Abstract
Under starvation conditions myxobacteria form multicellular fruiting bodies in which vegetative cells differentiate into heat- and desiccation-resistant myxospores. Myxobacteria in general are a rich source of secondary metabolites that often exhibit biological activities rarely found in nature. Although the involvement of a yellow compound in sporulation and fruiting body formation of Myxococcus xanthus was described almost 30 years ago, the chemical principle of the pigment remained elusive. This work presents the isolation and structure elucidation of a unique class of pigments that were named DKxanthenes (DKX). The corresponding biosynthetic gene cluster was identified, and DKX-negative mutants were constructed to investigate the physiological role of DKX during development. In these mutants, fruiting body formation was delayed. Moreover, severely reduced amounts of viable spores were observed after 120 h of starvation, whereas no viable spores were formed at all after 72 h. The addition of purified DKX to the mutants resulted in the formation of viable spores after 72 h. Even though an antioxidative activity could be assigned to DKX, the true biochemical mechanism underlying the complementation remains to be elucidated.
Keywords: fruiting body formation, natural product, biosynthetic gene cluster
Myxobacteria are soil-inhabiting Gram-negative bacteria that can undergo a complex developmental program in response to nutrient limitation (1, 2). Under starvation conditions myxobacteria form fruiting bodies, which can have a tree-like appearance in some species and which can be seen with the naked eye (3). In these fruiting bodies vegetative cells undergo differentiation into heat- and desiccation-resistant myxospores that ensure the survival of the colony. The formation of fruiting bodies and mature spores require precise coordination and communication processes that are so far best characterized in the myxobacterial model organism Myxococcus xanthus (4).
Furthermore, myxobacteria are a rich source of several secondary metabolites with often unusual structural features and new or rare modes of action (5, 6). More than 100 basic structures with overall ≈500 derivatives have been isolated from different myxobacteria including several Myxococcus species. Despite the bright yellow color of most M. xanthus strains, the model strain M. xanthus DK1622 was not thought to be a producer of secondary metabolites at all until its genome was sequenced and annotated recently (www.tigr.org). Approximately 8% of the genome are dedicated to secondary metabolism and at least 18 biosynthesis gene clusters for the production of polyketides, nonribosomally made peptides or hybrids thereof have been detected in the genome (7). For unknown reasons, most of these gene clusters are located between 3.2 and 5.8 Mbp of the circular genome in such close proximity that it is hard to define their borders. This unexpected genetic diversity makes M. xanthus DK1622 a very potent secondary metabolite producer comparable to sequenced streptomycetes (8, 9) that has stimulated further research to isolate the corresponding compounds. So far myxochromides, myxovirescins, myxochelins, and myxalamides have been isolated from DK1622. All of these compounds have been previously isolated from other myxobacteria or represent new derivatives of these compounds (refs. 10 and 11 and unpublished results).
Here, we report the isolation of the DKxanthenes (DKX), a family of new secondary metabolites from M. xanthus DK1622 that represent the major yellow pigment of strain DK1622 and several other Myxococcus species analyzed. Because earlier studies (12, 13) indicated an involvement of the pigment in morphological differentiation, we compared mutants defective in its biosynthesis with the wild-type strain and show that they are required for mature spore formation during fruiting body formation.
Results
Isolation and Structure Elucidation of DKX from M. xanthus DK1050.
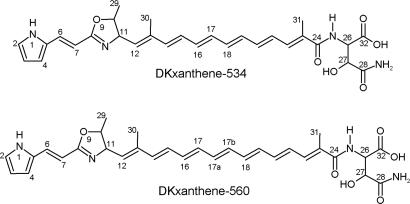
In the course of identifying the secondary metabolites of M. xanthus DK1622 we initiated the isolation of the yellow pigments. However, because of the well known phase variation between yellow and beige (tan) variants we started our analysis with strain DK1050, which is a stable yellow derivative of strain FB that itself is the parent strain of DK1622 (14). DK1050 was grown in rich medium with the adsorber resin Amberlite XAD-16 (1%) and pure DKxanthene-534 and DKxanthene-560 (Fig. 1) were isolated from the cells after extraction with ethylacetate-methanol followed by extensive column chromatography. The molecular formula of the main product DKxanthene-534 was determined from high-resolution electrospray ionization MS analysis as C29H35N4O6, (m/z 535.25557). 13C NMR analysis confirmed the number of carbons with 14 carbon atoms between 108 and 138 ppm indicating the presence of several double bonds. Four signals showed characteristic chemical shifts (δC 163–176 ppm) of ester or amide carbons and three additional carbons could be assigned to methyl groups (12–20 ppm). Three strong signals in 1H NMR confirmed the presence of these methyl-groups, two of them appeared as singlets (δH 1.84 and 1.90) and one as a doublet (δH 1.32, J = 6.0 Hz). Through 1H-1H-COSY experiments, determination of coupling constants, and heteronuclear multiple bond correlations (HMBC) (Fig. 2 and Table 1), the methyl group at δH 1.32 could be assigned to a 5-methyl-oxazoline ring that most likely is biosynthetically derived from threonine. The coupling constants (3J = 7.5 Hz) as well as missing NOESY correlations between H-10 and H-11 strongly indicate a trans configuration of both protons as observed for other l-threonine-derived oxazoline ring-systems (15). Furthermore, a cis configuration of these protons would result in a much larger coupling constant of ≈10 Hz (16).
