Abstract
Peroxisome proliferator-activated receptor (PPAR) δ is a member of the nuclear hormone receptor superfamily. PPARδ may ameliorate metabolic diseases such as obesity and diabetes. However, PPARδ's role in colorectal carcinogenesis remains controversial. Here, we present genetic and pharmacologic evidence demonstrating that deletion of PPARδ decreases intestinal adenoma growth in ApcMin/+ mice and inhibits tumor-promoting effects of a PPARδ agonist GW501516. More importantly, we found that activation of PPARδ up-regulated VEGF in colon carcinoma cells. VEGF directly promotes colon tumor epithelial cell survival through activation of PI3K–Akt signaling. These results not only highlight concerns about the use of PPARδ agonists for treatment of metabolic disorders in patients who are at high risk for colorectal cancer, but also support the rationale for developing PPARδ antagonists for prevention and/or treatment of cancer.
Keywords: apoptosis, colorectal cancer
Elevated dietary fat intake is an environmental factor that exacerbates some diseases. Recent studies have shown that peroxisome proliferator-activated receptors (PPARs) can act as lipid sensors to regulate nutrient metabolism and energy homeostasis. PPARs are members of the nuclear hormone receptor superfamily and are ligand-dependent transcription factors. Three members of the PPAR family include PPARα, PPARδ/β, and PPARγ. Genetic and pharmacologic studies reveal that PPARδ is important for enhancing fat metabolism, decreasing weight gain, improving insulin sensitivity, and elevating high-density lipoprotein (HDL) levels (1–4). These findings suggest that PPARδ agonists are potential agents for the treatment of dyslipidemias, obesity, and Type 2 diabetes. In this respect, a PPARδ agonist GW501516 is currently under evaluation in phase III clinical trials for these kinds of indications. However, our previous studies showing that GW501516 accelerates intestinal adenoma growth in ApcMin/+ mice (5) raise concerns about developing PPARδ agonists for human use, especially in people who are at a high risk for developing colorectal cancer. Clarifying this issue is of critical importance to avoid harmful effects in patients who may be considered for treatment with these agents.
PPARδ's role in colorectal carcinogenesis remains ambiguous. The first evidence linking PPARδ to carcinogenesis emerged from colorectal cancer studies. PPARδ expression is elevated in the majority of human colorectal cancers, ApcMin/+ mice, and azoxymethane-treated rats (6, 7). PPARδ is up-regulated by both Wnt/APC/β-catenin and oncogenic K-Ras (6, 8), and PPARδ activity is induced by PGE2 (9). These signaling pathways are active during development of colorectal cancer. Moreover, PPARδ is also a potential target of nonsteroidal antiinflammatory drugs (NSAIDs) (6), and the protective effect of NSAIDs against colorectal adenomas was reported to be modulated by a polymorphism in the PPARδ gene (10). The disruption of both PPARδ alleles in human HCT-116 colon carcinoma cells inhibits tumor growth in xenograft studies, suggesting that PPARδ promotes tumor progression (11). Although we had previously shown that a PPARδ agonist is proneoplastic (5), it was not known whether PPARδ, in fact, mediates this effect. More importantly, PPARδ's involvement in colorectal cancer is now being hotly debated because of conflicting reports in the literature. Although one study shows that the loss of PPARδ does not affect intestinal polyp multiplicity in ApcMin/+ mice (12), two other reports reveal that the disruption of PPARδ increased polyp formation in ApcMin/+ mice in the absence of exogenous PPARδ stimulation (13, 14). These two studies implicate PPARδ, like PPARγ, as a potential tumor-suppressor gene. Thus, the role of PPARδ in colon carcinogenesis has become controversial, necessitating further in-depth studies.
Apoptosis, proliferation, and angiogenesis are essential cellular processes for human cancer progression. The PPARδ agonist GW501516 has been reported to stimulate proliferation of human breast, prostate, and hepatocellular carcinoma cells (15, 16). PPARδ was also shown to play an important role in promoting cell survival in the kidney after hypertonic stress (17) and in the skin after wound injury (18, 19). However, little is known regarding the role of PPARδ in these cellular processes during colorectal cancer progression.
VEGF stimulates endothelial cell proliferation and prevents apoptosis in the endothelial cells of newly formed vessels (20). Although the role of VEGF in stimulating tumor-associated angiogenesis through binding to VEGFRs on endothelial cells is well documented, emerging data suggest that VEGFRs are expressed in liquid and solid tumor cells including hematopoietic malignancies (21, 22), non-small-cell lung carcinomas (23), prostate cancer (24), melanoma (25), and breast cancer (26). These findings imply a potential role for the VEGF/VEGFR autocrine loop in cancer biology.
