Abstract
Influenza virus remains a global health threat, with millions of infections annually and the impending threat that a strain of avian influenza may develop into a human pandemic. Despite its importance as a pathogen, little is known about the virus structure, in part because of its intrinsic structural variability (pleiomorphy): the primary distinction is between spherical and elongated particles, but both vary in size. Pleiomorphy has thwarted structural analysis by image reconstruction of electron micrographs based on averaging many identical particles. In this study, we used cryoelectron tomography to visualize the 3D structures of 110 individual virions of the X-31 (H3N2) strain of influenza A. The tomograms distinguish two kinds of glycoprotein spikes [hemagglutinin (HA) and neuraminidase (NA)] in the viral envelope, resolve the matrix protein layer lining the envelope, and depict internal configurations of ribonucleoprotein (RNP) complexes. They also reveal the stems that link the glycoprotein ectodomains to the membrane and interactions among the glycoproteins, the matrix, and the RNPs that presumably control the budding of nascent virions from host cells. Five classes of virions, four spherical and one elongated, are distinguished by features of their matrix layer and RNP organization. Some virions have substantial gaps in their matrix layer (“molecular fontanels”), and others appear to lack a matrix layer entirely, suggesting the existence of an alternative budding pathway in which matrix protein is minimally involved.
Keywords: envelope glycoproteins, matrix protein, ribonucleoprotein particles, virus assembly, virus structure
Influenza virus belongs to the orthomyxoviridae, a family of enveloped viruses with segmented genomes of single-stranded negative-sense RNA (1). Although high-resolution structures have been determined by x-ray crystallography for several viral components or fragments thereof (2–4), information about the 3D structure(s) of complete virions has remained scanty. Current models, based primarily on electron microscopy of negatively stained samples (5–7), envisage an envelope containing the trimeric hemagglutinin (HA), tetrameric neuraminidase (NA) (8), and M2 (9) glycoproteins, lined with a continuous layer of matrix protein, enclosing multiple ribonucleoprotein (RNP) complexes (10). An RNP complex consists of a segment of genomic RNA coated by the nucleocapsid protein, with a loop at one end, whereas the other end has a duplex formed by base pairing of the termini to which the viral polymerase is bound (11–14).
Influenza virions assemble as they bud from the surface of infected cells. In this process, the glycoproteins accumulate in lipid rafts, where they interact with the underlying matrix protein. Host factors also participate, particularly in the final stage of pinching off (15). During assembly, a dilemma common to all viruses with segmented genomes must be resolved: how to assign the various segments appropriately to nascent virions? In transverse thin-section electron micrographs of budding influenza virions, a commonly observed motif is a ring with seven features, each thought to be an RNP, surrounding a central such feature (16, 17). Their total number, eight, matches the number of distinct RNA segments in the influenza virus genome. However, it has not been determined whether each virion receives one copy of each segment or a random selection, nor how RNPs are recruited to the budding site (15).
To date, influenza virus has been refractory to 3D structural analysis, in large part because its pleiomorphy has precluded visualization by image reconstruction of electron micrographs by procedures that rely on averaging many identical particles, which have been applied successfully to many icosahedrally symmetric viruses (18). The virion structure is of interest not only in the context of virus assembly but also in light of the possibility that pleiomorphic variations may correlate with infectivity and/or pathogenicity. As a step toward addressing these questions, we have used cryoelectron tomography, a technique capable of rendering the 3D structures of individual macromolecular particles in their native states (19–25), to visualize influenza virions of the type A egg-adapted X-31 strain (26).
Results
Influenza Virus Morphology.
Density maps of 110 virions were compiled from six tomograms. Their resolution is 5.5 nm in-plane, according to the NLOO2D criterion. A central slice through one of the tomograms is shown in Fig. 1. The majority of virions (n = 78) may be described as spherical (axial ratio <1.2), the remainder are oval or kidney-shaped with axial ratios up to 1.4 (n = 17) or more elongated with axial ratios as high as 7.7 (n = 15). The outer diameters of the spherical virions ranged from 84 to 170 nm (mean, 120 nm). The average short diameter of elongated virions was fairly uniform and lower, at ≈100 nm.