Fig. 1.
Structures of DKxanthene-534 and -560.
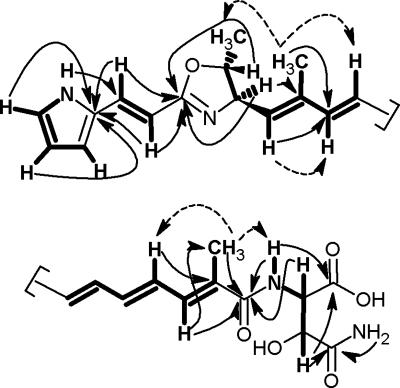
Fig. 2.
Selected 1H-1H-COSY (bold lines), HMBC (arrows), and NOESY (broken arrows) correlations of DKxanthene-534.
Table 1.
13C NMR and 1H NMR spectral data of DKxanthene-534 and -560 in [D6]DMSO at 500 MHz (1H) and 125.7 MHz (13C)
| Number | DKxanthene-534 |
DKxanthene-560 |
||||
|---|---|---|---|---|---|---|
| 13C | 1H | J, Hz | 13C | 1H | J, Hz | |
| 1 | 11.41 br s | 11.37 br s | ||||
| 2 | 122.4 | 6.93 ddd | 1.5/3.0/3.0 | 122.1 | 6.93 m | |
| 3 | 109.8 | 6.10 ddd | 1.5/3.0/3.0 | 109.6 | 6.10 ddd | 1.5/3.0/3.5 |
| 4 | 113.0 | 6.41 m | 112.7 | 6.41 m | ||
| 5 | 128.8 | 128.6 | ||||
| 6 | 130.0 | 7.10 d | 16.0 | 129.8 | 7.10 d | 16.0 |
| 7 | 108.4 | 6.30 d | 16.0 | 108.2 | 6.30 d | 16.0 |
| 8 | 163.0 | 162.8 | ||||
| 10 | 81.0 | 4.27 dq | 6.0/7.5 | 80.8 | 4.27 m | |
| 11 | 70.9 | 4.52 dd | 7.5/9.0 | 70.6 | 4.52 dd | 7.5/9.0 |
| 12 | 133.7 | 5.45 d | 9.0 | 133.4 | 5.45 d | 9.0 |
| 13 | 135.8 | 135.6 | ||||
| 14 | 137.8 | 6.38 d | 15.03 | 137.5 | 6.38 d | 15.0 |
| 15 | 133.2 | 6.46 dd | 15.0/11.04 | 128.7* | 6.37 m | |
| 16 | 131.1* | 6.42–6.45 | 130.8* | 6.46 m | ||
| 17 | 134.8* | 6.42–6.45 | 133.0* | 6.42 m | ||
| 17a† | 133.1* | 6.47 m | ||||
| 17b† | 134.2* | 6.41 m | ||||
| 18 | 135.4* | 6.42–6.45 | 134.6* | 6.43 m | ||
| 19 | 128.9 | 6.37 dd | 15.0/11.05 | 135.2* | 6.46 m | |
| 20 | 137.6 | 6.47 dd | 11.0/15.04 | 137.4 | 6.47 dd | 11.0/15.0 |
| 21 | 128.6 | 6.60 dd | 9.5/15.04 | 128.4 | 6.60 dd | 9.5/15.0 |
| 22 | 132.6 | 6.88 d | 9.5 | 132.5 | 6.90 d | 9.5 |
| 23 | 133.1 | 132.9 | ||||
| 24 | 166.6 | 166.6 | ||||
| 25 | 7.16 d | 8.0 | 7.10 m | |||
| 26 | 54.8 | 4.18 dd | 4.0/8.0 | 54.7 | 4.27 m | |
| 27 | 71.2 | 4.05 d | 4.0 | 70.66 | 4.18 m | |
| 28 | 175.9 | 174.2 | ||||
| 28-NH2 | 7.04 s/7.13 s | 7.10 s/7.20 s | ||||
| 29 | 19.9 | 1.32 d | 6.0 | 19.7 | 1.32 d | 6.0 |
| 30 | 12.9 | 1.84 s | 12.7 | 1.84 s | ||
| 31 | 13.0 | 1.90 s | 12.9 | 1.91 s | ||
| 32 | 172.3 | 172.2 | ||||
s, singlet; d, doublet; m, multiplet; dd, doublet of doublet; ddd, doublet of doublet of doublet; q, quartet; br, broad.