The aim of this study was to investigate the role of PPARδ in modulating intestinal adenoma growth. We used genetic and pharmacologic approaches to evaluate in vivo effects of PPARδ activation. Our results demonstrate that deletion of PPARδ resulted in a decrease in polyp number and size in ApcMin/+ mice. Moreover, the PPARδ agonist GW501516 is not effective in accelerating intestinal adenoma growth in PPARδ-deficient ApcMin/+ mice. More importantly, we demonstrate that the effects of PPARδ are mediated, at least in part, through VEGF, which, in turn, promotes epithelial tumor cell survival. This autocrine activity of VEGF on carcinoma cells works through activation of the PI3K–Akt pathway. Collectively, these results reveal that PPARδ activation promotes tumor growth by inhibiting epithelial tumor cell apoptosis through activation of a VEGF autocrine signaling loop.
Results and Discussion
PPARδ Accelerates Intestinal Adenoma Growth in Apc Min/+ Mice.
APC is a well characterized tumor suppressor-gene. APC mutations are involved in the initiation of both hereditary and sporadic colorectal cancer. ApcMin/+ mice bearing a germ-line mutation in the APC gene develop multiple polyps in the small intestine and have been used widely to study intestinal polyposis. Because information regarding PPARδ's role in colorectal cancer is limited and highly controversial, we critically evaluated the functional consequence of the lack of PPARδ in ApcMin/+ mice. Mice at the age of 13 weeks from each experimental group were killed, and then polyp number and size was determined. As shown in Fig. 1, both male (Fig. 1A) and female (Fig. 1C) PPARδ-deficient ApcMin/+ mice (PPARδ−/−/ApcMin/+) exhibited a 3-fold reduction in small intestinal polyps as compared with control ApcMin/+ mice (PPARδ+/+/ApcMin/+) on the identical genetic background. Notably, deletion of PPARδ results in an ≈10-fold decrease in the number of large polyps (>1 mm) in both male and female ApcMin/+ mice. A similar trend is observed in the large intestines of these mice (Fig. 1 B and D). Although heterozygous deletion of PPARδ (PPARδ+/−/ApcMin/+) does not significantly reduce the total number of small and large intestinal polyps in male mice, this disruption significantly diminishes the number of small intestinal polyps that were >1 mm (Fig. 1A). Histological analysis revealed that large, medium, or small polyps from different genotypes are all adenomas (Fig. 1E). These results provide genetic evidence showing that PPARδ accelerates polyp growth.
Fig. 1.
The effect of PPARδ deletion on intestinal polyp number and size. (A–D) Both male (A and B) and female (C and D) mice with different genotypes at the age of 13 weeks were killed to quantitate polyp number and size in the small intestine (A and C) and large intestine (B and D). Data are expressed as mean ± SE (∗, P < 0.05; Bonferroni test). (E) Representative H&E-stained sections from male PPARδ−/−/ApcMin/+ and PPARδ+/+/ApMin/+ mice are shown (Scale bar, 500 μm.)
Our results differ from previous reports by other laboratories (13, 14). One explanation for these disparate results may be due to differences in the genetic background of ApcMin/+ mice, animal breeding, or possibly to differences in the specific targeting strategy used to delete PPARδ. For example, the average number of polyps in 13-week old ApcMin/+ mice on a C57BL/6 genetic background is ≈50, whereas the polyp number in ApcMin/+ mice on a mixed-genetic background (C57BL/6 × 129/SV) is ≈120. Our results also show that the breeding strategy affects the number and size of polyps in mice even on the same genetic background. Mice generated by breeding female PPARδ−/−/ApcMin/+ with male PPARδ−/−/ Apc+/+ exhibit increased adenoma number with a larger average size than those obtained by breeding female PPARδ−/−/Apc+/+ with male PPARδ−/−/ApcMin/+ (data not shown). Finally, the PPARδ null mice we studied were obtained from Beatrice Desvergne (University of Lausanne, Switzerland). These mice were generated by deleting exons 4 and 5 encoding the DNA-binding domain (27), whereas Peters group (28) generated the PPARδ knockout mice by inserting a neomycin-resistance cassette into the last exon (exon 8). It has been suggested that the strategy used to disrupt PPARδ by the Peters group might have led to a hypomorphic allele, which retains some aporeceptor function, thus making it difficult to correctly interpret their results. Indeed, conflicting results in the context of embryonic lethality have also been observed from these two PPARδ mutant mouse strains (27, 28).
PPARδ Mediates the Effect of GW501516 in Promoting Intestinal Polyp Growth.
