Abstract
Using a Cre-Lox conditional knockout strategy, we generated a germ cell-specific androgen receptor (AR) knockout mouse (G-AR−/y) with normal spermatogenesis. Sperm count and motility in epididymis from AR−/y mice are similar to that of WT (G-AR+/y) mice. Furthermore, fertility tests show there was no difference in fertility, and almost 100% of female pups sired by G-AR−/y males younger than 15 weeks carried the deleted AR allele, suggesting the efficient AR knockout occurred in germ cells during meiosis. Together, these data provide in vivo evidence showing male mice without AR in germ cells can still have normal spermatogenesis and fertility, suggesting the essential roles of AR during spermatogenesis might come from indirect cell–cell communication in a paracrine fashion. We then compared the consequences of AR loss in the spermatogenesis and fertility of G-AR−/y mice with two other testicular cell-specific AR−/y mice and total AR knockout male mice. The results provide clear in vivo evidence that androgen/AR signaling in Sertoli cells plays a direct important role in spermatogenesis and in Leydig cells plays an autocrine regulatory role to modulate Leydig cell steroidogenic function. Total AR knockout male mice have the most severe defects among these mice. These contrasting data with G-AR−/y mice suggest AR might have different roles in the various cells within testis to contribute to normal spermatogenesis and male fertility in mice.
Keywords: germ cell, knockout, Sertoli cells, Leydig cells
Spermatogenesis (exocrine) and androgen biosynthesis (endocrine) are major functions of mammalian testes. Both functions are complicated and highly regulated. Early studies indicated that initiation, maintenance, and reinitiating of spermatogenesis depend absolutely on androgen action (1). These androgen effects on spermatogenesis were well documented in hypophysectomized and Leydig cell-destroyed animals (2). The testis contains at least four different types of cells (Sertoli, Leydig, and peritubular myoid cells and germ cells at different spermatogenic stages), and spermatogenesis depends highly on autocrine and paracrine communication among different types of testicular cells. The role of the androgen receptor (AR) in each cell type within the testis remains unclear and controversial.
Before the availability of AR-specific antibodies and AR cDNA probes, the distribution of AR was studied by using [3H]-labeled androgen-binding assays. These results found that androgen-binding activity was located in the germ cells and other testicular cells involved in spermatogenesis (3, 4). Since the isolation of AR cDNA (5) and the generation of AR-specific antibodies, much effort has been made to determine which cells in testes express functional AR (6, 7). There is general agreement that AR can be detected in Sertoli cells, peritubular myoid cells, and cells in the interstitial spaces, including Leydig cells (3, 6, 8–11). However, the localization of AR using antibodies in male germ cells remains controversial. Several studies indicated that AR is present in germ cells in different species (8, 10–13); however, other reports show there is little AR staining in the germ cells (3, 9, 14–18). Interestingly, results from another study reveal that the expression of AR in male germ cells is stage-specific, expressing only at the stage of elongated spermatid (10). Previous animal studies conducted in rats also found an AR-specific coregulator, SNURF/RNF4, is expressed in elongated spermatids (19). Together, these studies suggest that AR might play a direct role in germ cells at the elongated spermatid stage.
Synaptonemal complex protein 1 (Sycp1) is expressed specifically at the leptotene–zygotene stage of male germ cells (20). The expression of the Sycp1-Cre transgene is able to excise the loxP-floxed DNA fragment during meiosis in testes without disturbing fertility (20). Here, we report a germ cell AR−/y mouse model (G-AR−/y) through mating our floxed AR/AR (fAR/AR) female mice with Sycp1-Cre transgenic male mice to study direct AR functions in male germ cells. We also compare the testis phenotype of G-AR−/y mice with our total AR knockout (T-AR−/y), Sertoli cell-specific AR knockout (S-AR−/y), and Leydig cell-selective AR knockout (L-AR−/y) mice to dissect the distinct roles of AR in different types of testicular cells involved in spermatogenesis.
Results
Generation of T-AR−/y, S-AR−/y, L-AR−/y, and G-AR−/y Mice.