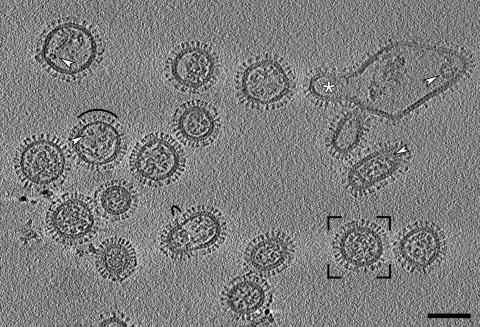
Fig. 1.
Section through a cryotomogram of a field of influenza virions. Three planes were averaged to reduce noise, giving a section 4.7 nm thick. White arrowheads mark typical RNPs: at left, transverse C-shaped sections; at right, longitudinal sections through relatively straight as well as curved RNPs. Black arcs mark areas with matrix layer gaps and missing or lower density packings of glycoprotein spikes. The framed virion is shown in greater detail in Fig. 2a. The large irregular particle at top right was probably generated during cell disruption; it contains at least one budding virus (white asterisk). (Scale bar, 100 nm.)
Despite their pleiomorphy, most virions have three features in common. First, they are covered with glycoprotein spikes, protruding from the virion surface. Second, underlying the spikes at the virion periphery are two layers of approximately equal density and thickness. These layers are closely apposed with a center-to-center spacing of 5.5 nm. We interpret the outer layer as the lipid bilayer in which the transmembrane segments of the glycoproteins are implanted and the inner layer as a monolayer of matrix protein. Third, they contain RNPs whose appearance varies according to their disposition relative to a given tomographic slice, but which typically present as curved or straight filaments (arrowheads, right side, Fig. 1) or, in cross-section, as C-shaped densities (arrowheads, left side, Fig. 1).
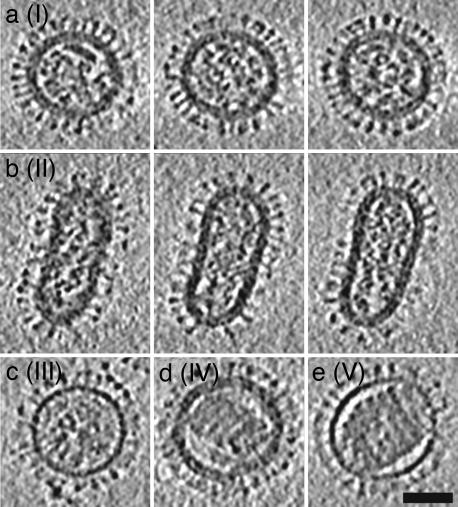
Based on their appearance in tomograms, virions were sorted into five classes, I–V, listed according to relative abundance: I (n = 88), II (n = 15), III (n = 5), IV (n = 1), and V (n = 1) (Fig. 2). Class II comprises elongated particles; the other four classes emerged from further sorting of the spherical particles. The criteria applied were the presence or absence of a discernible matrix layer and RNP organization. Class I has an evident matrix layer and a somewhat disordered arrangement of RNPs; class III lacks a discernible matrix layer and also has jumbled RNPs. Classes IV (with matrix) and V (no matrix) have condensed cores that appear in longitudinal slices (Fig. 2 d and e) as parallel arrays of seven to eight rods; however, tracking them through the tomograms revealed that these cores are single solenoids of seven to eight coils with a pitch of ≈10 nm (Fig. 7c). The contour length of the coil was measured on a class V virion to be ≈1.26 μm. The elongated class II particles share the features of class I, except that their RNPs tend to be approximately aligned with the long axis of the particle (Fig. 2b). The possibility remains that elongated counterparts of the rarer classes III, IV, and V may be detected in a larger sampling of virion populations. Indeed, in cryomicrographs, we did observe some elongated virions with solenoid-like contents (data not shown).
Fig. 2.
Examples of the five morphological classes (I–V) of influenza virions. For the more abundant class I and II particles, three serial slices, 10.9 nm apart and 4.7 nm thick, are shown in a and b, respectively, whereas single near-central slices are shown for classes III-V in c–e. The class I virion (a) is the framed particle in Fig. 1. (Scale bar, 50 nm.)
Fig. 7.