To determine whether PPARδ mediates the tumor-promoting effects of the PPARδ agonist GW501516, PPARδ+/+/ApcMin/+, and PPARδ−/−/ApcMin/+ mice were treated with 0.5% carboxymethylcellulose (CMC) solution containing GW501516 or vehicle alone. After 7 weeks of treatment with GW501516, male PPARδ+/+/ApcMin/+ mice exhibit a 2- to 3.6-fold increase in tumor number in the small intestine and colon, respectively, as compared with controls (Fig. 2A and B). GW501516 treatment mainly increased the number of large polyps (>1 mm) in both small and large intestine, suggesting that PPARδ activation primarily affects the rate of polyp growth. Unlike in PPARδ+/+/ApcMin/+ mice, the administration of GW501516 fails to affect small and large intestinal polyp burden in both male and female PPARδ−/−/ApcMin/+ mice (Fig. 2). Interestingly, the number and size of intestinal polyps in PPARδ-deficient ApcMin/+ mice treated with CMC were significantly less than those ApcMin/+ mice treated with CMC (Fig. 2). However, this was not observed in the colons of female mice. Moreover, PPARδ+/+/ApcMin/+ mice treated with CMC exhibit lower polyp number and size than untreated mice (Figs. 1 and 2), suggesting that CMC by itself has some inhibitory influence on tumor growth. These results demonstrate that PPARδ is critical for the tumor-promoting effects of GW501516.
Fig. 2.
PPARδ mediates the effect of the GW501516 in promoting intestinal polyp growth. Both PPARδ−/−/ApcMin/+ and PPARδ+/+/ApcMin/+ male (A and B) and female (C and D) mice at the age of 6 weeks were treated with vehicle or GW501516 for 7 weeks as described in Methods. At the end of the experimental period, the polyp numbers and sizes in small (A and C) and large (B and D) intestine were quantitated as described in Fig. 1.
Activation of PPARδ Induces VEGF Expression.
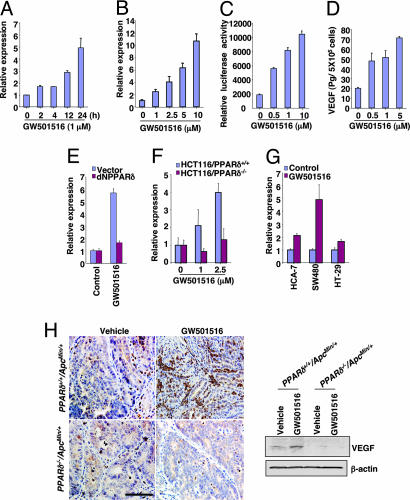
To investigate the molecular mechanism by which PPARδ activation promotes tumor growth, we initially screened the downstream target genes of PPARδ by microarray analysis. VEGF was identified as one of the potential targets for GW501516 in both human LS-174T and HCT-116 colon carcinoma cells (data not shown). To confirm the microarray results, we performed real-time quantitative PCR, VEGF promoter reporter, and ELISAs in LS-174T cells. We found that GW501516 induced VEGF mRNA levels (Fig. 3A and B), promoter activity (Fig. 3C), and protein secretion (Fig. 3D) in a dose-dependent manner. In contrast, blocking PPARδ signaling by overexpressing a dominant negative PPARδ (dNPPARδ) in LS-174T cells or deletion of PPARδ in HCT-116 cells inhibits GW501516-induced VEGF mRNA expression (Fig. 3 E and F), demonstrating that PPARδ mediates the effects of GW501516 to induce VEGF. GW501516 also induced VEGF in other colorectal cancer (CRC) cells (Fig. 3G). We next examined VEGF expression in adenomas from mice treated with GW501516. Our immunostaining (Left) and Western blot (Right) analyses show that VEGF is up-regulated after GW501516 treatment in PPARδ+/+/ApcMin/+ mice but not in PPARδ−/−/ApcMin/+ mice (Fig. 3H). Diffuse cytoplasmic staining (brown) for VEGF was observed in both epithelial and stromal cells of intestinal polyps. Taken together, these results demonstrate that PPARδ activation up-regulates VEGF transcription, expression, and release in epithelial tumor cells.
Fig. 3.
Activation of PPARδ up-regulates VEGF expression in CRC cell lines. (A and B) After serum-free starvation for 24 h, the LS-174T cells were treated with 1 μM GW501516 for the indicated times (A) or indicated concentration (B) of GW501516 for 24 h. Quantitative real-time PCR assays were performed as described in Methods. The relative expression of target gene represents an average of triplicates normalized against the transcript levels of hβ-Actin. Data are represented as the mean ± SE of the relative expression from three independent experiments. (C) The LS-174T cells were transiently transfected with VEGF luciferase reporter and pRL-SV40 plasmids, followed by treatment with GW501516 for 24 h. The dual-luciferase assays were performed as described in Methods. Data are presented as the mean ± SE of relative luciferase activity from three independent experiments. (D) LS-174T cells were treated with GW501516 as described in Methods. The levels of VEGF in cell supernatants were determined by ELISA. Three independent experiments with duplicates were performed. (E) The polyclonal dNPPARδ or empty vector LS-174T cells were treated with 1 μM GW501516 for 24 h after serum-free starving for 24 h, and quantitative real-time PCR assays were carried out as noted in Fig. 3 A and B. (F) The wild-type or PPARδ−/− HCT116 cells were treated as described in E, and quantitative real-time PCR assays were carried out. (G) Quantitative real-time PCR analysis of the mRNA level of VEGF in indicated CRC cell lines treated with 1 μM GW501516 for 24 h after serum-free starving for 24 h. (H) A representative section shows VEGF immunoreactive staining (brown) in the intestinal polyp taken from male mice treated with vehicle and GW501516 for 7 weeks. (Scale bar, 200 μm.) VEGF expression was determined by Western blot analysis (Right). Each sample included 60 polyps collected from three animals for each experimental group.
