Abstract
The human CMV UL37x1-encoded protein, also known as the viral mitochondria-localized inhibitor of apoptosis, traffics to the endoplasmic reticulum and mitochondria of infected cells. It induces the fragmentation of mitochondria and blocks apoptosis. We demonstrate that UL37x1 protein mobilizes Ca2+ from the endoplasmic reticulum into the cytosol. This release is accompanied by cell rounding, cell swelling, and reorganization of the actin cytoskeleton, and these morphological changes can be substantially blocked by a Ca2+ chelating agent. The UL37x1-mediated release of Ca2+ from the endoplasmic reticulum likely has multiple consequences, including induction of the unfolded protein response, modulation of mitochondrial function, induction of mitochondrial fission, and protection against apoptotic stimuli.
Keywords: actin cytoskeleton, apoptosis, cytopathic effect, mitochondrial fission, viral mitochondria-localized inhibitor of apoptosis
Human CMV (HCMV) is a widespread β-herpesvirus that takes its name from the striking morphological changes that it induces in infected cells. As infection progresses, cells become round, increase in volume, and accumulate distinctive inclusion bodies in the nucleus and cytoplasm. HCMV infection is generally asymptomatic in healthy individuals, but the virus is a life-threatening adventitious pathogen in immunocompromised individuals and a serious problem during pregnancy, where it can cause birth defects (1).
Upon infection of a permissive cell, HCMV expresses its genes in a regulated cascade, and immediate-early genes are expressed first. Their products induce subsequent waves of viral gene expression and prepare the cell for infection in a variety of ways, such as modulating cell cycle progression, blocking apoptosis, and interfering with the presentation of viral antigens to the immune system. The UL37 transcription unit is first expressed during the immediate-early phase (2, 3). Its primary transcript is processed to generate at least 10 spliced mRNAs as well as one unspliced mRNA (4), and three encoded proteins have been identified. The unspliced RNA ends at a poly(A) site located part way through the transcription unit, and, as a consequence, it comprises the first of three coding exons in the unit. Its encoded protein, pUL37x1, contains an N-terminal, hydrophobic signal peptide that is not cleaved from the mature protein (5). It traffics from endoplasmic reticulum (ER) to mitochondria, accumulating in both locations (6–8).
UL37x1 is highly conserved among HCMV strains (9, 10), and mutations within the ORF interfere with efficient replication of AD169 strains (11, 12) but not the TownevarATCC strain of HCMV (13). The protein blocks apoptosis and is often referred to as the viral mitochondria-localized inhibitor of apoptosis (14, 15). Although pUL37x1 shares no sequence homology with antiapoptotic Bcl-2 family members, it acts similarly by sequestering Bax and Bak and preventing release of cytochrome c from the mitochondria (7, 16–18). Interestingly, pUL37x1 disrupts the mitochondrial network, suggesting that it modifies the balance of fission/fusion that normally controls mitochondrial structure (18–20).
We demonstrate that pUL37x1 induces the release of Ca2+ from the ER into the cytosol. This release is accompanied by cell rounding, swelling, and disruption of the actin cytoskeletal network, and these morphological changes can be substantially blocked by a calcium chelating agent.
Results
A pUL37x1-Deficient Mutant Fails to Induce Normal HCMV Cytopathic Effect.
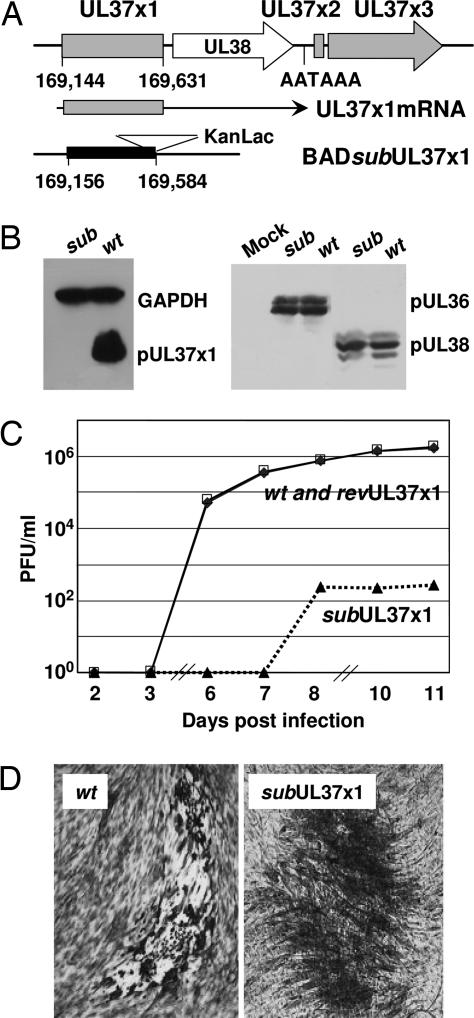
An HCMV mutant, BADsubUL37x1, lacking most of the UL37x1 coding region, and a revertant, BADrevUL37x1, were generated in the AD169 strain of HCMV (Fig. 1A). As expected, the mutant virus failed to accumulate pUL37x1 (Fig. 1B Left), and, because the sequence encoded by the deleted exon is present in the other known UL37 proteins, the mutation disrupts their synthesis as well. The mutation did not interfere with production of the nearby UL36- and UL38-coded proteins (Fig. 1B Right).
Fig. 1.