*Assignments are interchangeable; coupling constants were determined in CD3OD3, [D6]benzene/CD3OD 1:14, and [D6]benzene/CD3OD 9:15.
†17a and 17b represent the additional double bond of DKxanthene-560.
The pyrrole ring moiety was readily identified by its characteristic shifts in 1H and 13C NMR and the characteristic signal for the NH-group at δH = 11.41 (br s) in the proton NMR with strong 1H-1H-COSY correlations to all ring protons.
The hydroxyasparagine moiety could be elucidated by a combination of 1H-1H-COSY and HMBC correlations (Fig. 2). However, its stereochemistry could not be deduced from the NMR data.
1H-1H-COSY and HMBC data of the two remaining methyl groups (δH 1.84 and 1.90) showed correlations to protons of a polyene moiety. Structure elucidation of this polyene chain was hampered because of signal overlap between 5.45 and 7.10 ppm resulting in no clear assignment of coupling partners and coupling constants for H-14 to H-19 in [D6]DMSO. By using CD3OD and mixtures of CD3OD and [D6]benzene as solvent the configuration of all double bonds except H-16 and H-17 could be assigned as trans. Calculation of the theoretical chemical shifts of protons H-16 and H-17 in cis- or trans-configuration [see supporting information (SI) Fig. 6] strongly indicates an all-trans-configuration for DKxanthene-534 (Fig. 1). Further evidence for this configuration is the observed UV spectrum with an absorption maximum of λmax = 379 nm. This is in very good agreement with the theoretical maximum of λmax = 380 nm, calculated by the increment system using 200 nm as basic value for an amide and 30 nm added for each conjugated trans-double bond (17).
NOESY experiments revealed further insights into the stereochemistry of DKxanthene-534 (Fig. 2). The trans-configuration between H-12 and CH3-30 (δH 1.84) was assigned through strong correlations of H-30 to H-11, H-15, and H-29, whereas no correlations could be observed to H-10, H-12, and H-14. Accordingly, the configuration between H-22 and CH3-31 (δH 1.90) could be assigned as trans through correlations of H-31 to H-21 and H-25.
In DKxanthene-560 (C31H37N4O6, high-resolution electro-spray ionization MS m/z 561.27142), almost identical chemical shifts and correlations were observed for the pyrrole and asparagine part of the molecule in the different NMR experiments (Table 1). However, two additional signals in the 1H NMR (δH 6.46 and 6.41) with strong 1H-1H-COSY correlations to further polyene protons indicated the presence of an additional double bond. Through heteronuclear sequential quantum correlations the additional 1H NMR signals were assigned to signals at 134.2 and 130.8 ppm in 13C NMR; HMBC correlations confirmed their localization within the polyene chain. Again, the observed absorption maximum of λmax = 408 nm matches well with the calculated maximum of λmax = 410 nm in an all-trans-configuration of the polyene (17).
The structures of additional DKXs were identified through detailed feeding experiments followed by the analysis of the MS/MS-fragmentation to confirm substitution pattern and fragment identity (Fig. 3 and Table 2). [13C4,15N1]threonine was used as a marker because its incorporation resulted in a +5-shift of the molecular ion and several MS/MS fragments of all DKXs containing the complete oxazoline moiety, whereas a +3 shift was observed in specific fragments in the derivatives after the elimination of ethanol (O9-C10-C29) from the oxazoline ring. Incorporation of [D6]propionic acid led to +3, +6 or, +9 shifts in the molecular ion, according to the different methylation patterns with up to three methyl groups derived from methylmalonyl-CoA incorporation as found in several other myxobacterial metabolites (18, 19).
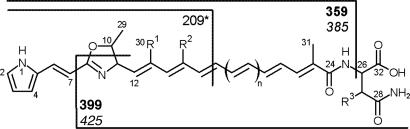
Fig. 3.
Selected MS/MS fragments exemplified for DKxanthene-534 (numbers given in bold) and DKxanthene-560 (numbers shown in italics). For better readability, only the masses of nonlabeled fragments are shown. The 209 fragment (*) has additionally lost EtOH from the oxazoline moiety. The numbering is shown for DKxanthene-534 (n = 1; see Table 2). For assignment of R1–R3, see Table 2.
Table 2.