Elevated VEGF correlates well with tumor progression and poor prognosis in many human tumors, including colorectal carcinomas (29, 30). VEGF expression is regulated by a number of factors, including hypoxia, COX-2, growth factors, cytokines, oncogenes, or tumor-suppressor genes (31). In addition, there is evidence that activation of PPARα inhibits VEGF expression in CRC cells (32), whereas PPARγ activation up-regulates its expression in human vascular smooth muscle cells and macrophages (33, 34). Our in vitro and in vivo results represent evidence showing that activation of PPARδ induces VEGF expression in intestinal epithelial tumor cells. The precise mechanism by which PPARδ regulates VEGF expression warrants further investigation.
VEGF Promotes Epithelial Cell Survival by Activating Antiapoptotic Factor Akt.
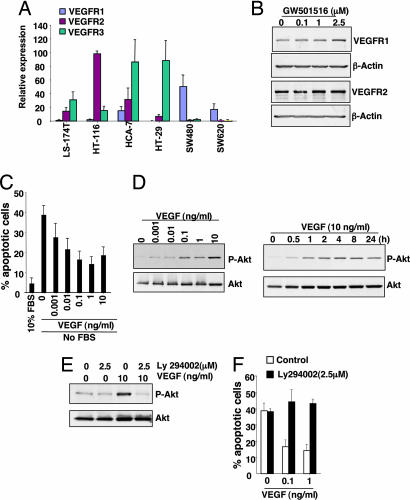
Recent evidence shows that VEGF signaling has important roles in nonendothelial cells. For example, VEGF displays prosurvival activity for neuron, chondrogenic and osteogenic cells, and hematopoietic stem cells (35). To establish the functional significance of VEGF in epithelial cancer cells, we determined whether VEGF's receptors are expressed in human CRC cell lines. Analysis of real-time quantitative PCR shows that each cell line exhibits different VEGF receptor expression profiles (Fig. 4A). We also examined whether a PPARδ ligand regulates VEGFR expression in LS-174T cells. Western blot analysis reveals that treatment of LS174T cells with GW501516 had no effect on VEGFR1–2 expression (Fig. 4B). Because epithelial tumor cells express VEGF receptors, we postulated that VEGF induces cell proliferation and promotes cell survival. LS-174T cells were treated with increasing concentrations of VEGF after serum deprivation. VEGF only slightly induces cell proliferation (data not shown). However, treatment with VEGF significantly attenuated apoptosis induced by serum deprivation (Fig. 4C). Because recent genetic evidence shows that Akt is critical for VEGF-mediated in vivo angiogenesis (36) and because VEGF can promote endothelial cell survival via the PI3K–Akt pathway (37, 38), we hypothesized that VEGF promotes epithelial cell survival through this pathway. As shown in Fig. 4D, VEGF induced Akt phosphorylation in LS-174T cells in a dose- and time-dependent manner. VEGF-induced activation of Akt is completely blocked by treatment with a specific PI3K inhibitor, LY294002 (2.5 μM) (Fig. 4E). This inhibitor also reversed the ability of VEGF to promote epithelial cell survival (Fig. 4F). Similar results were obtained in the HCT-116 cells (data not shown). These results show that VEGF promotes intestinal epithelial cancer cell survival through PI3-Akt signaling.
Fig. 4.
VEGF promotes CRC cell survival and induces Akt activation. (A) Quantitative real-time PCR analysis of VEGFR mRNA were performed as described in Fig. 3 A and B. (B) LS-174T cells were treated as described in Fig. 3D. VEGFR1–2 protein levels were analyzed by Western blotting. (C) LS-174T cells were treated with VEGF as described in Methods. The number of apoptotic cells was determined by flow cytometry using an annexin V-FITC kit. Data are expressed as the mean + SE of percent of apoptotic cells from three separate experiments. (D) LS-174T cells were treated with the indicated concentration of VEGF for 2 h (Left) or 10 ng/ml VEGF for the indicated times (Right) after serum starvation for 24 h. The level of phosphorylated Akt was detected by Western blotting using anti-phospho-Akt (Ser-473) antibody. The blots were reprobed with Akt antibody to monitor the loading of samples. (E) LS-174T cells were pretreated with the inhibitor for 1 h after serum starvation for 24 h and then incubated with 1 ng/ml VEGF for 2 h. Akt activation was measured by following the same approach as mentioned above. B, D, and E are representative of three different experiments that showed similar results. (F) LS-174T cells were pretreated with the inhibitor for 1 h and then treated with VEGF for 2 days in serum-free media. The percent of apoptotic cells was measured as noted above.