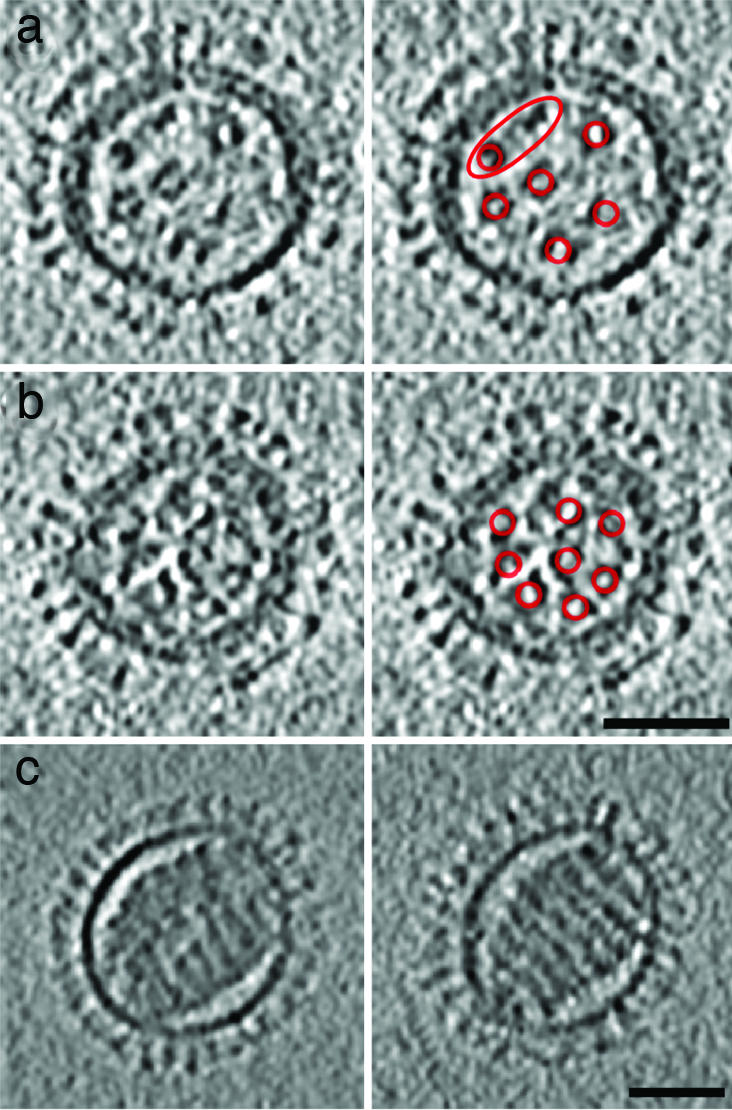
Central slices through influenza virions containing (7 + 1)-like and solenoid RNP configurations. a and b show two examples of (7 + 1)-like configurations in spherical class I virions. In each case, a slice is shown in Left and, on Right, it is indexed with red rings around the RNPs, which are mostly C-shaped in cross-section. In virion (a), which has an incomplete matrix layer, only six peripheral RNPs are clearly visible (two are enclosed in the red ellipse). Weaker density may denote a seventh peripheral RNP. (c) Two serial slices through the solenoidal core of a class V virion, which has no discrenible matrix layer. The slices are 18.7 nm apart and 4.7 nm thick. (Scale bars, 50 nm.)
Glycoprotein Spikes.
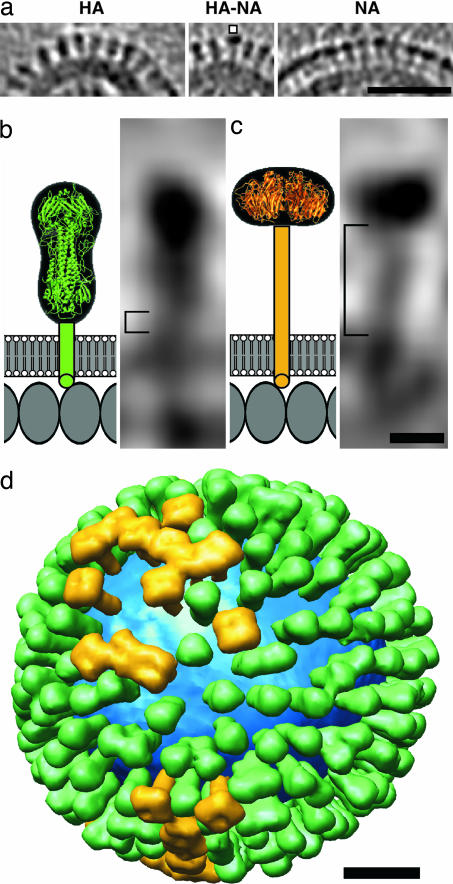
Except in a few low-density patches (e.g., arcs, Fig. 1), the glycoproteins are packed closely but irregularly, with an average center-to-center spacing of ≈11 nm. Both HA and NA extend radially from the membrane to terminate in bulbous heads (Fig. 3a). NA is slightly longer than HA and may be distinguished in longitudinal sections by its shorter head (HA has a characteristically bilobed “peanut” shape) and longer stem (Fig. 3 b vs. c) and in transverse sections by its square profile, as opposed to the triangular profile of HA (Fig. 4).