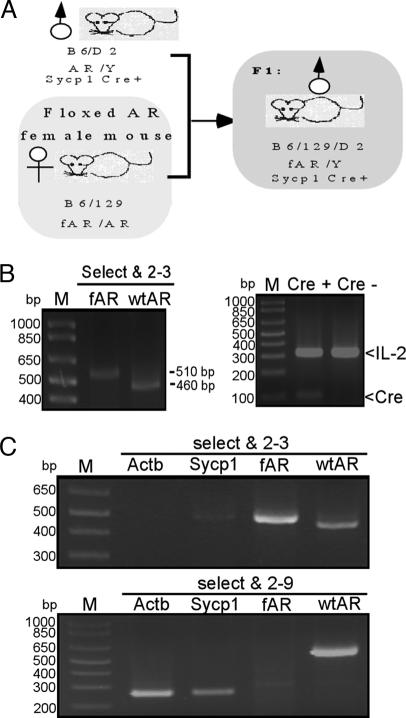
To generate total or cell-specific AR knockout mice, we have created the B6/129SVEV loxP-fAR mice (21). According to testicular histology from testicular feminized (22, 23) or Actb-Cre T-AR−/y mice (21), the germ cells still can develop to the pachytene stage, suggesting that meiosis after the pachytene stage might depend highly on the function of androgen/AR. Therefore, we designed an experiment to excise the fAR gene in leptotene–zygotene spermatocytes by the Sycp1-Cre transgene (20). After mating Sycp1-Cre transgenic males with homozygous fAR females, we produced G-AR−/y mice (Fig. 1A), with ar/Y in germ cells and fAR/Y in other cells. To examine AR recombination, we used PCR primers “select and 2–3” for fAR allele and WT AR allele and primers “select and 2–9” for deleted allele (ar) and WT AR allele, as shown (21). The genotype of G-AR−/y was characterized in tail genomic DNA by PCR with primers “select and 2–3.” Because the tail of the G-AR−/y mouse does not express Cre recombinase protein, we expect our PCR results to show the fAR allele with ≈510 bp (Fig. 1B). We also applied PCR to confirm the Cre transgene by using IL-2 as the internal control. Testicular genomic DNA was isolated from T-AR−/y, G-AR−/y, fAR, and WT males. Using primers “select and 2–3” to examine fAR and WT AR alleles, we obtained PCR products with ≈510 and ≈460 bp, respectively. In testes from G-AR−/y mice, we found a relatively strong ≈270-bp PCR product (recombinant AR, ar) by primers “select and 2–9” (Fig. 1C Lower), and a very weak ≈510-bp PCR product equal to fAR in size by primers “select and 2–3” (Fig. 1C Upper). This minor ≈510-bp product amplified from the testis of G-AR−/y indicates the lack of recombination in nongerm cells (somatic cells), which do not express Sycp1 and represent <10% cell composition of the testis. Together, these results indicated that the AR allele of germ cells in G-AR−/y was deleted by Cre recombinase. Using similar strategies, we also examined tail and testis genomic DNA of T-AR−/y, S-AR−/y, and L-AR−/y mice to confirm the AR allele of target cells in testis was deleted by Cre recombinase, as shown [see refs. 21 and 24 and Q. Xu (personal communication) for characterization of L-AR−/y].
Fig. 1.
Generation and genotyping of germ cell-specific AR knockout (G-AR−/y) mice. (A) Mating strategy and study design. The mouse marked by the gray box is a Sycp1-Cre conditional AR knockout male (G-AR−/y) mouse. (B) Genotyping of tail genomic DNA of G-AR−/y mice. The WT allele of the AR gene generated a PCR product ≈460 bp in size by primers “select and 2–3.” The fAR gene generated a ≈510-bp product with the same primers. The presence of the Sycp1-Cre and internal control IL-2 genes was confirmed by PCR. (C) (Upper) Genotyping of testis genomic DNA of G-AR−/y mice using primers “select and 2–3.” The PCR product amplified by “select and 2–3” can distinguish WT and fAR alleles with PCR products ≈460 and ≈510 bp, respectively. (Lower) Genotyping of testis genomic DNA of G-AR−/y mice using primers “select and 2–9.” The WT allele generated a ≈600-bp product. fAR with neo cassette was too large to get a PCR product under these conditions. With the fAR recombined by Sycp1-Cre, the recombinant allele produced a ≈270-bp product using the same primers.
Examining the Knockout Efficiency of G-AR−/y Mice by Female Offspring Genotype.
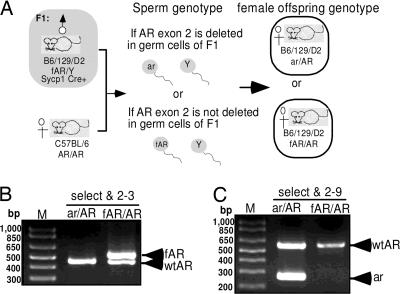
Because the spermatocytes and spermatids are functionally syncytial and diploidy with the intercellular bridges (25), there might be a concern that if Sycpl-Cre AR knockout efficiency is not 100%, some rare spermatids produced from G-AR−/y male still get their functional AR by escaping from the Cre recombination through intercellular bridges. Therefore, it is important to confirm our knockout efficiency to demonstrate the AR function in germ cells by Sycp1-Cre AR knockout strategy. The AR genotypes of 57 female pups, sired by male G-AR−/y mice mating with female AR+/+ (B6) mice, were checked by PCR to identify the ratio of female mice possessing the deleted allele to total female mice. The female genotype fertilized by sperm carrying deleted AR allele would produce ar/AR heterozygous female offspring, and those fertilized by sperm carrying fAR allele would produce the fAR/AR female genotype (Fig. 2). All 16 female progeny from three individual G-AR−/y male mice (mice A–C in Table 1) at not more than 15 weeks of age, were genotyped as ar/AR heterozygous genotype. Five of 10 female progeny were genotyped ar/AR heterozygous when sired by 16-week-old G-AR−/y male mice. Only 4 of 12 and 1 of 5 female offspring were genotyped ar/AR when sired by G-AR−/y male mice at 17 and 18 weeks of age, respectively. When G-AR−/y male mice are older than 20 weeks, all their female progenies were fAR/AR genotype and there were no detectable recombinant AR allele (ar). Together, these data suggests that Sycp1-Cre transgene can almost completely excise loxP-floxed sequence in postmitotic germ cells of fAR male mice at a young reproductive age, but Cre-induced recombination is gradually decreased in male germ cells starting at 16 weeks of age and is completely lost after 20 weeks of age.
Fig. 2.
Determining the efficiency of AR knockout in germ cells by genotyping of female offspring. (A) Two different genotypes of female offspring from G-AR−/y male mating with B6 AR+/+ female mice. (B and C) Designed primers, “select,” “2–9,” and “2–3” were used to determine genotype by PCR.
Table 1.