VEGF Mediates PPARδ-Induced Akt Activation and Colon Carcinoma Cell Survival.
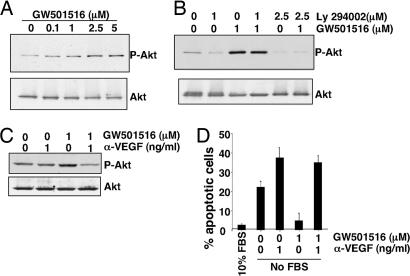
As a first step in determining whether VEGF mediates the antiapoptotic effects of PPARδ on LS-174T cells, we examined the ability of PPARδ to activate Akt. As shown in Fig. 5A, GW501516 induced Akt phosphorylation in a dose-dependent manner. The activation of Akt was completely inhibited by the PI3K inhibitor LY294002 (Fig. 5B). We next investigated whether VEGF mediates PPARδ-induced Akt activation and epithelial tumor cell survival. We found that treatment with a VEGF neutralizing antibody inhibits Akt phosphorylation induced by GW501516 (Fig. 5C) and attenuates the antiapoptotic effects of GW501516 in LS-174T cells (Fig. 5D). These results demonstrate that VEGF mediates the antiapoptotic effects of PPARδ in intestinal epithelial tumor cells by activating the PI3K–Akt cell survival pathway. Taken together, our results demonstrate that a VEGF autocrine loop plays an important role in CRC cell survival.
Fig. 5.
VEGF mediates the effects of PPARδ on Akt activation and inhibition of apoptosis. (A) LS-174T cells were treated with the indicated concentration of GW501516 for 24 h after serum starvation for 24 h. Akt activation was measured as noted above. (B) LS-174T cells were pretreated with the inhibitor for 1 h after serum starvation for 24 h and then incubated with 1 μM GW501516 for 24 h. (C) LS-174T cells were pretreated with 1 μg/ml anti-hVEGF neutralizing antibody for 1 h and then treated with GW501516 for 24 h after serum starvation for 24 h. The above figures are representative of three different experiments with similar results. (D) LS-174T cells were pretreated with 1 μg/ml anti-hVEGF neutralizing antibody for 1 h and then treated with GW501516 for 4 days under serum-free conditions. The percentage of apoptotic cells was measured as noted above.
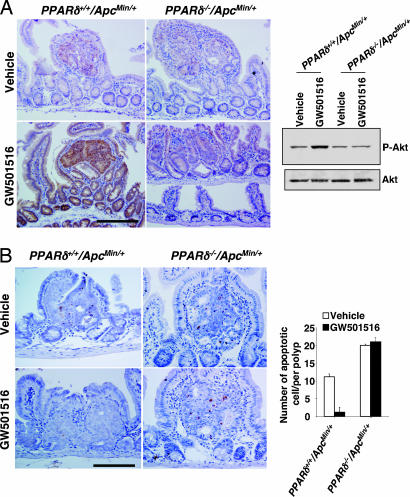
In determining the true biological significance of a novel pathway, it is always important to confirm in vitro results in an in vivo context. Thus, we evaluated the effect of GW501516 on Akt activation in vivo. As demonstrated in Fig. 6A, GW501516 treatment results in a dramatic increase in Akt phosphorylation in intestinal adenomas taken from PPARδ+/+/ApcMin/+ but not PPARδ−/−/ApcMin/+ mice by immunohistochemistry (Fig. 6A Left) and Western blot (Fig. 6A Right) analysis. To further evaluate whether GW501516 promotes tumor cell survival in vivo, TUNEL assays were performed to detect apoptotic cells within intestinal adenomas. The number of apoptotic cells is markedly reduced in polyps from PPARδ+/+/ApcMin/+ mice treated with GW501516 compared with that seen in control (vehicle-treated) mice (Fig. 6B). In contrast, GW501516 fails to affect apoptotic rates in intestinal adenomas of PPARδ−/−/ApcMin/+ mice, demonstrating that PPARδ mediates the antiapoptotic effects of GW501516 (Fig. 6B). These observations indicate that the antiapoptotic effect of PPARδ correlates well with induction of VEGF in vivo. In addition to the effect of VEGF on promoting epithelial cell survival, it is possible that activation of PPARδ accelerates tumor growth through VEGF by increasing vascular permeability.
Fig. 6.
GW501516 activates Akt in ApcMin/+ mouse polyps. (A) Phospho-Akt immunostaining was performed in sections of small intestine from both PPARδ+/+/ApcMin/+ and PPARδ−/−/ApcMin/+ male mice treated with vehicle or GW501516 for 7 weeks. A representative section shows phospho-Akt immunoreactive staining (brown) in the epithelial cells of polyps. (Scale bar, 100 μm.) Phospho-Akt in polyps was determined by Western blot analysis as described in Fig. 3H Right). (B) TUNEL staining of small intestinal adenomas from both PPARδ+/+/ApcMin/+ and PPARδ−/−/ApcMin/+ mice treated with vehicle or GW501516. A representative section shows that apoptotic nuclei are stained brown by the DeadEnd colorimetric TUNEL system as described in Methods. (Scale bar, 100 μm.) The bar graph represents mean + SE of apoptotic cells per polyp from 30 polyps taken from three mice for each experimental group (Right).