Characterization of a UL37x1-deficient HCMV. (A) Diagram of the genomic region encoding pUL37x1. The top line represents viral DNA with the location and relative orientation of UL37 coding exons (shaded) and the UL38 ORF represented by boxes and arrows. The nucleotide positions of the first and last base pairs encoding UL37x1 are indicated, as is the polyadenylation motif located within the UL37 unit. The middle line shows the structure of the mRNA encoding pUL37x1, and the bottom line diagrams the substitution mutation in BADsubUL37x1 with a solid rectangle marking the deletion and a triangle depicting the KanLac marker insert. The coordinates of the first and last base pairs deleted are indicated. (B) Western blot assays at 24 hpi with BADsubUL37x1 (sub) or BADwt (wt). (Left) Antibodies to pUL37x1 and GAPDH (loading control). (Right) Antibodies to pUL36 and pUL38. (C) Growth kinetics of BADwt (♦), BADsubUL37x1 (▴), and BADrevUL37x1 (|B%). Fibroblasts were infected at a multiplicity of 0.1 pfu per cell, and virus yields were determined by plaque assay. (D) Plaques produced by BADwt and BADsubUL37x1 after staining with methylene blue.
The mutant virus exhibited a severe growth defect in human fibroblasts (Fig. 1C), generating a yield that was reduced by a factor of >1,000, and the revertant grew like wild-type virus, arguing that the mutant phenotype was a consequence of the intended alteration and not a spurious mutation at another location. When virus yields were determined by plaque assay, it was evident that the pUL37x1-deficient mutant failed to induce the normal HCMV cytopathic effect (Fig. 1D). Whereas BADwt-infected cells became rounded, BADsubUL37x1-infected fibroblasts maintained their normal spindle shape.
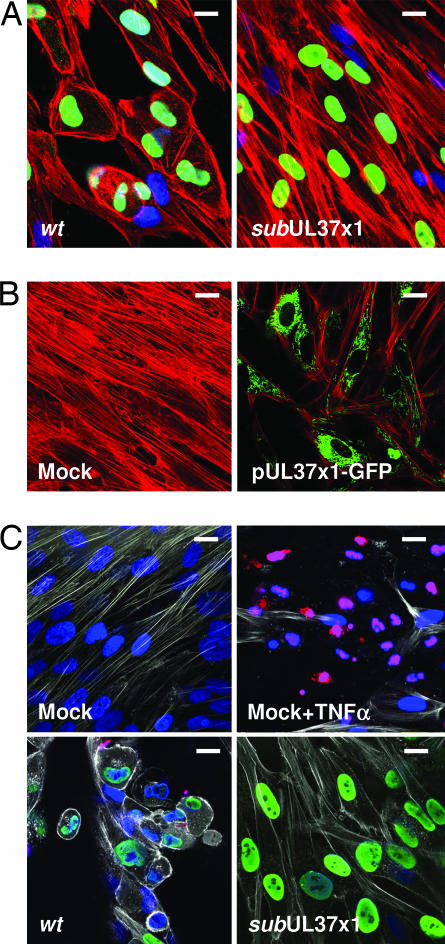
To investigate the basis for the altered cell shape, we examined the structure of the actin cytoskeleton by immunofluorescence. As expected for the wild-type virus, the cytoskeleton's organization was markedly disrupted by 24 h postinfection (hpi) with loss of filamentous actin (F-actin) and accumulation of cortical actin at the plasma membrane (Fig. 2A). In contrast, the F-actin network was maintained after infection with the mutant (Fig. 2A), and remained indistinguishable from that in mock-infected cells (data not shown). The actin reorganization is mediated entirely by pUL37x1, because cells receiving a plasmid expressing a pUL37x1-GFP fusion protein exhibited similar alterations as were seen after infection with BADwt (Fig. 2B). Because some pUL37x1-deficient mutants have been reported to induce apoptosis after infection of fibroblasts, we performed a TUNEL assay to ask whether BADsubUL37x1-infected cells undergo apoptosis (Fig. 2C). Although the assay readily detected cell death after treatment with TNFα, no apoptosis was evident in cultures infected with wild-type or mutant viruses. It is not clear why this mutant does not induce apoptosis, because it was constructed in an AD169 strain that lacks functional pUL36 (10), which has been proposed to mask pUL37x1 defects (13). Nevertheless, we can conclude that the change in F-actin occurs in response to pUL37x1 without the induction of apoptosis.
Fig. 2.
F-actin is reorganized by pUL37x1. Fibroblasts were infected at a multiplicity of ≈1 pfu per cell. (A) Immunofluorescence at 24 hpi with BADwt or BADsubUL37x1 using Alexa Fluor 546 phalloidin to detect actin (red), antibody to HCMV IE1 protein (green), and DAPI to stain DNA (blue). (B) Immunofluorescence at 24 h after mock treatment or electroporation of fibroblasts with a plasmid expressing pUL37x1-GFP using Alexa Fluor 546 phalloidin to detect actin (red) and monitoring GFP fluorescence (green). (C) TUNEL assay for apoptosis after mock infection, after treatment of mock-infected cells with TNFα, or 3 days after infection at a multiplicity of 3 pfu per cell with wt or subUL37x1. Actin was detected with Alexa Fluor 633 phalloidin (white), and IE1 protein (green) was detected by using an antibody. TUNEL-positive cells and cell fragments are red, and DNA was stained with DAPI (blue). (Scale bars: ≈10 μm.)