Results of genotyping for female offspring derived from G-AR−/y males mating with AR+/+ female mice
| G-AR−/y male |
Genotype of female offspring |
Total number of female offspring | Predicted knockout efficiency, % | ||
|---|---|---|---|---|---|
| Mice ID | Mating age, weeks | ar/AR | fAR/AR | ||
| A, B, C | ≦15 | 16 | 0 | 16 | 100 |
| A, D | 16 | 5 | 5 | 10 | 50 |
| A, B | 17 | 4 | 8 | 12 | 33 |
| E | 18 | 1 | 4 | 5 | 20 |
| A, B | ≥20 | 0 | 14 | 14 | 0 |
Epididymal Sperm Count and Fertility Test.
To compare the number and motility of sperm between AR+/y, G-AR−/y, L-AR−/y, and S-AR−/y mice, the cauda epididymis was removed and sperms were released and suspended in culture medium for 30 min at 37°C in a 5% CO2 incubator. After counting, our results indicated that there was no sperm existing in the epididymis of S-AR−/y and L-AR−/y mice, and there was no significant difference in sperm count for G-AR−/y mice as compared with AR+/y (Table 2). The motility of sperm was found similar between G-AR−/y and AR+/y mice (45–55%). To further characterize the fertility of AR+/y, G-AR−/y, L-AR−/y, and S-AR−/y mice, four male mice of each genotype and four AR+/y male mice at 8 weeks of age were mated with AR+/+ B6 female mice. Although vaginal plugs were always present in female mice the morning after mating, S-AR−/y and L-AR−/y male mice (Table 2) failed to impregnate their mates in three successive sets of 2-week pairings, with six different AR+/+ female mice. However, each individual G-AR−/y and AR+/y male mouse can produce five to six litters from six mating of different AR+/+ female mice. There were also no significant differences in the pup numbers produced by G-AR−/y and AR+/y male mice (Table 2; results from four male mice with 21–24 litters for each genotype).
Table 2.
Fertility test and epididymal sperm content analysis
| Genotype | Litter no. | Vaginal plug | Sperm count/epidiymis | Motility |
|---|---|---|---|---|
| AR+/y | 7.7 ± 1.2 | + | 26 × 106 | Normal |
| G-AR−/y | 7.8 ± 1.0 | + | 25 × 106 | Normal |
| L-AR−/y | 0 | + | 0 | 0 |
| S-AR−/y | 0 | + | 0 | 0 |
| T-AR−/y | 0 | − | No epididymis |
Fertility test and epididymal sperm content analysis in 14-week-old AR+/y, G-AR−/y, S-AR−/y, L-AR−/y, and T-AR−/y mice (n = 4 for each group).
Testis Size and Serum Testosterone Level Between AR+/y, G-AR−/y, T-AR−/y, L-AR−/y, and S-AR−/y Mice.
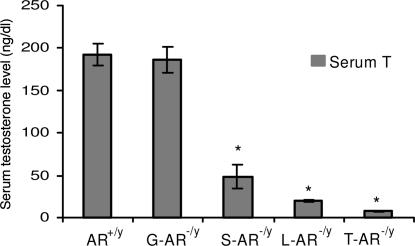
As in our previous publications (21), the testes of T-AR−/y mice were small in size (reaching only 7% of AR+/y testis size) and cryptorchid in the low abdominal area close to the internal inguinal ring, which is similar in humans with complete androgen insensitivity syndrome. The testes of S-AR−/y and L-AR−/y were normally descended as AR+/y, but the size of testes decreased and reached 23.4% and 31.1% of AR+/y testis size, respectively (Table 3). The testes of G-AR−/y mice showed normal position and similar size compared with AR+/y testes (Table 3). Serum testosterone levels of T-AR−/y, S-AR−/y and L-AR−/y were significantly decreased as compared with AR+/y (Fig. 3). Serum testosterone levels of G-AR−/y mice were within normal range, although there were wide variations between animals.
Table 3.
Body and testis weight (mean ± SEM)
| Group | n | BW (g) (% of WT) | TW (mg) (% of WT) |
|---|---|---|---|
| G-AR−/y | 10 | 27.94 ± 0.78 (93.4%) | 105.4 ± 5.1 (96.3%) |
| S-AR−/y | 10 | 26.84 ± 0.78 (96.2%) | 26.5 ± 2.0 (23.4%)* |
| L-AR−/y | 10 | 28.52 ± 0.49 (94.7%) | 35.8 ± 3.4 (31.1%)* |
| T-AR−/y | 10 | 27.76 ± 0.50 (92.0%) | 7.7 ± 0.2 (6.9%)* |
*Significant difference (P < 0.05). BW, body weight; TW, testis weight.
Fig. 3.
Serum testosterone levels (mean ± SEM) in 14-week-old AR+/y, G-AR−/y, S-AR−/y, L-AR−/y, and T-AR−/y mice. ∗, Significant difference (P < 0.05).
Morphology and Histological Analyses of AR+/y, G-AR−/y, T-AR−/y, S-AR−/y, and L-AR−/y Testes.