In conclusion, this study reveals that deletion of PPARδ attenuates the intestinal adenoma burden in ApcMin/+ mice and presents compelling evidence showing that VEGF, up-regulated by PPARδ in CRC cells, can act in an autocrine fashion to promote epithelial cell survival through activation of PI3K–Akt signaling. Our results not only support the rationale for developing PPARδ antagonists for use in cancer prevention and/or treatment but also establish a potential molecular basis for understanding of the epidemiologic association between obesity and the relative risk of colorectal cancer.
Methods
Animals.
PPARδ-null ApcMin/+ mice (PPARδ−/−/ApcMin/+) and the control (PPARδ+/+/ApcMin/+) were derived from same-litter mates by breeding PPARδ−/−/Apc+/+ on a mixed-genetic background (C57BL/6 × 129/SV) with PPARδ+/+/ApcMin/+ on a C57BL/6 genetic background (The Jackson Laboratory, Bar Harbor, ME) and fed with standard mouse diet in the Animal Care Facility according to National Institutes of Health and institutional guidelines. At the age of 13 weeks, these mice were killed by CO2. For GW501516 treatment experiments, PPARδ−/−/ApcMin/+ (n = 34) and PPARδ+/+/ApcMin/+ mice (n = 30) at the age of 6 weeks were randomly grouped into two groups treated with 150 μl of 0.5% CMC or 0.5% CMC solution containing GW501516 (10 mg/kg of body weight) by daily gavage feeding. After treatment for 7 weeks, mice were killed, and polyp size and number was measured as described (9). After the number of tumors was counted, intestinal tissues were embedded in paraffin. For histological analysis, sections 5 μm in thickness were stained with H&E to examine polyp morphology from all groups. The unstained sections were subjected to TUNEL assays and immunohistochemical staining.
Cell Culture and Reagents.
LS-174T, HCT-116, HCA-7, SW480, HT-29, and SW620 cells were maintained in McCoy's 5A medium with 10% FBS. PPARδ-null HCT-116 cells were a gift from K. W. Kinzler (Johns Hopkins School of Medicine, Baltimore, MD) (11). GW501516 was obtained from Ramidus AB (Lund, Sweden). Ly 294002 was obtained from Calbiochem (La Jolla, CA).
Real-Time Quantitative PCR.
VEGF, VEGFR1, VEGFR2, and VEGFR3 mRNA was quantified by real-time quantitative PCR using iCycler (Bio-Rad, Hercules, CA) and iQ SYBR green Supermix (Bio-Rad). The assay was conducted previously described (39). Primers for VEGF, VEGFR1, VEGFR2, VEGFR3, and Actin genes were chosen by using the Beacon Designer 4 program.
Transfection and Reporter Activity Assay.
VEGF promoter reporter construct was generated by subcloning a full-length VEGF promoter into pGL3 by standard techniques. The LS-174T cells (2 × 105) were transiently cotransfected with 0.4 μg of VEGF (−2,274 to +50) and 5 ng of pRL-SV40 plasmids by using LipofectAMINE Plus reagent according to the manufacturer's instructions (Life Technologies, Rockville, MA). After transfection, the cells were treated with either vehicle or GW501516 for 24 h. Luciferase activity was measured by using a Dual Luciferase kit (Promega, Madison, WI) with a Monolight 3010 luminometer (BD Biosciences/Pharmingen, San Diego, CA). The relative luciferase activity was determined by normalized to Renilla luciferase.
ELISA.
VEGF production from cell-free supernatants was measured by using a human VEGF Quantikine ELISA kit (R & D Systems, Minneapolis, MN) according to the manufacturer's instructions. Briefly, LS-174T cells (5 × 105) were cultured in serum-free medium for 16 h. Then cells were treated with vehicle or indicated concentration of GW501516 for 24 h. The supernatants were subjected to ELISA.
Immunohistochemical Staining.
Tissue sections (n = 5 per animal) were stained with a VEGF antibody (LAB Vision, Fremont, CA) and a phospho-Akt antibody (Ser-473) at a dilution of 1:250 (Cell Signaling Technology, Beverly, MA). The immunohistochemical staining was completed by using a Zymed-Histostain-SP Kit (Zymed, South San Francisco, CA) as described (9).
Western Blot Analysis.
Whole-cell extracts were prepared from cells or polyps treated with vehicle, LY294002, VEGF, or/and GW501516 at the indicated times and dose after serum starvation for 24 h. Western blots were performed as described (40). A phospho-Akt antibody (Ser 473) (Cell Signaling Technology), VEGF antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and VEGFR1–2 antibodies (LAB Vision) were used in a 1:500 dilution. The blots were stripped and then reprobed with Akt (Cell Signaling Technology) or β-Actin antibody.