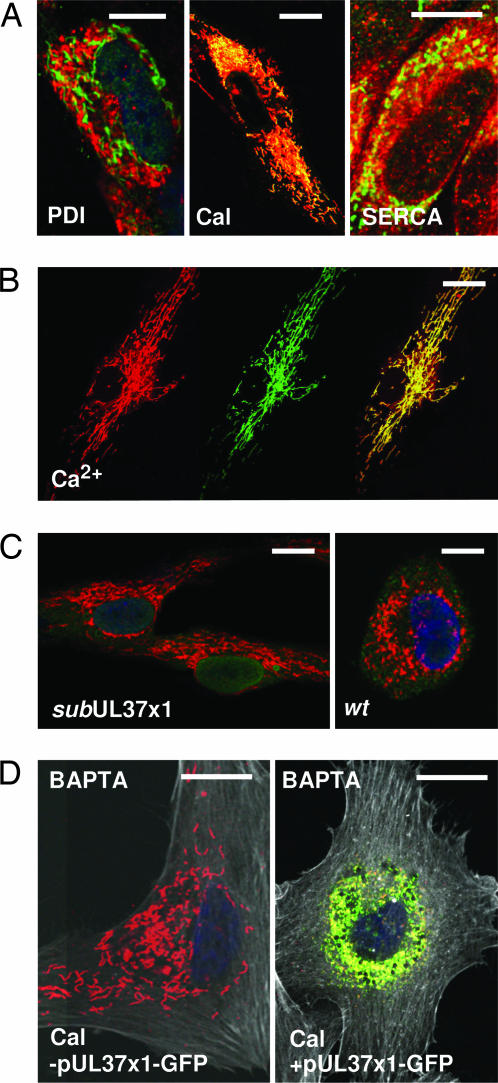
Because the mitochondrial location of UL37-coded proteins has been implicated in their antiapoptotic function (6–8), we explored the possibility that the portion of the UL37 proteins located in the ER might influence F-actin structure. Accordingly, we tested for colocalization of pUL37x1 with several ER proteins. The viral protein did not colocalize by immunofluorescence with protein disulfide isomerase, which resides in the ER lumen; but it did partially colocalize with calnexin and the sarcoendoplasmic reticulum Ca2+-ATPase (SERCA), two ER membrane proteins (Fig. 3A). We also found that pUL37x1-GFP colocalized with Ca2+ stores in the ER of fibroblasts receiving an expression plasmid (Fig. 3B). In some cells, the ER remained filamentous at 24 h after electroporation (Fig. 3B) and in other cells the ER became compacted near the nucleus (data not shown). The ER was generally collapsed near the nucleus at 24 hpi with BADwt, but not after infection with pUL37x1-deficient virus (Fig. 3C).
Fig. 3.
pUL37x1 associates with ER membrane constituents. Fibroblast infections were at a multiplicity of ≈1 pfu per cell. (A) Immunofluorescent assays at 24 hpi with BADwt for colocalization of pUL37x1 (green) with protein disulfide isomerase, SERCA, or calnexin (red). Yellow indicates overlapping signals. (B) Colocalization of Ca2+ detected with calcium orange (Left, red) and pUL37x1 (Center, green) at 24 h after electroporation with a pUL37x1-GFP plasma. (Right) The merged images. Yellow indicates overlapping signals. (C) Immunofluorescent image of ER calnexin (red) at 24 hpi with mutant or wild-type virus. DNA is stained with DAPI (blue). (D) Immunofluorescent image of ER calnexin (red) in cells treated with BAPTA (10 μM) for 1 h. Actin was visualized (white) by using Alexa Fluor 633 phalloidin. Cells were mock-treated (Left) or electroporated with an expression plasmid (Right). Overlapping calnexin and pUL37x1 signals are yellow. (Scale bars: ≈10 μm.)
Given its association with the ER membrane and Ca2+ stores, we suspected that pUL37x1 might induce the release of Ca2+ from the ER and this, in turn, might induce the actin reorganization. To test this idea, we treated fibroblasts with 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid-acetoxymethyl ester (BAPTA) (10 μM), which enters cells and chelates Ca2+. BAPTA prevented the alteration in F-actin in response to pUL37x1-GFP (Fig. 3D, compare -pUL37x1 and +pUL37x1 panels, F-actin is white), while allowing the ER to collapse in response to the viral protein. This experiment argues that Ca2+ is directly responsible for the pUL37x1-mediated change in F-actin, and it shows that the structure of the ER can be modified by pUL37x1 without a visible change in F-actin, provided that Ca2+ is chelated.
Ca2+ Is Released from the ER in Response to pUL37x1.