Fig. 3.
Distributions and shape-based differentiation of HA and NA spikes. (a) HA cluster (Left); single NA (marked) in a cluster of HAs (Center); and cluster of mainly NA spikes (Right). (Scale bar, 50 nm.) (b and c) The stem lengths of HA and NA (square brackets in b and c, respectively) were measured as described in Materials and Methods. The structures of the stems, transmembrane segments, and endodomain tails are not known, and they are shown schematically. Molecules in the matrix layer are inferred to be packed in a monolayer with a spacing of ≈4 nm (our unpublished results). (Scale bar, 5 nm.) (d) Model of the distribution of glycoprotein HA (green) and NA (gold) on a single influenza virion. The lipid bilayer is blue. (Scale bar, 20 nm.)
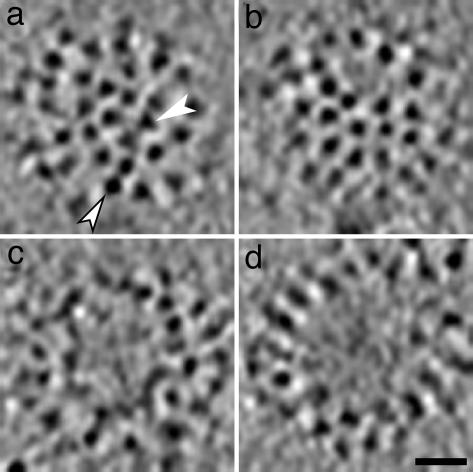
Fig. 4.
Patches of glycoprotein spikes depicted in tangential sections (4.7 nm thick) through virions in cryotomograms. In most cases, triangular HA spikes (e.g., white arrowhead in a) are distinguishable from square NA spikes (e.g., white arrowhead with black border in a). Spikes are close-packed, without lateral order, apart from occasional bare patches of variable size. Some examples of bare patches are shown in b–d. (Scale bar, 20 nm.)
The packing density of spikes was assessed by counting them within typical well resolved patches; on average, an area of 40 × 40 nm was found to contain 13 spikes. No difference in this respect was observed between spherical and elongated virions. Thus a spherical virion of average diameter (120 nm) should have ≈375 spikes. This figure was confirmed by counting all of the spikes on two virions of average diameter whose entire surfaces were visualized clearly enough for this to be possible, yielding 351 (301 HA plus 50 NA) and 328 (290 HA plus 38 NA) spikes per virion. Allowing for a few bare patches (such patches were observed on ≈40% of spherical virions, e.g., Fig. 4 b–d), these quantitations are consistent. They are also in reasonable agreement with prior biochemical estimates of 340–400 HA trimers per virion (10).
We used molecular modeling to exploit our ability to identify individual spikes in the tomograms as HA or NA (Fig. 3). The HA ectodomain solved by x-ray crystallography is ≈12 nm in depth (2) and that of NA, ≈6 nm (ref. 3; Fig. 3 b and c). When the HA ectodomain is docked into tomographic densities for HA spikes, its outer tip is ≈14 nm from the membrane surface (i.e., there is a 2-nm stem), whereas for NA spikes, this spacing is ≈16 nm (i.e., a 10-nm stem). Between the ectodomain sequences and the predicted transmembrane regions are linkers of 10 (HA; ref. 27) and 51 (NA; ref. 28) amino acids, respectively. If they are predominantly α-helical, this would correspond to stem lengths of 1.5 and 7.6 nm, respectively, in general accord with our measurements.
The distributions of HA and NA over the virion surface appear not to be entirely random; congregations of the more abundant HA spikes are the pattern most frequently observed; then single NA spikes surrounded by HA, and local clusters of NA (Fig. 3a). A tomographic model showing the distribution of spikes over the surface of a single virion is presented in Fig. 3d.