Apoptosis Assays.
LS-174T cells (2.5 × 105) were incubated in serum-free media containing 10% FBS, vehicle, LY294002, or/and VEGF for 2 days. For the VEGF antibody experiments, the cells were treated with vehicle, anti-VEGF neutralizing antibody (R & D Systems, Minneapolis, MN), or/and GW501516 for 4 days. The number of apoptotic cells was determined by flow cytometry using TACS Annexin V-FITC Apoptosis Detection kit according to the manufacturer's instructions (R & D Systems).
TUNEL Assays.
The fragmented DNA of apoptotic cells in tissue sections was end-labeled by using the Dead-End colorimetric TUNEL system according to the manufacturer's instructions (Promega, Madison, WI).
Statistical Analysis.
A post hoc test (ANOVA) was used to calculate P values for experiments in Figs. 1 and 2.
Acknowledgments
This work was supported, in part, by National Institutes of Health (NIH) Grants R01DK 62112, P01-CA-77839, R37-DK47297, and P30 CA068485 (to R.N.D.) and R37-HD12304 and HD33994 (to S.K.D.) and the Mary Kay Ash Charitable Foundation (S.K.D.). R.N.D. is the B. F. Byrd Professor of Molecular Oncology. R.N.D. received NIH MERIT Award R37-DK47297. We also thank the T. J. Martell Foundation and the National Colorectal Cancer Research Alliance for generous support (to R.N.D.).
Glossary
Abbreviations
- CMC
carboxymethylcellulose
- CRC
colorectal cancer
- PPAR
peroxisome proliferator-activated receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Wang YX, Lee CH, Tiep S, Yu RT, Ham J, Kang H, Evans RM. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, Watanabe M, Magoori K, Ioka RX, Tachibana K, et al. Proc Natl Acad Sci USA. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans RM, Barish GD, Wang YX. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 4.Oliver WR, Jr, Shenk JL, Snaith MR, Russell CS, Plunket KD, Bodkin NL, Lewis MC, Winegar DA, Sznaidman ML, Lambert MH, et al. Proc Natl Acad Sci USA. 2001;98:5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta RA, Wang D, Katkuri S, Wang H, Dey SK, DuBois RN. Nat Med. 2004;10:245–247. doi: 10.1038/nm993. [DOI] [PubMed] [Google Scholar]
- 6.He TC, Chan TA, Vogelstein B, Kinzler KW. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RA, Tan J, Krause WF, Geraci MW, Willson TM, Dey SK, DuBois RN. Proc Natl Acad Sci USA. 2000;97:13275–13280. doi: 10.1073/pnas.97.24.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao J, Sheng H, DuBois RN. Cancer Res. 2002;62:3282–3288. [PubMed] [Google Scholar]
- 9.Wang D, Wang H, Shi Q, Katkuri S, Walhi W, Desvergne B, Das SK, Dey SK, DuBois RN. Cancer Cell. 2004;6:285–295. doi: 10.1016/j.ccr.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Siezen CL, Tijhuis MJ, Kram NR, van Soest EM, de Jong DJ, Fodde R, van Kranen HJ, Kampman E. Pharmacogenet Genomics. 2006;16:43–50. doi: 10.1097/01.fpc.0000182778.03180.f3. [DOI] [PubMed] [Google Scholar]
- 11.Park BH, Vogelstein B, Kinzler KW. Proc Natl Acad Sci USA. 2001;98:2598–2603. doi: 10.1073/pnas.051630998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM. Proc Natl Acad Sci USA. 2002;99:303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harman FS, Nicol CJ, Marin HE, Ward JM, Gonzalez FJ, Peters JM. Nat Med. 2004;10:481–483. doi: 10.1038/nm1026. [DOI] [PubMed] [Google Scholar]
- 14.Reed KR, Sansom OJ, Hayes AJ, Gescher AJ, Winton DJ, Peters JM, Clarke AR. Oncogene. 2004;23:8992–8996. doi: 10.1038/sj.onc.1208143. [DOI] [PubMed] [Google Scholar]
- 15.Stephen RL, Gustafsson MC, Jarvis M, Tatoud R, Marshall BR, Knight D, Ehrenborg E, Harris AL, Wolf CR, Palmer CN. Cancer Res. 2004;64:3162–3170. doi: 10.1158/0008-5472.can-03-2760. [DOI] [PubMed] [Google Scholar]
- 16.Glinghammar B, Skogsberg J, Hamsten A, Ehrenborg E. Biochem Biophys Res Commun. 2003;308:361–368. doi: 10.1016/s0006-291x(03)01384-6. [DOI] [PubMed] [Google Scholar]