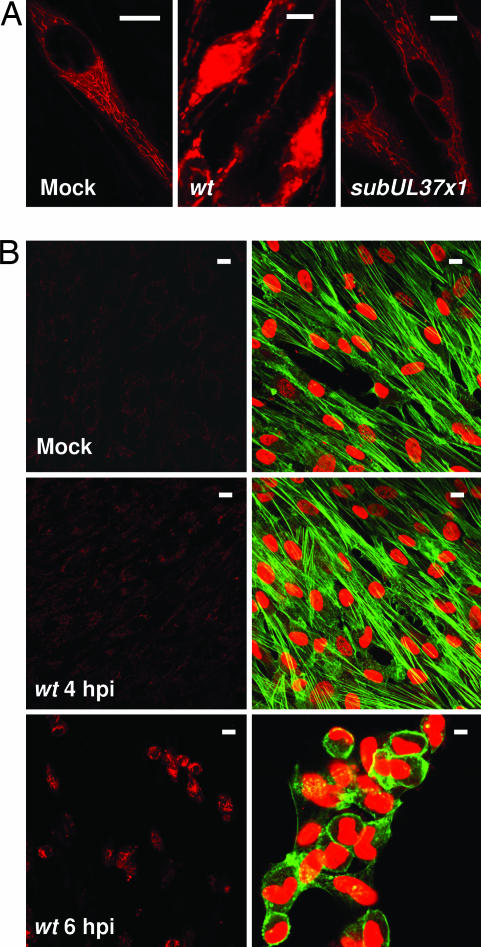
To directly assay for release of Ca2+ into the cytosol, we used calcium orange, a fluorescent indicator that binds Ca2+ in living cells (Fig. 4A). In mock-infected fibroblasts or at 24 hpi with BADsubUL37x1, Ca2+ was detected in filamentous structures indicative of an ER location. In contrast, BADwt-infected cells exhibited a much more intense signal that was no longer restricted to the filamentous pattern, suggesting that Ca2+ was present in the cytosol. The intense signal could be seen at 6 hpi at a multiplicity of ≈3 pfu per cell, but not at 4 hpi, and the change in actin structure correlated with detection of the intense Ca2+ signal (Fig. 4B). The release of Ca2+ and actin reorganization were delayed for ≈1.5 h when the infection was performed at a multiplicity of ≈0.5 pfu per cell (data not shown).
Fig. 4.
Altered Ca2+ localization correlates with F-actin reorganization. Infections were at a multiplicity of ≈1 pfu per cell, and living fibroblasts were imaged immediately after adding calcium orange (red signal). (A) Assays were performed at 24 h after mock, BADwt, or BADsubUL37x1 infection. (B) Calcium orange assays (Left) were performed at 4 h after mock infection or 4 or 6 h after BADwt infection. (Right) In a duplicate sample, DNA (red) and actin (green) were visualized. (Scale bars: ≈10 μm.)
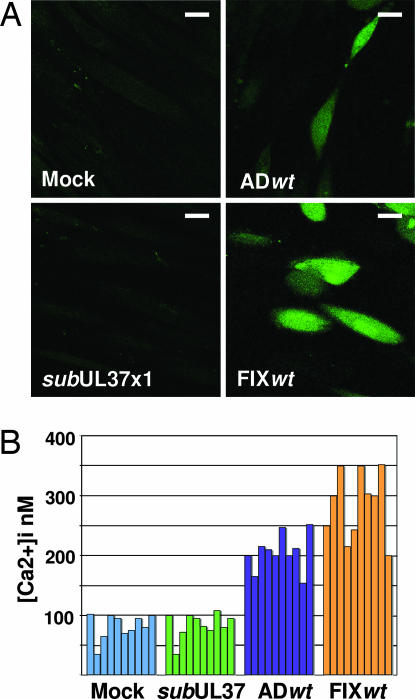
To confirm that Ca2+ was released to the cytosol, fibroblasts were loaded with Fluo-3, a high-affinity Ca2+ indicator that binds free Ca2+ in the cytoplasm, but not Ca2+ bound by storage proteins in the ER. The fluorescent signal was considerably more intense in cells receiving wild-type virus than after mock or BADsubUL37x1 infection (Fig. 5A). Fluorescent signals were measured in multiple individual cells and the concentration of free intracellular Ca2+, [Ca2+]i, was calculated (Fig. 5B). Mock- and BADsubUL37x1-infected cells contained similar amounts of [Ca2+]i, ranging from 30–100 nM. In contrast, BADwt-infected fibroblasts contained 150–250 nM [Ca2+]i. Because BADwt is a laboratory strain of HCMV, we also assayed FIXwt, a clinical isolate of the virus, and it induced [Ca2+]i to 200–350 nM, confirming that the release of Ca2+ to the cytosol is a general consequence of infection by different HCMV strains.
Fig. 5.
pUL37x1 increases [Ca2+]i. (A) Fibroblasts were mock-infected or infected with BADwt, FIXwt, or BADsubUL37x1 at a multiplicity of ≈1 pfu per cell. Twelve hours later, cells were loaded with Fluo-3 and fluorescence was imaged. (Scale bars: ≈10 μm.) (B) [Ca2+]i was estimated in 10 individual cells.
Discussion
It has long been known that HCMV infection rapidly disrupts the actin cytoskeleton (21) and induces an increase in free [Ca2+]i (22), and now we can assign these changes to the UL37x1 coding region. pUL37x1 traffics to the ER (6, 8), colocalizes with proteins and Ca2+ stores at the ER membrane (Fig. 3 A and B), and induces the release of Ca2+ to the cytosol (Figs. 4 and 5). The release is detected between 4 and 6 hpi (Fig. 4B), distinguishing it from the transient Ca2+ flux that occurs when many viruses, including HCMV and herpes simplex virus, first interact with the cell (23, 24). Treatment of infected cells with thapsigargin or clotrimazole, which induce the release of Ca2+ from the ER, induces a profound block to viral replication (25). Because pUL37x1 normally mobilizes ER Ca2+ during the HCMV replication cycle, it must do so in a regulated fashion that does not simply mimic the action of drugs that disrupt calcium homeostasis in the ER.