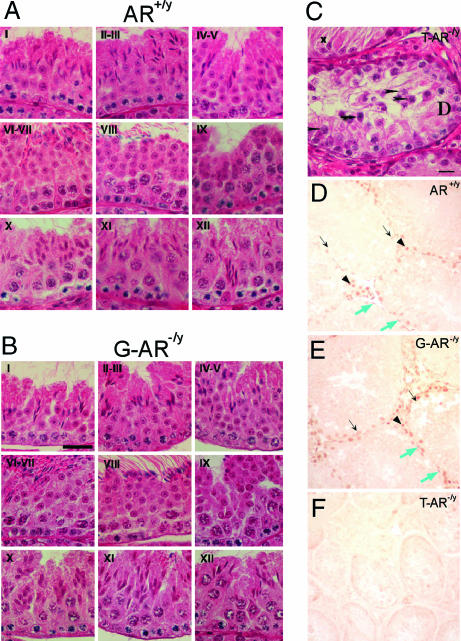
In comparison with AR+/y mice, testes from G-AR−/y mice have normal spermatogenesis at every spermatogenic stage (Fig. 4A and B). No maturation arrest or disturbance in spermatid differentiation was seen. The testicular histology of 14-week-old T-AR−/y mice showed no lumen formation in seminiferous tubules, decreases in diameter, germ cell hypoplasia, and germ development stopped at pachytene spermatocyte stage of the first meiosis division (Fig. 4C; ref. 21); S-AR−/y mice showed no lumen formation in seminiferous tubules as well as decreased diameter, with germ cell development stopped at pachytene/diplotene primary spermatocyte stage of the first meiosis division (Fig. 5E; ref. 24); L-AR−/y mice showed no lumen formation in seminiferous tubules as well as decreased diameter, with germ cell development stopped at the stage around secondary spermatocyte to round spermatid, and no mature and elongated spermatid or spermatozoa can be found (Fig. 5H; Q. Xu, personal communication).
Fig. 4.
Histological analyses and AR staining in testis of AR+/y, G-AR−/y, and T-AR−/y mice. Histology of testes by H&E staining (A–C) and immunostaining (D–F) of AR protein in testicular sections from 14-week-old AR+/y, G-AR−/y, and T-AR−/y mice. Four to six 14-week-old mice from individual groups were killed, and testes were excised for histology section. Three of the G-AR−/y mice have been confirmed with the 100% AR knockout in germ cells by genotyping of female offspring (Table 1). I–XII in each image in A and B indicate the specific stage of spermatogenesis. There is no difference in testicular morphology between AR+/y and G-AR−/y mice. In contrast, testes from T-AR−/y mice (C) showed maturation arrest at pachytene spermatocytes (arrowheads) and apoptotic-like bodies (arrows) in some seminiferous tubules and Sertoli cells only (with degeneration, x) in the other tubules. (Scale bars: A and B, 30 μm; C, 15 μm.) In testes from both AR+/y (D) and G-AR−/y (E) mice, the AR protein was located in the nuclei of Sertoli cells (black arrows), Leydig cells (arrowheads), and peritubular myoid cells (blue arrows). The testis from the T-AR−/y mouse (F) was used as a negative control to confirm the specificity of AR staining. (Scale bar: 30 μm.)
Fig. 5.
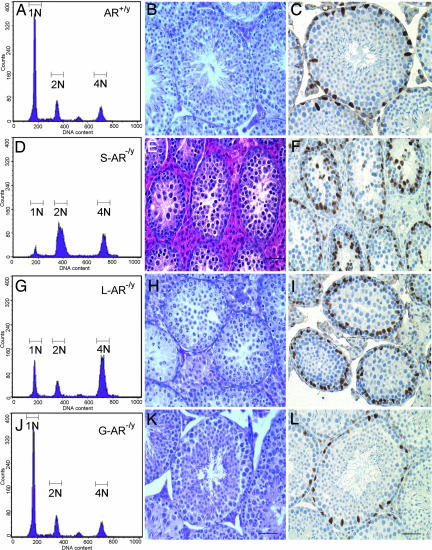
Flow cytometric analyses, morphology analyses, and GATA-1 staining in testes of AR+/y, S-AR−/y, and T-AR−/y. (A, D, G, and J) Analysis of germ cell DNA content of AR+/y, S-AR−/y, L-AR−/y, and G-AR−/y mice by using flow cytometry. N1 represents haploid cells, N2 represents diploid cells, and N3 represents tetraploid cells. Compared with the AR+/y testis (A), the S-AR−/y testis showed a 3-fold increase in diploid cells, a 2-fold increase in tetraploid cells, and an 11-fold reduction in haploid cells (D); the L-AR−/y testes showed a 4-fold increase in tetraploid cells and a 2.8-fold reduction in haploid cells (G). There were similar distributions of DNA content histogram peaks between the G-AR−/y (J) and AR+/y testis. (B, E, H, and K) Testicular morphology analysis of 14-week-old mice. Compared with the AR+/y testis (B), the S-AR−/y testis showed no lumen formation in seminiferous tubules, and germ cell development stopped at the pachytene/diplotene primary spermatocyte stage of the first meiosis division (E); L-AR−/y testes showed no lumen formation in seminiferous tubules, and germ cell development stopped at the stage around the secondary spermatocyte to round spermatid; no elongated spermatid or released spermatozoa can be found (H); the G-AR−/y testis (K) showed relatively comparable seminiferous tubule diameters and a full range of germ cell development. (C, F, I, and L) Pathomicrographs illustrating the distribution of Sertoli (GATA-1 immunopositive; brown staining) cell nuclei in a 14-week-old mouse testis. GATA-1-immunopositive Sertoli cell nuclei located at the basal portion of the seminiferous tubules in AR+/y, L-AR−/y, and G-AR−/y testes (C, I, and L). However, the locations of GATA-1-immunopositive Sertoli cell nuclei are disorganized and dislocalized in S-AR−/y testes (F). (Scale bar, 50 μm.)
AR Staining in Testes of AR+/y, G-AR−/y, and T-AR−/y Mice.
