-CA078461 and by a grant from the Breast Cancer Research Foundation.
Abbreviations
- EMT
epithelial–mesenchymal transition
- HMEC
human mammary epithelial cell
- MDCK
Madin–Darby canine kidney
- IDC
invasive ductal carcinoma
- ADH
atypical ductal hyperplasia
- DCIS
ductal carcinoma in situ.
Footnotes
The authors declare no conflict of interest.
References
- 1.Fidler IJ. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 2.Thiery JP. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 3.Niehrs C. Nat Rev Genet. 2004;5:425–434. doi: 10.1038/nrg1347. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert SF. Developmental Biology. Sunderland, MA: Sinauer; 1997. pp. 209–252. [Google Scholar]
- 5.Thiery JP, Sleeman JP. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 6.Shook D, Keller R. Mech Dev. 2003;120:1351–1383. doi: 10.1016/j.mod.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Grunert S, Jechlinger M, Beug H. Nat Rev Mol Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 9.DeRobertis EM. In: Gastrulation, from Cells to Embryo. Stern CD, editor. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2004. pp. 581–590. [Google Scholar]
- 10.Blum M, Gaunt SJ, Cho KW, Steinbeisser H, Blumberg B, Bittner D, De Robertis EM. Cell. 1992;69:1097–1106. doi: 10.1016/0092-8674(92)90632-m. [DOI] [PubMed] [Google Scholar]
- 11.Blumberg B, Wright CV, De Robertis EM, Cho KW. Science. 1991;253:194–196. doi: 10.1126/science.1677215. [DOI] [PubMed] [Google Scholar]
- 12.Cho KW, Blumberg B, Steinbeisser H, De Robertis EM. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niehrs C, Keller R, Cho KW, De Robertis EM. Cell. 1993;72:491–503. doi: 10.1016/0092-8674(93)90069-3. [DOI] [PubMed] [Google Scholar]
- 14.Watabe T, Kim S, Candia A, Rothbacher U, Hashimoto C, Inoue K, Cho KW. Genes Dev. 1995;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- 15.Moon RT, Kimelman D. BioEssays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 16.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, et al. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Going JJ, Anderson TJ, Battersby S, MacIntyre CC. Am J Pathol. 1988;130:193–204. [PMC free article] [PubMed] [Google Scholar]
- 18.Harland R, Gerhart J. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 19.Barth AI, Pollack AL, Altschuler Y, Mostov KE, Nelson WJ. J Cell Biol. 1997;136:693–706. doi: 10.1083/jcb.136.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki M, Hecht A, Kruse U, Kemler R, Vogt PK. Proc Natl Acad Sci USA. 1999;96:139–144. doi: 10.1073/pnas.96.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 22.Wieser R, Wrana JL, Massague J. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 24.Ku M, Sokol SY, Wu J, Tussie-Luna MI, Roy AL, Hata A. Mol Cell Biol. 2005;25:7144–7157. doi: 10.1128/MCB.25.16.7144-7157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price JE, Polyzos A, Zhang RD, Daniels LM. Cancer Res. 1990;50:717–721. [PubMed] [Google Scholar]
- 26.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 27.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 28.Savagner P, Yamada KM, Thiery JP. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holbro T, Civenni G, Hynes NE. Exp Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 30.Menard S, Fortis S, Castiglioni F, Agresti R, Balsari A. Oncology. 2001;61(Suppl 2):67–72. doi: 10.1159/000055404. [DOI] [PubMed] [Google Scholar]
- 31.Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ, et al. Mol Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- 32.Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. Nat Genet. 2005;37:1047–1054. doi: 10.1038/ng1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, et al. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Danilov V, Blum M, Schweickert A, Campione M, Steinbeisser H. J Biol Chem. 1998;273:627–635. doi: 10.1074/jbc.273.1.627. [DOI] [PubMed] [Google Scholar]
- 35.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark EA, Golub TR, Lander ES, Hynes RO. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 38.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.