pUL37x1 is necessary (Fig. 2A) and sufficient (Fig. 2B) to induce the early aspects of HCMV cytopathology, i.e., cell rounding and swelling with loss of F-actin and accumulation of cortical actin. This observation is consistent with a recent report (26) that an independently derived pUL37x1-deficient mutant also fails to induce normal HCMV cytopathic effect. The disruption of F-actin results directly from the release of Ca2+ into the cytosol, because BAPTA, which chelates Ca2+, can block cell rounding and actin reorganization (Fig. 3D). The mechanism by which Ca2+ release mediates actin reorganization in infected cells is not known. Ca2+-dependent actin-severing proteins, such as gelosin (27), might be activated and contribute to the process. It also is conceivable that the Rho family of GTPases, which commonly control actin rearrangements (28), cooperate with Ca2+ to disrupt F-actin.
BAPTA treatment also revealed that the ER can collapse into a perinuclear position, even when cells do not round and actin is not reorganized (Fig. 3D). However, in the absence of BAPTA, the ER was often observed in its normal configuration for a short time after the release of Ca2+ and F-actin loss (data not shown). Consequently, it appears that the ER often collapses after F-actin reorganization, even though the collapse can occur if F-actin does not undergo a visible change.
When cells undergo hypotonic swelling, stretch-activated calcium channels trigger responses that restore osmotic homeostasis and cell shape (29, 30). Actin reorganization accompanies an elevation in [Ca2+]i in both HCMV-infected cells and cells undergoing hypoosmotic stimulation (29), but the control of cell volume appears to be different in the two cases. Normally, elevated intracellular calcium activates Ca2+-dependent K+ and Cl− channels that reduce cell volume (31). In HCMV-infected cells, however, the pUL37x1-dependent increase in [Ca2+]i triggers cell swelling. It is intriguing that cells expressing pUL37x1-GFP did not increase significantly in volume (data not shown), even though Ca2+ was released from the ER and F-actin was rearranged. This observation implies that, although pUL37x1 is required for changing the shape and volume of infected cells, one or more additional viral proteins might short circuit activation of the Ca2+-activated ion channels that normally maintain proper cell volume.
In addition to impacting signaling processes that regulate cell shape and volume, the release of Ca2+ from the ER would potentially reduce the activity of Ca2+-dependent ER chaperones, causing the accumulation of unfolded proteins in the ER and inducing the unfolded protein response (32). HCMV activates this response (33), and it is likely that pUL37x1 is responsible at least in part for the induction. Even though the unfolded protein response is activated, many of its consequences are blocked by HCMV (33), probably by the action of additional viral gene products.
The release of Ca2+ from the ER generates local high concentrations of the ion at mitochondria (34), and mitochondrial Ca2+ uptake then results from an electrochemical potential gradient (35). Ca2+ uptake modulates energy metabolism within mitochondria. Key mitochondrial processes are mediated by Ca2+-dependent metabolite carriers (36) and enzymes (37, 38), and, consequently, Ca2+ uptake has the potential to stimulate aerobic metabolism (39) and enhance ATP production (40). Given the large demands of viral replication for energy, one can rationalize why HCMV would modulate the availability of Ca2+ to mitochondria. A failure to generate sufficient energy resources could in part account for the markedly reduced yield of the pUL37x1-deficient mutant (Fig. 1C). However, the effect on mitochondrial function is remains uncertain. Constitutive pUL37x1 expression in HeLa or 3T3 cells reduced the uptake of phosphate into isolated mitochondria or mitochondria in permeabilized cells and caused a reduction in the level of ATP (26). Perhaps regulated pUL37x1 expression or the presence of additional viral gene products will prove to distinguish virus-infected cells from cells expressing the protein outside the context of infection.
It has long been established that pUL37x1 blocks apoptosis (14, 15). Does its ability to release Ca2+ stores from the ER relate to its antiapoptotic activity? Ca2+ plays a central role in apoptosis (41). Elevated Ca2+ induces opening of the mitochondrial permeability transition pore complex (42), causing release of cytochrome c from the mitochondria and activation of caspase 9 in the cytosol in cells exposed to apoptotic stimuli (43). However, the Ca2+ content of the ER determines the cell's sensitivity to apoptotic stimuli. Ablation of calreticulin or overexpression of plasma membrane ATPases decreases ER Ca2+ stores and protects against apoptosis (44, 45). In a similar vein, overexpression of Bcl2 stimulates the exit of Ca2+ from the ER and inhibits apoptosis (46, 47), and sensitivity to apoptotic stimuli can be restored by overexpression of SERCA, which pumps Ca2+ into mitochondria (45). Furthermore, cells from double knockout mice deficient in the proapoptotic Bax and Bak proteins accumulate reduced Ca2+ levels in their ER, and exhibit reduced sensitivity to a range of apoptotic stimuli (48). Consequently, the release of Ca2+ from the ER by pUL37x1, could be expected to protect cells from apoptosis, and we propose that this is at least an element of the viral protein's protective action.
Fission of the mitochondrial network is induced by pUL37x1 (19, 20, 26). Mitochondrial morphology is normally in flux with continual fusion and fission of the population, and the balance between fragmented and elongated organelles can influence mitochondrial function (49). The release of Ca2+ from the ER can induce fission of mitochondria (50). More recent work (51) has led to the proposal that ER Ca2+ release coupled to mitochondrial uptake leads to relocalization of Drp1 from the cytosol to mitochondria, where it functions as a component of the machinery that induces fission. This model supports the idea that pUL37x1-mediated release of Ca2+ from the ER induces mitochondrial fission.