To confirm the cell-specific knockout of AR in testicular cells, we performed immunostaining of AR to check the expression of AR in testis. The AR+/y testes showed staining of the nuclei of Sertoli cells, peritubular myoid cells, and Leydig cells (Fig. 4D). This pattern is similar to most of the published results. In the T-AR−/y testes, only weak diffuse AR staining, even in pachytene spermatocytes, was detected, and no obvious nuclear staining was found in any cell type (Fig. 4F). The results from AR+/y and T-AR−/y testes confirmed the specificity for AR staining. We then stained the G-AR−/y testes and found the nuclear expression of AR in the Sertoli, Leydig, and peritubular myoid cells (Fig. 4E). We also performed immunostaining of GATA-1 to check the localization of Sertoli cells in seminiferous tubules in different cell-specific AR knockout mice. GATA-1 is a zinc-finger transcription factor that is immunoexpressed in the nuclei of Sertoli cells and the levels of its expression in the adult testes depend on changes in the spermatogenic cycle (26). Sertoli cells are tall columnar epithelial cells, which have irregularly shapes and are extended from the basal to the adluminal compartment of the seminiferous tubules (27). Our results showed that GATA-1 immunopositive Sertoli cell nuclei are located at the basal portion of the seminiferous tubules in AR+/y, L-AR−/y, and G-AR−/y testes (Fig. 5 C, I, L, and O). However, the location of GATA-1 immunopositive Sertoli cell nuclei in S-AR−/y testis (Fig. 5F) showed disorganization and dislocalization. This result indicates loss of functional AR in Sertoli cell will deteriorate its morphological organization.
Flow Cytometric Analyses of Testicular Contents of AR+/y, G-AR−/y, T-AR−/y, L-AR−/y, and S-AR−/y Mice.
Using flow cytometric analyses of propidium iodide-labeled cells, we detected three main histogram peaks of DNA content, which represented haploid (1N, round spermatids and spermatozoa), diploid (2N, spermatogonia, preleptotene primary spermatocytes, secondary spermatocytes, and somatic cells) and tetraploid cells (4N, spermatogonia, leptotene, zygotene, pachytene, and diplotene primary spermatocytes). Compared with AR+/y testes, S-AR−/y testes showed a 3-fold increase in diploid cells, a 2-fold increase in tetraploid cells, and an 11-fold reduction in haploid cells (Fig. 5D); L-AR−/y testes showed a 4-fold increase in tetraploid cells and a 2.8-fold reduction in haploid cells (Fig. 5G). There were similar distributions of DNA content histogram peaks between G-AR−/y and AR+/y testes. These results correlate well with the findings of testis morphology analysis, where S-AR−/y testes showed no lumen formation in seminiferous tubules as well as germ cell development stopped at pachytene/diplotene primary spermatocyte stage of first meiosis division (Fig. 5E). L-AR−/y testes showed no lumen formation in seminiferous tubules; they also showed that germ cell development stopped at the round spermatid stage. No mature and elongated spermatid or spermatozoa can be found (Fig. 5H).
Discussion
Hormonal regulation of the spermatogenic process occurs across three phases: initiation, maintenance, and reinitiation (28). Androgens alone have been shown to initiate qualitatively complete spermatogenesis in the gonadotropin-deficient mouse (29), but follicle-stimulating hormone alone fails to rescue spermatogenesis beyond the meiotic stages (30, 31). Furthermore, earlier studies using classic hormone-withdrawal experiments in rats provide additional supportive evidence that androgen is necessary for the completion of meiosis and the differentiation of round spermatids into the spermatozoa. Removal of androgen from adult rats by hypophysectomy is displayed initially as loss of midstage round spermatids and elongated spermatids (32). After long-term hypophysectomy and elimination of residual testosterone activity by flutamide or ethane dimethanesulphonate treatment, spermatogenesis rarely proceeds beyond meiosis, only few round spermatids can be observed, and no elongated spermatids are present (33, 34). Recent publications clearly demonstrated that AR is essential for the completion of meiosis and the development of spermatozoa in T-AR−/y (21) and S-AR−/y mouse models (24, 35, 36).
T-AR−/y Mice Confirmed the Necessity of AR Signaling for both External and Internal Male Phenotype Development.
Our published data show that male T-AR−/y mice displayed feminized external genitalia, markedly decreased serum testosterone levels (96% decreased as compared with AR+/y), and increased leuteinizing hormone levels (21). Internally, the accessory sex organs of the T-AR−/y mice were agenic, and the testes were cryptorchid and had significantly reduced cell numbers of Leydig, Sertoli, and germ cells as compared with AR+/y testes (21). In contrast to the full composition of germ cells in various stages of maturation, including late-stage elongated spermatids observed in the AR+/y testis, some tubules in the T-AR−/y sections lack germ cells and others contained few germ cells. The complete absence of round spermatids, elongated spermatids, and spermatozoa throughout the T-AR−/y testis is similar to that of the testicular feminized mice testis, suggesting that spermatogenic arrest occurs at the pachytene spermatocyte stage in these mice (21, 37).
G-AR−/y Mice Showed Comparable Seminiferous Tubule Diameters and the Full Range of Germ Cell Development Compared with AR+/y Littermates.
Although it was suggested that the presence of functional AR in germ cells was not essential for development into sperm in a chimera study by Lyon et al. (38) and in a recent spermatogonial transplantation study (2), our studies clearly show that the quality and quantity, as well as the fertility, of spermatogenesis are normal, whereas the AR gene is deficient in nearly all of germ cells beyond the pachytene stage. The major concern about the chimera study (38) is that many germ cells from the pachytene stage to spermatids (the obvious androgen-dependent period during spermatogenesis) still contain an intact AR gene that might support spermatogenesis through autocrine or paracrine effects. The spermatogonial transplantation from testicular feminized mice to AR+/y testes with an intact AR gene could produce colonies with normal spermatogenesis but could not reestablish fertility (22). These data suggest that AR expression in germ cells might not play essential roles for germ cell meiosis and round spermatid differentiation. Moreover, our study concludes that the deletion of the functional AR gene in germ cells has no effect on sperm motility, efficiency of sperm production, and normal fertility of mature sperm. Although the fAR allele was not excised in spermatogonia and early spermatocytes of G-AR−/y mice, no AR expression could be found in spermatogonia and spermatocytes in any previous studies (10). Testicular morphology from T-AR−/y and testicular feminized mice also suggests that AR has no effect on this stage of spermatogenesis, because the testes of these mice contained spermatogonia, and spermatocytes reached the pachytene stage (21, 22).