Matrix Protein Layer.
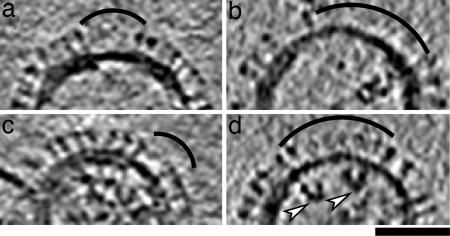
In virions of classes I, II, and IV, a matrix layer is visible under the viral membrane and closely apposed to it (Figs. 1–3). Their center-to-center separation is ≈5.5 nm, close to our resolution limit. The matrix layer has the same apparent thickness as the membrane. Contacts between them, presumably by the connecting densities that are seen in the tomograms (e.g., Fig. 3 a and b) but are at our resolution limit, should involve interactions of the matrix protein with the small endodomain tails of the glycoproteins (29), predicted sizes, 6 (NA; ref. 28) and 10 residues (HA; ref. 27). The existence of direct interactions of the glyco-proteins with the matrix layer is further supported by the correlation of gaps in the matrix layer with membrane regions in which the glycoproteins are either absent or less abundant than elsewhere (Fig. 5a and b).
Fig. 5.
Influenza virus particles with gaps in their matrix layer. Virions containing matrix gaps tend to have a decreased density of glycoproteins in the envelope regions overlying the gaps, which are indexed with arcs. The white arrowheads in d points to C-shaped transverse sections through two RNPs. For each particle, a 4.7-nm-thick slice is shown. (Scale bar, 50 nm.)
Of the particles with a matrix layer (102/110), ≈20% contained a visible gap. This is probably an underestimate, because gaps oriented perpendicular to the untilted specimen plane should be less reliably detected because of lower resolution in this dimension caused by the incomplete tilt range covered (the so-called “missing wedge” effect; ref. 30). Some particles had more than one gap; specifically, 13 had 1 gap, 5 had 2 gaps, and 2 had 3 gaps. The gaps ranged in size up to ≈80 nm. Virions of classes III and V, amounting to 6% of the data set, appear to lack a matrix layer entirely (Fig. 2 c and e).
RNPs.
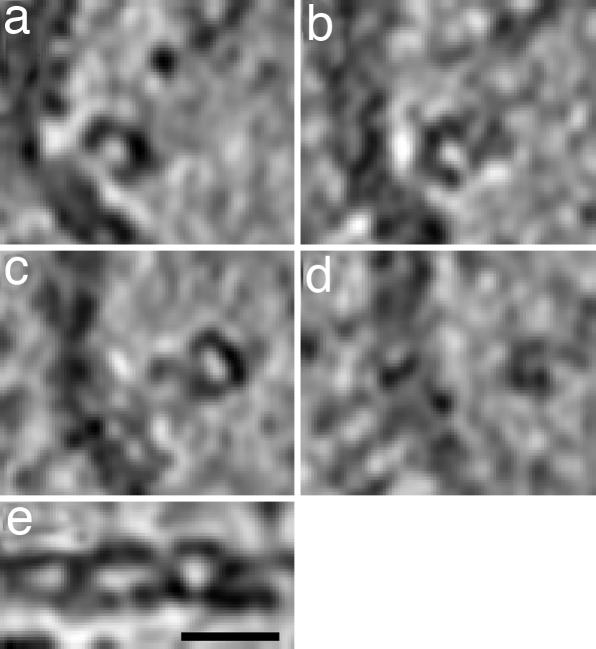
The RNPs are packed quite densely in most virions, making it difficult to delineate individual complexes. Nevertheless, we could make the following observations, based on the more abundant class I and II particles (Fig. 2 a and b). RNPs are ≈14 nm thick, and in some cases, mostly in class II virions (Fig. 2b), we were able to trace individual complexes, whose lengths were found to vary from ≈24 to ≈110 nm (n = 40). Each RNP makes contact with the matrix layer at one or both ends. Most RNPs exhibit some curvature. They typically present a C-shaped motif in cross-section (e.g., arrowheads, left side; Figs. 1 and 5d). This motif tends to rotate between successive sections, suggesting a loosely ordered but basically helical structure (ref. 11; Fig. 6).