Excess mitochondrial fission is often associated with early stages of apoptosis and inhibition of fission can retard apoptosis (52), but pUL37x1-mediated fission does not induce apoptosis (19, 20). Importantly, fission is not required for apoptosis (53). How then might pUL37x1 induce fission while protecting against apoptosis? Szabadkai et al. (54) overexpressed Drp1, a component of the machinery that normally sponsors fission, to fragment the mitochondria of HeLa cells. They found that, although the treatment made cells more susceptible to staurosporine-induced apoptosis, they were relatively resistant to several agents, including ceramide, that trigger mitochondrial outer membrane permeabilization and subsequent apoptosis through the induction of intra-mitochondrial “Ca2+ waves.” This experiment was interpreted to indicate that fragmentation reduced the luminal connectivity of the mitochondrial network, breaking the wave of Ca2+ uptake required for optimal proapoptotic signaling. This is consistent with a study showing that Ca2+ entering mitochondria can spread in an explosive wave within a tubular network, whereas propagation was uncoordinated after fragmentation of the network (55). It is possible, then, that pUL37x1-induced mitochondrial fission contributes to its antiapoptotic activity.
Materials and Methods
Viruses, Cells, and Plasmids.
BADwt is derived from a BAC clone of the HCMV AD169 strain pAD/Cre (56). A pUL37x1-deficient derivative of BADwt, BADsubUL37x1, was generated from pAD/Cre by allelic exchange (56). It lacks the AD169 sequence from 169,144 through 169,631 (10) and includes kanamycin resistance and LacZ markers. A revertant, BADrevUL37x1, was prepared from the BAC clone of BADsubUL37x1. The gross structures of BAC DNAs were monitored by restriction enzyme analysis, and the boundaries of alterations were confirmed by sequence analysis. FIXwt is derived from a BAC clone (57) of the HCMV VR1814 clinical isolate (58). Virus was propagated and studied in primary human foreskin fibroblasts maintained in medium with 10% FBS. Infectious virus was quantified by plaque assay on fibroblasts. A derivative of pEGFP-N1 expressing pUL37x1-GFP was a gift of A. Watson (Children's National Medical Center).
Analysis of Proteins.
Western blots used murine monoclonal antibodies to pUL36 (59), pUL38 (M. C. Silva and T.S., unpublished observations), and pUL37x1. A pUL37x1-specific hybridoma (4B6-B) was produced by immunization of mice with a fusion protein containing amino acids 27–40 of pUL37x1.
Indirect immunofluorescence used murine monoclonal antibodies to pUL37x1, IE1 (60), protein disulfide isomerase (Nventa, San Diego, CA), SERCA (Affinity BioReagents, Golden, CO), and calnexin (Abcam, Cambridge, MA), together with Alexa Fluor 568-conjugated anti-mouse secondary antibody (Invitrogen, Carlsbad, CA). Actin was detected with Alexa Fluor phalloidin (Invitrogen). Nuclear DNA was stained with DAPI (Invitrogen). Fluorescent images were captured with a Zeiss LSM510 confocal microscope.
Assay for Apoptosis.
Apoptosis was assayed by using TUNEL (Roche, Indianapolis, IN). DNA was stained with DAPI.
Measurement of Free [Ca2+]i.
The location of total Ca2+ (bound plus free) in fibroblasts was assayed by fluorescence by using calcium orange (4 μM), and free intracellular Ca2+ was localized by fluorescence by using Fluo-3 (4 μM) (Invitrogen).
The concentration of free intracellular Ca2+, [Ca2+]i, was determined according to the method of Grynkiewicz et al. (61). Briefly, confluent fibroblasts were mock-infected or infected at a multiplicity of ≈1 pfu per cell. After 12 h, cells were washed with prewarmed HBSS containing 25 mM Hepes (pH 7.45), 2.5 mM probenecid, and 0.5% BSA (HBSS+) and loaded with Fluo-3 (4 μM) in HBSS+ for 30 min at 37°C. Nine to 20 optical slices per cell were collected with a 0.75-μm Z step interval, by using a PerkinElmer RS3 Spinning Disk mated to a Nikon TE200S microscope with a Plan Neofluor ×40, 1.3 N.A. objective. Optical slices were projected along the z axis, and fluorescent intensity (F) was plotted versus area. Next, to measure the minimal fluorescent intensity (Fmin) after Ca2+ depletion, ionomycin (10 μM), thapsigargin (1 μM), and EGTA (2 mM) were added to the cultures to release intracellular Ca2+ stores and chelate free Ca2+, and fluorescent intensity was determined as described above. Finally, to measure the maximal Ca2+ fluorescent intensity (Fmax), the cells received Ca2+ (20 mM) in HBSS+, and fluorescent intensity was again determined. Fluorescent intensities were converted to an estimate of [Ca2+]i by using the following formula: [Ca2+]i = KdFluo-3[(F − Fmin)/(Fmax − F)], where KdFluo-3 = 390 nM.