Because spermatocytes and spermatids are functionally syncytial through intercellular bridges, the argument might be raised that the residual AR protein is still present in most spermatids if the efficiency of the gene knockout is not 100%. The Sycp1-Cre transgene has been used to delete the floxed gene with a germ cell specificity and very high efficiency (almost 100%; ref. 20). In our offspring genotype study, we also found that in young male G-AR−/y mice, the knockout efficiency was 100%. At least in the majority of G-AR−/y mice, exon 2 of the AR gene was almost completely deleted by Cre recombinase. Therefore, the expression of AR in postmitotic germ cells was nearly absent in G-AR−/y mice. Our PCR result for testicular genomic DNA was also consistent with this conclusion, because the recombinant allele was the predominant form of AR in the testes of G-AR−/y mice. However, our study strongly suggests that recombination in G-AR−/y mice is gradually lost in an age-dependent manner. One possible explanation is that the copy number of Sycp1-Cre transgene was diluted or eliminated after rapid proliferation of spermatogonia. When the copy number or expression level of Cre-transgene falls below a certain threshold, the fAR gene can no longer be excised. Furthermore, the methylation of loxP sites in the testes of the transgenic animal is another possible mechanism underlying this phenomenon (39).
Our results suggest that the functional AR in germ cells is not essential for spermatogenesis and normal fertility of mice. However, the animal model might not be exactly the same as the human condition. The testicular morphology of men with complete androgen insensitivity syndrome showed Sertoli cells only and very scant spermatogonia in seminiferous tubules (40, 41). In the mouse counterpart, the testicular histology of T-AR−/y revealed maturation arrest, with the development of germ cells stopping at the pachytene stage of spermatocytes (21). Thus, the role of AR in human spermatogenesis might be more critical than in mice. Manipulating the AR in the laboratory developed human embryonic gonadal cells (42) or culturing of human spermatogonia might help us further clarify the role of AR in human germ cells during spermatogenesis.
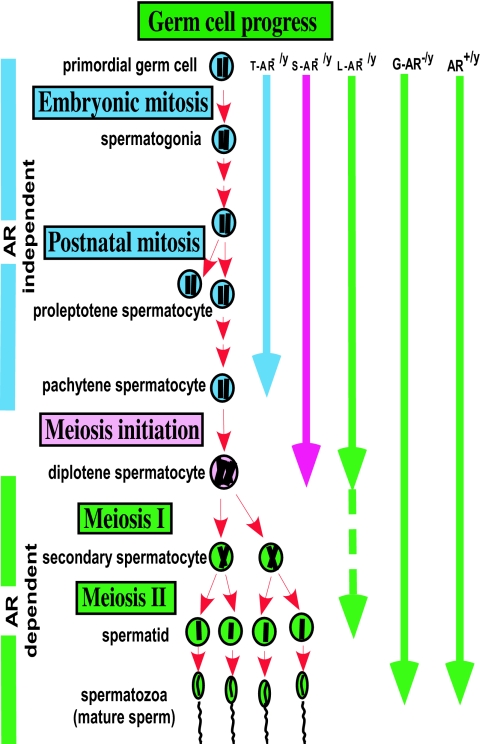
Our results showed clear in vivo evidence that androgen/AR signaling in different testicular cell types has different impacts on spermatogenesis (Fig. 6). Apparently, androgen/AR signaling in Sertoli cells plays the most important role in spermatogenesis, and testes from S-AR−/y mice show the poorest phenotype. On the other hand, our results confirmed previous in vitro findings (43) that androgen/AR signaling in Leydig cells displays autocrine regulation. As mentioned previously, lacking functional AR in Leydig cells has a major influence on Leydig cell steroidogenic function leading to spermatogenesis arrest, predominately at the round spermatid stage. Although debatable, our results clearly demonstrate that functional AR in germ cells is not essential for spermatogenesis and normal fertility of mice.
Fig. 6.
Diagram of germ cell progression in AR+/y, G-AR−/y, L-AR−/y, S-AR−/y, and T-AR−/y testes. AR+/y and G-AR−/y testes can achieve full germ cell progression. However, spermatogenesis in the T-AR−/y, S-AR−/y, and L-AR−/y testes ceases predominately at the pachytene, pachytene/diplotene, and around the secondary spermatocyte to round spermatid stages, respectively.
Taken together, using cell-specific knockout of AR in various testicular cells, our data indicate that AR has differential roles in the different testicular cells responsible for spermatogenesis and male fertility.
Materials and Methods
Generation of T-AR−/y, S-AR−/y, L-AR−/y, and G-AR−/y Mice.