Fig. 6.
Distributions of density perpendicular to and along influenza virus ribonucleoprotein particles are illustrated in tomographic slices. (a–d) Four serial slices, 4.7 nm thick and 9.4 nm apart, through part of an influenza virion. An RNP segment is oriented approximately perpendicular to the viewing direction. (e) A central longitudinal section of the same RNP complex, after computational straightening, suggests a double helical structure. (Scale bar, 20 nm.)
The arrangement of the RNPs varies with the morphology and size of the virion. In elongated particles, they form a near-parallel bundle in which all RNPs associate with the matrix at the same end of the virion (Fig. 2b). In a few instances, we were able to distinguish eight such complexes, usually one at the center surrounded by seven. Some spherical particles exhibit a similar arrangement in at least one plane (e.g., Fig. 7a and b), albeit with the RNPs not so well aligned. Smaller particles (<100 nm in diameter; ≈10% of the data) tend to have fewer than eight segments. The minimum number detected was two. In large spherical particles (>130 nm), the arrangement of RNPs was less orderly. In spherical particles, the contact points of the various RNP complexes on the matrix layer were not clustered but were distributed around the interior surface.
Discussion
The present observations afford insight into the molecular architecture of influenza virions, whose diverse forms represent the outcome of complex patterns of interactions among viral components: the glycoproteins, matrix protein, and the RNPs; and host factors (15). Because different viral strains propagated in the same cells produce different distributions of virion shapes (e.g., elongated vs. spherical), it follows that plasticity in the interactions among viral components is a major contributor to pleiomorphy. In this study, we have used electron tomography to make a systematic account of pleiomorphic variability in one well studied strain of the virus.
The Matrix Layer.
In assembly, the matrix protein has the dual roles of concentrating the glycoproteins at the budding site (there appear to be no regular interactions between neighboring spikes; Fig. 4) and of interacting with the virion's complement of RNPs. If, as seems likely, the matrix layer involves a regular packing of subunits (ref. 7; our unpublished observations), this packing must be adaptable enough to allow the formation of differently sized and shaped virions.
Some virions have substantial gaps in their matrix layer. We suspect that these “molecular fontanels” may represent sites where the virions pinched off in budding from the host cell. It is not apparent whether the gaps have functional significance, but the close association of the matrix protein with the membrane suggests this association might impede membrane fusion, and a matrix-free area of envelope might offer a favorable site to initiate the fusion process.
We also observed some virions, classes III and V, that appear to lack a matrix layer entirely (Fig. 2 c and e). This observation implies that either they have only a few molecules of matrix protein, enough to play its role in budding but not enough to be perceived as a continuous layer in the tomograms, or they assemble by a different budding mechanism.
We have considered some possible relationships between spherical virions and elongated ones; for instance, that spherical virions may bud in the same way but simply have pinched off earlier. However, this hypothesis does not square with the observation that most spherical virions have larger diameters (120 vs. 100 nm). Alternatively, could spherical virions be derived from elongated ones by an in vitro rearrangement in overall structure accompanied by a disordering of the RNPs? This hypothesis, although not ruled out, invokes a major reorganization of the matrix layers, after budding.
RNPs.
EM studies of stained plastic sections of budding influenza virions have presented evidence for a (7 + 1) configuration of aligned RNPs (17). Our observations of complete ice-embedded elongated virions (class II) are consistent with this model. Although we observed that smaller virions tend to have fewer than eight RNPs, we also observed some spherical virions whose RNPs were similarly arranged (e.g., Fig. 7 a and b). More commonly, we could count eight RNPs, but they were not aligned. These observations suggest there may be specialized attachment points for each RNP on the matrix layer, with these points being closely clustered or even merged in elongated virions and less regularly arranged in spherical virions. However, we have yet to detect structural features in the matrix layer that are specific to these sites.
The condensed core of our class IV and V particles (Fig. 2 d and e) has been observed before, also in rare occurrence (31). Our tomograms reveal that these cores are solenoidal windings of a filament that is much longer than any individual RNP that we observed and whose diameter is about half that of an RNP. One possible explanation is that the solenoid may represent an immature precursor form in which the RNPs are initially packaged as a single concatenated filament that is subsequently dispersed and reorganized. Although speculative, this proposition would afford a mechanism whereby a designated consignment of multiple segments could be incorporated into a given viral particle.