Acknowledgments
We thank M. Steinberg (Princeton University) and D. Spector (Pennsylvania State University College of Medicine, Hershey, PA) for helpful insights, R. A. Rebres (University of California, San Francisco, CA) for advice on measurement of intracellular calcium, and A. Watson (Children's National Medical Center) for the gift of a pUL37x1-GFP expression plasmid. This work was supported by National Institutes of Health Grant CA-085786 (to T.S.) and a supplement to that grant to promote reentry into biomedical research careers (to R.S.-F.). A.M.C.-P. was supported in part by National Institutes of Health Grant AI-057906.
Abbreviations
- ER
endoplasmic reticulum
- SERCA
sarcoendoplasmic reticulum Ca2+-ATPase
- BAPTA
1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid-acetoxymethyl ester
- hpi
hours postinfection
- HCMV
human CMV.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gerna G, Baldanti F, Revello MG. Hum Immunol. 2004;65:381–386. doi: 10.1016/j.humimm.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T, Bankier AT, Satchwell SC, Preddy E, Barrell BG. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- 3.Tenney DJ, Colberg-Poley AM. J Virol. 1991;65:6724–6734. doi: 10.1128/jvi.65.12.6724-6734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adair R, Liebisch GW, Colberg-Poley AM. J Gen Virol. 2003;84:3353–3358. doi: 10.1099/vir.0.19404-0. [DOI] [PubMed] [Google Scholar]
- 5.Mavinakere MS, Williamson CD, Goldmacher VS, Colberg-Poley AM. J Virol. 2006;80:6771–6783. doi: 10.1128/JVI.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Barazi HO, Colberg-Poley AM. J Virol. 1996;70:7198–7208. doi: 10.1128/jvi.70.10.7198-7208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldmacher VS, Bartle LM, Skaletskaya A, Dionne CA, Kedersha NL, Vater CA, Han JW, Lutz RJ, Watanabe S, Cahir McFarland ED, et al. Proc Natl Acad Sci USA. 1999;96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colberg-Poley AM, Patel MB, Erezo DP, Slater JE. J Gen Virol. 2000;81:1779–1789. doi: 10.1099/0022-1317-81-7-1779. [DOI] [PubMed] [Google Scholar]
- 9.Hayajneh WA, Colberg-Poley AM, Skaletskaya A, Bartle LM, Lesperance MM, Contopoulos-Ioannidis DG, Kedersha NL, Goldmacher VS. Virology. 2001;279:233–240. doi: 10.1006/viro.2000.0726. [DOI] [PubMed] [Google Scholar]
- 10.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. Proc Natl Acad Sci USA. 2003;100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brune W, Nevels M, Shenk T. J Virol. 2003;77:11633–11643. doi: 10.1128/JVI.77.21.11633-11643.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reboredo M, Greaves RF, Hahn G. J Gen Virol. 2004;85:3555–3567. doi: 10.1099/vir.0.80379-0. [DOI] [PubMed] [Google Scholar]
- 13.McCormick AL, Meiering CD, Smith GB, Mocarski ES. J Virol. 2005;79:12205–12217. doi: 10.1128/JVI.79.19.12205-12217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldmacher VS. Apoptosis. 2005;10:251–265. doi: 10.1007/s10495-005-0800-z. [DOI] [PubMed] [Google Scholar]
- 15.Andoniou CE, Degli-Esposti MA. Immunol Cell Biol. 2006;84:99–106. doi: 10.1111/j.1440-1711.2005.01412.x. [DOI] [PubMed] [Google Scholar]
- 16.Arnoult D, Bartle LM, Skaletskaya A, Poncet D, Zamzami N, Park PU, Sharpe J, Youle RJ, Goldmacher VS. Proc Natl Acad Sci USA. 2004;101:7988–7993. doi: 10.1073/pnas.0401897101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poncet D, Larochette N, Pauleau AL, Boya P, Jalil AA, Cartron PF, Vallette F, Schnebelen C, Bartle LM, Skaletskaya A, et al. J Biol Chem. 2004;279:22605–22614. doi: 10.1074/jbc.M308408200. [DOI] [PubMed] [Google Scholar]
- 18.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 19.McCormick AL, Smith VL, Chow D, Mocarski ES. J Virol. 2003;77:631–641. doi: 10.1128/JVI.77.1.631-641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roumier T, Szabadkai G, Simoni AM, Perfettini JL, Paulau AL, Castedo M, Metivier D, Badley A, Rizzuto R, Kroemer G. Cell Death Differ. 2006;13:348–351. doi: 10.1038/sj.cdd.4401750. [DOI] [PubMed] [Google Scholar]
- 21.Jones NL, Lewis JC, Kilpatrick BA. Eur J Cell Biol. 1986;41:304–312. [PubMed] [Google Scholar]
- 22.Nokta M, Eaton D, Steinsland OS, Albrecht T. Virology. 1987;157:259–267. doi: 10.1016/0042-6822(87)90268-6. [DOI] [PubMed] [Google Scholar]
- 23.Keay S, Baldwin BR, Smith MW, Wasserman SS, Goldman WF. Am J Physiol. 1995;269:C11–C21. doi: 10.1152/ajpcell.1995.269.1.C11. [DOI] [PubMed] [Google Scholar]