The strategy to generate fAR gene-targeting mice has been described (21). Because the AR gene is located on the X chromosome in mice, and thus there is a single allele of AR in a male mouse, we mated male Actb-Cre (Cre under the control of the β-actin) mice with fAR/AR female mice to produce T-AR−/y male mice. Mating male AMH-Cre transgenic mice with fAR/AR female mice produced male S-AR−/y male mice (AMH-ar/Y in Sertoli cells and fAR/Y in the other cells; ref. 24). Mating male AMHRII-Cre transgenic mice with fAR/AR female mice produced male L-AR−/y mice (AMHRII-ar/Y in Leydig cells and partially in Sertoli cells, fAR/Y in the other cells; Q. Xu, personal communication). Mating male Sycp1-Cre transgenic mice with fAR/AR female mice produced G-AR−/y male mice (Cre under the control of the Sycp1 gene promoter, Sycp1-ar/Y in germ cells, and fAR/Y in the other cells; ref. 20) (Fig. 1A). Male WT (AR+/y) B6 or fAR/Y male mice without the Cre transgene were used as controls. Each type of transgenic mice expresses fAR and Cre alleles in tail genomic DNA. We genotyped 21-day-old pups from tail snips by PCR, as described (21).
Female Offspring Genotyping.
To determine AR knockout efficiency during spermatogenesis, female offspring from G-AR−/y mice in fertility tests were studied by genotype with PCR. The genotype of male offspring from G-AR−/y male mating with WT B6 female (AR+/+) is not included, because the X chromosome (where AR gene is located) of male offspring is from the AR+/+ mother rather than the G-AR−/y male. The ratio of female offspring carrying recombinant AR allele (ar) to total female pups was used to determine AR knockout efficiency.
Other Methods.
Fertility assessment, evaluation of epididymal sperm, morphology assessment, immunohistochemistry with anti-AR antibody, propidium iodide staining and flow cytometry, and serum testosterone levels were described previously (21, 24).
Acknowledgments
These studies were supported by National Institutes of Health Grants DK60905 and DK60912 and by the George H. Whipple Professorship Endowment.
Abbreviations
- AR
androgen receptor
- fAR
floxed AR
- T-AR−/y
total AR knockout
- S-AR−/y
Sertoli cell-specific knockout
- L-AR−/y
Leydig cell-specific knockout
- G-AR−/y
germ cell AR knockout.
Footnotes
The authors declare no conflict of interest.
References
- 1.Roberts KP, Zirkin BR. Ann NY Acad Sci. 1991;637:90–106. doi: 10.1111/j.1749-6632.1991.tb27303.x. [DOI] [PubMed] [Google Scholar]
- 2.Johnston DS, Russell LD, Friel PJ, Griswold MD. Endocrinology. 2001;142:2405–2408. doi: 10.1210/endo.142.6.8317. [DOI] [PubMed] [Google Scholar]
- 3.Anthony CT, Kovacs WJ, Skinner MK. Endocrinology. 1989;125:2628–2635. doi: 10.1210/endo-125-5-2628. [DOI] [PubMed] [Google Scholar]
- 4.Sanborn BM, Steinberger A, Meistrich ML, Steinberger E. J Steroid Biochem. 1975;6:1459–1465. doi: 10.1016/0022-4731(75)90197-1. [DOI] [PubMed] [Google Scholar]
- 5.Chang CS, Kokontis J, Liao ST. Science. 1988;240:324–326. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- 6.Sar M, Lubahn DB, French FS, Wilson EM. Endocrinology. 1990;127:3180–3186. doi: 10.1210/endo-127-6-3180. [DOI] [PubMed] [Google Scholar]
- 7.Takeda H, Chodak G, Mutchnik S, Nakamoto T, Chang C. J Endocrinol. 1990;126:17–25. doi: 10.1677/joe.0.1260017. [DOI] [PubMed] [Google Scholar]
- 8.Kimura N, Mizokami A, Oonuma T, Sasano H, Nagura H. J Histochem Cytochem. 1993;41:671–678. doi: 10.1177/41.5.8468448. [DOI] [PubMed] [Google Scholar]
- 9.Bremner WJ, Millar MR, Sharpe RM, Saunders PT. Endocrinology. 1994;135:1227–1234. doi: 10.1210/endo.135.3.8070367. [DOI] [PubMed] [Google Scholar]
- 10.Vornberger W, Prins G, Musto NA, Suarez-Quian CA. Endocrinology. 1994;134:2307–2316. doi: 10.1210/endo.134.5.8156934. [DOI] [PubMed] [Google Scholar]
- 11.Zhou X, Kudo A, Kawakami H, Hirano H. Anat Rec. 1996;245:509–518. doi: 10.1002/(SICI)1097-0185(199607)245:3<509::AID-AR7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Janssen PJ, Brinkmann AO, Boersma WJ, Van der Kwast TH. J Histochem Cytochem. 1994;42:1169–1175. doi: 10.1177/42.8.8027537. [DOI] [PubMed] [Google Scholar]
- 13.Arenas MI, Royuela M, Lobo MV, Alfaro JM, Fraile B, Paniagua R. J Anat. 2001;199:465–472. doi: 10.1046/j.1469-7580.2001.19940465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galena HJ, Pillai AK, Terner C. J Endocrinol. 1974;63:223–237. doi: 10.1677/joe.0.0630223. [DOI] [PubMed] [Google Scholar]