Materials and Methods
Virus Propagation and Purification.
X-31 [A/Aichi/68 (H3N2)] influenza virus, grown in embryonated chicken eggs and step gradient-purified according to standard procedures, at ≈2 mg/ml protein, was purchased from Charles River Laboratories (North Franklin, CT). For electron microscopy, the virus was further concentrated by pelleting (137,000 × g for 2 h) and resuspended at ≈6 mg/ml protein in 20 mM Hepes/20 mM Mes/130 mM NaCl, pH 7.5.
Tomographic Data Collection.
Virus was mixed 1:4 with a suspension of 10-nm colloidal gold particles (Ted Pella, Redding, CA). Four-microliter drops were applied to holey carbon films, thinned by blotting, and vitrified by plunging into liquid ethane before transfer into a liquid-N2-cooled specimen holder (model 626; Gatan, Warrendale, PA). A Tecnai-12 electron microscope (FEI, Hillsboro, OR) operating at 120 keV was used to record single-axis tilt series at 1° steps, covering ranges of approximately −64° to +72°. The microscope was equipped with an energy filter (Gatan GIF 2002) that was operated in the zero-energy-loss mode with a slit width of 20 eV. Images were recorded on a 2,048 × 2,048-pixel CCD camera (Gatan) at ×38,500 magnification (0.78-nm pixels) and 4–6 μm defocus, corresponding to first zeros of the contrast transfer function at (3.7 nm)−1 − (4.5 nm)−1. Data were recorded under low-dose conditions, using SerialEM to conduct automatic tilting, tracking, focusing, and image acquisition (32). The cumulative electron dose was ≈55 electrons/Å2.
Image Processing.
Projections in a tilt series were mutually aligned by using the gold particles as fiducial markers and the tomogram calculated by the weighted back-projection method, using IMOD (33). The final analyses were performed on binned tomograms with a voxel size of 1.56 nm. The tomograms were denoised by bilateral filtering (34), with the following parameters: 1 iteration; σ1 = 1 pixel; σ2 = 3–5 times the standard deviation of the tomogram; and a kernel size of 5. These parameters were chosen to provide a moderate reduction in noise, while preserving information at relatively high resolution. Their resolution was estimated by the NLOO-2D method as implemented in ELECTRA (35). For each tomogram, resolution was assessed as the average of three particles at the threshold 0.3. Cryoelectron microscopy of virions was performed as reported (36).
Mapping the Distribution of Glycoprotein Spikes.
The lengths of the glycoprotein stems were measured after aligning their respective ectodomain crystal structures and a model lipid bilayer, taken to be 4 nm thick, with the corresponding densities in the tomograms. To facilitate processing of the glycoprotein layer, the viral membrane and the extraviral region were masked out. A model was generated in which each pair of pseudoatoms was connected by a pseudobond fitted by hand into each density identified as a glycoprotein. Templates of the ectodomains were generated at 5.0-nm resolution from the crystal structures (3HMG for HA and 1V0Z for NA). A 3-nm-wide stalk was added to the NA map. These templates were used to refine the positions of the glycoproteins by crosscorrelation (taking into account the missing wedge), as well as to classify them as HA or NA, and were also used to build a model of the glycoprotein layer. This processing was done in Bsoft (37). Visualization was done with Chimera (www.cgl.ucsf.edu/chimera).
Acknowledgments
We thank Charles S. Smith and Brittney Manvilla for help with electron microscopy. This work was supported in part by the Intramural Research Program of National Institute of Arthritis and Musculoskeletal and Skin Diseases and the National Institutes of Health (NIH) Intramural Targeted Antiviral Program (A.C.S.) and an NIH R01 award, AI22470 (to J.M.W.). M.B. was supported in part by an NIH training grant (5T32AI07047).