- 24.Cheshenko N, Del Rosario B, Woda C, Marcellino D, Satlin LM, Herold BC. J Cell Biol. 2003;163:283–293. doi: 10.1083/jcb.200301084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isler JA, Maguire TG, Alwine JC. J Virol. 2005;79:15388–15397. doi: 10.1128/JVI.79.24.15388-15397.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poncet D, Pauleau AL, Szabadkai G, Vozza A, Scholz SR, Le Bras M, Briere JJ, Jalil A, Le Moigne R, Brenner C, et al. J Cell Biol. 2006;174:985–996. doi: 10.1083/jcb.200604069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silacci P, Mazzolai L, Gauci C, Stergiopulos N, Yin HL, Hayoz D. Cell Mol Life Sci. 2004;61:2614–2623. doi: 10.1007/s00018-004-4225-6. [DOI] [PubMed] [Google Scholar]
- 28.Hall A. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- 29.Bibby KJ, McCulloch CA. Am J Physiol. 1994;266:C1639–C1649. doi: 10.1152/ajpcell.1994.266.6.C1639. [DOI] [PubMed] [Google Scholar]
- 30.Boudreault F, Grygorczyk R. J Physiol (London) 2004;561:499–513. doi: 10.1113/jphysiol.2004.072306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen O. Nature. 1987;330:66–68. doi: 10.1038/330066a0. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, Ackerman SL. Curr Opin Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Isler JA, Skalet AH, Alwine JC. J Virol. 2005;79:6890–6899. doi: 10.1128/JVI.79.11.6890-6899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinton P, Rizzuto R. Cell Death Differ. 2006;13:1409–1418. doi: 10.1038/sj.cdd.4401960. [DOI] [PubMed] [Google Scholar]
- 35.Duchen MR. Mol Aspects Med. 2004;25:365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Lasorsa FM, Pinton P, Palmieri L, Fiermonte G, Rizzuto R, Palmieri F. J Biol Chem. 2003;278:38686–38692. doi: 10.1074/jbc.M304988200. [DOI] [PubMed] [Google Scholar]
- 37.Denton RM, McCormack JG, Edgell NJ. Biochem J. 1980;190:107–117. doi: 10.1042/bj1900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robb-Gaspers LD, Burnett P, Rutter GA, Denton RM, Rizzuto R, Thomas AP. EMBO J. 1998;17:4987–5000. doi: 10.1093/emboj/17.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- 40.Jouaville LS, Pinton P, Bastianutto C, Rutter GA, Rizzuto R. Proc Natl Acad Sci USA. 1999;96:13807–13812. doi: 10.1073/pnas.96.24.13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demaurex N, Distelhorst C. Science. 2003;300:65–67. doi: 10.1126/science.1083628. [DOI] [PubMed] [Google Scholar]
- 42.Bernardi P, Petronilli V. J Bionenerg Biomembr. 1996;28:131–138. doi: 10.1007/BF02110643. [DOI] [PubMed] [Google Scholar]
- 43.Szalai G, Krishnamurthy R, Hajnoczky G. EMBO J. 1999;18:6349–6361. doi: 10.1093/emboj/18.22.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura K, Bossy-Wetzel E, Burns K, Fadel MP, Lozyk M, Goping IS, Opas M, Bleackley RC, Green DR, Michalak M. J Cell Biol. 2000;150:731–740. doi: 10.1083/jcb.150.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinton P, Ferrari D, Rapizzi E, Di Virgilio F, Pozzan T, Rizzuto R. EMBO J. 2001;20:2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinton P, Ferrari D, Magalhaes P, Schulze-Osthoff K, Di Virgilio F, Pozzan T, Rizzuto R. J Cell Biol. 2000;148:857–862. doi: 10.1083/jcb.148.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foyouzi-Youssefi R, Arnaudeau S, Borner C, Kelley WL, Tschopp J, Lew DP, Demaurex N, Krause KH. Proc Natl Acad Sci USA. 2000;97:5723–5728. doi: 10.1073/pnas.97.11.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 49.Chan DC. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Publicover SJ, Duncan CJ, Smith JL. Cell Tissue Res. 1977;185:373–385. doi: 10.1007/BF00220297. [DOI] [PubMed] [Google Scholar]
- 51.Breckenridge DG, Stojanovic M, Marcellus RC, Shore GC. J Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perfettini JL, Roumier T, Kroemer G. Trends Cell Biol. 2005;15:179–183. doi: 10.1016/j.tcb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Parone PA, James DI, Da Cruz S, Mattenberger Y, Donze O, Barja F, Martinou JC. Mol Cell Biol. 2006;26:7397–7408. doi: 10.1128/MCB.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R. Mol Cell. 2004;16:59–68. doi: 10.1016/j.molcel.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 55.Frieden M, James D, Castelbou C, Danckaert A, Martinou JC, Demaurex N. J Biol Chem. 2004;279:22704–22714. doi: 10.1074/jbc.M312366200. [DOI] [PubMed] [Google Scholar]
- 56.Yu D, Smith GA, Enquist LW, Shenk T. J Virol. 2002;76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hahn G, Khan H, Baldanti F, Koszinowski UH, Revello MG, Gerna G. J Virol. 2002;76:9551–9555. doi: 10.1128/JVI.76.18.9551-9555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grazia Revello M, Baldanti F, Percivalle E, Sarasini A, De-Giuli L, Genini E, Lilleri D, Labo N, Gerna G. J Gen Virol. 2001;82:1429–1438. doi: 10.1099/0022-1317-82-6-1429. [DOI] [PubMed] [Google Scholar]
- 59.Patterson CE, Shenk T. J Virol. 1999;73:7126–7131. doi: 10.1128/jvi.73.9.7126-7131.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu H, Shen Y, Shenk T. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grynkiewicz G, Poenie M, Tsien RY. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]