- 15.Grootegoed JA, Peters MJ, Mulder E, Rommerts FF, Van der Molen HJ. Mol Cell Endocrinol. 1977;9:159–167. doi: 10.1016/0303-7207(77)90117-4. [DOI] [PubMed] [Google Scholar]
- 16.Suarez-Quian CA, Martinez-Garcia F, Nistal M, Regadera J. J Clin Endocrinol Metab. 1999;84:350–358. doi: 10.1210/jcem.84.1.5410. [DOI] [PubMed] [Google Scholar]
- 17.Van Roijen JH, Van Assen S, Van Der Kwast TH, De Rooij DG, Boersma WJ, Vreeburg JT, Weber RF. J Androl. 1995;16:510–516. [PubMed] [Google Scholar]
- 18.Pelletier G, Labrie C, Labrie F. J Endocrinol. 2000;165:359–370. doi: 10.1677/joe.0.1650359. [DOI] [PubMed] [Google Scholar]
- 19.Yan W, Hirvonen-Santti SJ, Palvimo JJ, Toppari J, Janne OA. Mech Dev. 2002;118:247–253. doi: 10.1016/s0925-4773(02)00261-7. [DOI] [PubMed] [Google Scholar]
- 20.Vidal F, Sage J, Cuzin F, Rassoulzadegan M. Mol Reprod Dev. 1998;51:274–280. doi: 10.1002/(SICI)1098-2795(199811)51:3<274::AID-MRD6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 21.Yeh S, Tsai MY, Xu Q, Mu XM, Lardy H, Huang KE, Lin H, Yeh SD, Altuwaijri S, Zhou X, et al. Proc Natl Acad Sci USA. 2002;99:13498–13503. doi: 10.1073/pnas.212474399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy L, O'Shaughnessy PJ. J Endocrinol. 1991;131:443–449. doi: 10.1677/joe.0.1310443. [DOI] [PubMed] [Google Scholar]
- 23.O'Shaughnessy PJ, Murphy L. J Mol Endocrinol. 1993;11:77–82. doi: 10.1677/jme.0.0110077. [DOI] [PubMed] [Google Scholar]
- 24.Chang C, Chen YT, Yeh SD, Xu Q, Wang RS, Guillou F, Lardy H, Yeh S. Proc Natl Acad Sci USA. 2004;101:6876–6881. doi: 10.1073/pnas.0307306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun RE, Behringer RR, Peschon JJ, Brinster RL, Palmiter RD. Nature. 1989;337:373–376. doi: 10.1038/337373a0. [DOI] [PubMed] [Google Scholar]
- 26.de Kretser DM, Kerr JB. In: The Physiology of Reproduction. Knobil E, Neill JB, Ewing LL, Greenwald GS, Markert CL, Pfaff DW, editors. New York: Raven; 1988. pp. 837–932. [Google Scholar]
- 27.Mruk DD, Cheng CY. Endocr Rev. 2004;25:747–806. doi: 10.1210/er.2003-0022. [DOI] [PubMed] [Google Scholar]
- 28.Weinbauer GF, Nieschlag E. In: Molecular Biology of the Male Reproductive System. de Kretser D, editor. San Diego: Academic; 1993. pp. 101–142. [Google Scholar]
- 29.Singh J, O'Neill C, Handelsman DJ. Endocrinology. 1995;136:5311–5321. doi: 10.1210/endo.136.12.7588276. [DOI] [PubMed] [Google Scholar]
- 30.Singh J, Handelsman DJ. J Androl. 1996;17:382–393. [PubMed] [Google Scholar]
- 31.Haywood M, Spaliviero J, Jimemez M, King NJ, Handelsman DJ, Allan CM. Endocrinology. 2003;144:509–517. doi: 10.1210/en.2002-220710. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh S, Sinha-Hikim AP, Russell LD. Tissue Cell. 1991;23:613–630. doi: 10.1016/0040-8166(91)90018-o. [DOI] [PubMed] [Google Scholar]
- 33.Kerr JB, Maddocks S, Sharpe RM. Cell Tissue Res. 1992;268:179–189. doi: 10.1007/BF00338067. [DOI] [PubMed] [Google Scholar]
- 34.Franca LR, Parreira GG, Gates RJ, Russell LD. J Androl. 1998;19:335–340. discussion 341–342. [PubMed] [Google Scholar]
- 35.De Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, Devos A, Tan K, Atanassova N, Claessens F, Lecureuil C, et al. Proc Natl Acad Sci USA. 2004;101:1327–1332. doi: 10.1073/pnas.0308114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holdcraft RW, Braun RE. Development (Cambridge, UK) 2004;131:459–467. doi: 10.1242/dev.00957. [DOI] [PubMed] [Google Scholar]
- 37.O'Shaughnessy PJ, Johnston H, Willerton L, Baker PJ. J Cell Sci. 2002;115:3491–3496. doi: 10.1242/jcs.115.17.3491. [DOI] [PubMed] [Google Scholar]
- 38.Lyon MF, Glenister PH, Lamoreux ML. Nature. 1975;258:620–622. doi: 10.1038/258620a0. [DOI] [PubMed] [Google Scholar]
- 39.Rassoulzadegan M, Magliano M, Cuzin F. EMBO J. 2002;21:440–450. doi: 10.1093/emboj/21.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Justrabo E, Cabanne F, Michiels R, Bastien H, Dusserre P, Pansiot F, Cayot F. J Pathol. 1978;126:165–171. doi: 10.1002/path.1711260305. [DOI] [PubMed] [Google Scholar]
- 41.Rutgers JL, Scully RE. Int J Gynecol Pathol. 1991;10:126–144. doi: 10.1097/00004347-199104000-00002. [DOI] [PubMed] [Google Scholar]
- 42.Shamblott MJ, Axelman J, Wang S, Bugg EM, Littlefield JW, Donovan PJ, Blumenthal PD, Huggins GR, Gearhart JD. Proc Natl Acad Sci USA. 1998;95:13726–13731. doi: 10.1073/pnas.95.23.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hales DB, Sha LL, Payne AH. J Biol Chem. 1987;262:11200–11206. [PubMed] [Google Scholar]