Abbreviations
- NA
neuraminidase
- RNP
ribonucleoprotein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Lamb RA, Krug RM. In: Fields Virology. Knipe DM, Howley PM, editors. Vol 1. New York: Lippincott Williams and Wilkins; 2001. pp. 1487–1579. [Google Scholar]
- 2.Wilson IA, Skehel JJ, Wiley DC. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 3.Varghese JN, Laver WG, Colman PM. Nature. 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 4.Harris A, Forouhar F, Qiu S, Sha B, Luo M. Virology. 2001;289:34–44. doi: 10.1006/viro.2001.1119. [DOI] [PubMed] [Google Scholar]
- 5.Schulze IT. Virology. 1972;47:181–196. doi: 10.1016/0042-6822(72)90251-6. [DOI] [PubMed] [Google Scholar]
- 6.Wrigley NG, Brown EB, Skehel JJ. In: Electron Microscopy of Proteins: Viral Structure. Harris JR, Horne RW, editors. Vol 5. London: Academic; 1986. pp. 103–164. [Google Scholar]
- 7.Ruigrok RW, Calder LJ, Wharton SA. Virology. 1989;173:311–316. doi: 10.1016/0042-6822(89)90248-1. [DOI] [PubMed] [Google Scholar]
- 8.Laver WG. In: Perspectives in Medical Virology. Zuckerman AJ, Mushahwar IK, editors. Vol 7. Amsterdam: Elsevier; 2002. pp. 31–47. [Google Scholar]
- 9.Pinto LH, Holsinger LJ, Lamb RA. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 10.Inglis SC, Carroll AR, Lamb RA, Mahy BW. Virology. 1976;74:489–503. doi: 10.1016/0042-6822(76)90355-x. [DOI] [PubMed] [Google Scholar]
- 11.Compans RW, Content J, Duesberg PH. J Virol. 1972;10:795–800. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murti KG, Webster RG, Jones IM. Virology. 1988;164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 13.Klumpp K, Ruigrok RW, Baudin F. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Area E, Martin-Benito J, Gastaminza P, Torreira E, Valpuesta JM, Carrascosa JL, Ortin J. Proc Natl Acad Sci USA. 2004;101:308–313. doi: 10.1073/pnas.0307127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nayak DP, Hui EK, Barman S. Virus Res. 2004;106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oxford JS, Hockley DJ. In: Perspectives in Medical Virology: Animal Virus Structure. Nermut MV, Steven AC, editors. Vol 3. New York: Elsevier; 1987. pp. 213–232. [Google Scholar]
- 17.Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 18.Baker TS, Olson NH, Fuller SD. Microbiol Mol Biol Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucic V, Forster F, Baumeister W. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 20.Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 21.Cyrklaff M, Risco C, Fernandez JJ, Jimenez MV, Esteban M, Baumeister W, Carrascosa JL. Proc Natl Acad Sci USA. 2005;102:2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forster F, Medalia O, Zauberman N, Baumeister W, Fass D. Proc Natl Acad Sci USA. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. J Mol Biol. 2005;346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briggs JA, Grünewald K, Glass B, Forster F, Krausslich HG, Fuller SD. Structure (Cambridge, UK) 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Zhu P, Liu J, Bess J, Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 26.Lee KH, Youn JW, Kim HJ, Seong BL. Arch Virol. 2001;146:369–377. doi: 10.1007/s007050170181. [DOI] [PubMed] [Google Scholar]
- 27.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 28.Blok J, Air GM. Biochemistry. 1982;21:4001–4007. doi: 10.1021/bi00260a015. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J, Leser GP, Pekosz A, Lamb RA. Virology. 2000;269:325–334. doi: 10.1006/viro.2000.0228. [DOI] [PubMed] [Google Scholar]
- 30.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Oxford: Oxford Univ Press; 2006. pp. 218–219. [Google Scholar]
- 31.Booy FP, Ruigrok RW, van Bruggen EF. J Mol Biol. 1985;184:667–676. doi: 10.1016/0022-2836(85)90312-2. [DOI] [PubMed] [Google Scholar]
- 32.Mastronarde DN. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Kremer JR, Mastronarde DN, McIntosh JR. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 34.Jiang W, Baker ML, Wu Q, Bajaj C, Chiu W. J Struct Biol. 2003;144:114–122. doi: 10.1016/j.jsb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 35.Cardone G, Grünewald K, Steven AC. J Struct Biol. 2005;151:117–129. doi: 10.1016/j.jsb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Cheng N, Conway JF, Watts NR, Hainfeld JF, Joshi V, Powell RD, Stahl SJ, Wingfield PE, Steven AC. J Struct Biol. 1999;127:169–176. doi: 10.1006/jsbi.1999.4120. [DOI] [PubMed] [Google Scholar]
- 37.Heymann JB, Belnap DM. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.06.006. in press. [DOI] [PubMed] [Google Scholar]