- 17.Hao CM, Redha R, Morrow J, Breyer MD. J Biol Chem. 2002;277:21341–21345. doi: 10.1074/jbc.M200695200. [DOI] [PubMed] [Google Scholar]
- 18.Di-Poi N, Tan NS, Michalik L, Wahli W, Desvergne B. Mol Cell. 2002;10:721–733. doi: 10.1016/s1097-2765(02)00646-9. [DOI] [PubMed] [Google Scholar]
- 19.Di-Poi N, Michalik L, Tan NS, Desvergne B, Wahli W. J Steroid Biochem Mol Biol. 2003;85:257–265. doi: 10.1016/s0960-0760(03)00215-2. [DOI] [PubMed] [Google Scholar]
- 20.Alon T, Hemo I, Itin A, Pe'er J, Stone J, Keshet E. Nat Med. 1995;1:1024–1028. doi: 10.1038/nm1095-1024. [DOI] [PubMed] [Google Scholar]
- 21.Dias S, Hattori K, Heissig B, Zhu Z, Wu Y, Witte L, Hicklin DJ, Tateno M, Bohlen P, Moore MA, Rafii S. Proc Natl Acad Sci USA. 2001;98:10857–10862. doi: 10.1073/pnas.191117498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellamy WT, Richter L, Frutiger Y, Grogan TM. Cancer Res. 1999;59:728–733. [PubMed] [Google Scholar]
- 23.Decaussin M, Sartelet H, Robert C, Moro D, Claraz C, Brambilla C, Brambilla E. J Pathol. 1999;188:369–377. doi: 10.1002/(SICI)1096-9896(199908)188:4<369::AID-PATH381>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 24.Ferrer FA, Miller LJ, Lindquist R, Kowalczyk P, Laudone VP, Albertsen PC, Kreutzer DL. Urology. 1999;54:567–572. doi: 10.1016/s0090-4295(99)00156-9. [DOI] [PubMed] [Google Scholar]
- 25.Lacal PM, Failla CM, Pagani E, Odorisio T, Schietroma C, Falcinelli S, Zambruno G, D'Atri S. J Invest Dermatol. 2000;115:1000–1007. doi: 10.1046/j.1523-1747.2000.00199.x. [DOI] [PubMed] [Google Scholar]
- 26.Price DJ, Miralem T, Jiang S, Steinberg R, Avraham H. Cell Growth Differ. 2001;12:129–135. [PubMed] [Google Scholar]
- 27.Nadra K, Anghel SI, Joye E, Tan NS, Basu-Modak S, Trono D, Wahli W, Desvergne B. Mol Cell Biol. 2006;26:3266–3281. doi: 10.1128/MCB.26.8.3266-3281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters JM, Lee SS, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ. Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JC, Chow NH, Wang ST, Huang SM. Eur J Cancer. 2000;36:748–753. doi: 10.1016/s0959-8049(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi Y, Kitadai Y, Bucana CD, Cleary KR, Ellis LM. Cancer Res. 1995;55:3964–3968. [PubMed] [Google Scholar]
- 31.Hicklin DJ, Ellis LM. J Clin Oncol. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 32.Grau R, Punzon C, Fresno M, Iniguez MA. Biochem J. 2006;395:81–88. doi: 10.1042/BJ20050964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamakawa K, Hosoi M, Koyama H, Tanaka S, Fukumoto S, Morii H, Nishizawa Y. Biochem Biophys Res Commun. 2000;271:571–574. doi: 10.1006/bbrc.2000.2665. [DOI] [PubMed] [Google Scholar]
- 34.Jozkowicz A, Dulak J, Piatkowska E, Placha W, Dembinska-Kiec A. Acta Biochim Pol. 2000;47:1147–1157. [PubMed] [Google Scholar]
- 35.Coultas L, Chawengsaksophak K, Rossant J. Nature. 2005;438:937–945. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- 36.Ackah E, Yu J, Zoellner S, Iwakiri Y, Skurk C, Shibata R, Ouchi N, Easton RM, Galasso G, Birnbaum MJ, et al. J Clin Invest. 2005;115:2119–2127. doi: 10.1172/JCI24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerber HP, McMurtrey A, Kowalski J, Yan M, Keyt BA, Dixit V, Ferrara N. J Biol Chem. 1998;273:30336–30343. doi: 10.1074/jbc.273.46.30336. [DOI] [PubMed] [Google Scholar]
- 38.Fujio Y, Walsh K. J Biol Chem. 1999;274:16349–16354. doi: 10.1074/jbc.274.23.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Wang H, Brown J, Daikoku T, Ning W, Shi Q, Richmond A, Strieter R, Dey SK, Dubois RN. J Exp Med. 2006;203:941–951. doi: 10.1084/jem.20052124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Buchanan FG, Wang H, Dey SK, DuBois RN. Cancer Res. 2005;65:1822–1829. doi: 10.1158/0008-5472.CAN-04-3671. [DOI] [PubMed] [Google Scholar]