DKxanthene derivatives identified by NMR analysis and feeding experiments followed by detailed MS/MS analysis.
| n | R1 | R2 | R3 | |
|---|---|---|---|---|
| DKxanthene-492 | 0 | CH3 | H | H |
| DKxanthene-504 | 1 | H | H | H |
| DKxanthene-508 | 0 | CH3 | H | OH |
| DKxanthene-518 | 1 | CH3 | H | H |
| DKxanthene-520 | 1 | H | H | OH |
| DKxanthene-534 | 1 | CH3 | H | OH |
| DKxanthene-544 | 2 | CH3 | H | H |
| DKxanthene-548 | 1 | CH3 | CH3 | OH |
| DKxanthene-560 | 2 | CH3 | H | OH |
| DKxanthene-574 | 2 | CH3 | CH3 | OH |
| DKxanthene-586 | 3 | CH3 | H | OH |
For the positions of chain length variability (n) and substituents R1–R3, see Fig. 3.
The proposed substitution pattern could be confirmed by specific fragmentation of the different derivatives. The presence of the C-27 hydroxy group could be identified by comparison of the different fragments with or without the (hydroxy)asparagine moiety. Furthermore, an additional methyl group (R2) in DKxanthene-548 and -574 was obvious from a shift of the N1-C8-C11-C16 fragment from 209 to 223 atomic mass units, whereas its position at C-15 was deduced from the biosynthetic logic of polyketide synthesis. In polyketide assembly lines, either malonyl-CoA (R2 = H) or methylmalonyl-CoA (R2 = CH3) units are used as building blocks of the growing polyketide chain (20).
In addition to the 11 oxazoline DKXs ranging from 492 to 586 atomic mass units, further compounds with a loss of two mass units were detected in the LC-MS analysis as minor components (data not shown). These compounds might be oxazole DKXs, which can be derived from spontaneous or enzymatic oxidation of the parent oxazoline compounds.
Identification of the DKxanthene Biosynthesis Gene Cluster.
The DKxanthene biosynthesis gene cluster was identified in DK1050 by transposon mutagenesis using the transposon pMycoMar as described (21). Colonies with a beige (tan) color instead of the usual yellow color were isolated and the transposon insertion site was identified by vector recovery from genomic DNA and sequenced as described (21). Ten tan colonies have been observed from overall 3500 transposon mutants. Four of them were identified as being part of a complex hybrid PKS/NRPS biosynthetic gene cluster ≈47 kb in size. The involvement of this gene cluster in the DKX biosynthesis was confirmed by targeted gene inactivation via plasmid insertion according to published procedures (22). This gave rise to mutants M. xanthus PM1284 and M. xanthus PM1297 (both DKX−) carrying plasmid integrations in two separate genes of the DKX biosynthetic gene cluster [dkxA (MXAN_4305) encoding a NRPS adenylation domain and dkxO (MXAN_4290) encoding a thioesterase, respectively]. Further analysis of genes possibly involved in DKxanthene biosynthesis is currently under investigation. Additionally, all mutants were compared with the wild type by HPLC-MS analysis for loss of DKX production.
Phenotypic Analysis.
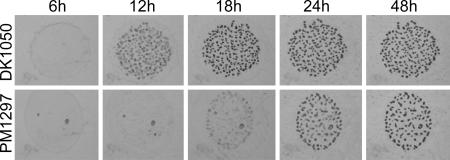
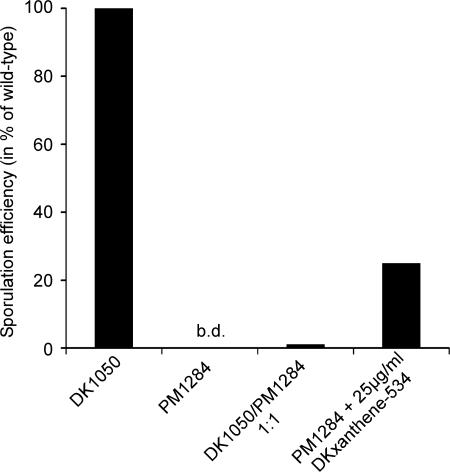
Fruiting body formation of DKX-negative mutants was compared with the parent strain DK1050. Whereas the wild type showed aggregates at 12 h after starvation, PM1297 and all other DKX− mutants were 6–12 h delayed (Fig. 4). However, after 3 d no difference between wild type and mutants was detectable. Fruiting body formation as well as the production of refractile myxospores was observed in both cases. Unexpectedly, when both strains were tested for viable spores after 3 d of development, no colonies could be observed for the mutants. It took up to day 7 until mature spores could be recovered from the mutant strains although at a reduced rate (18% of the wild-type level) indicating either a defect in spore maturation or germination. Only a slight increase in viable spores to 21% and 24% of the wild-type level could be observed for the mutant after 10 and 14 d of development, respectively. Complementation of developmental sporulation within 72 h could be obtained by either codevelopment with the wild type or by supplementation of the mutant with purified DKxanthene-534 (Fig. 5).
Fig. 4.
Fruiting body formation of M. xanthus DK1050 (wild type, DKX+) and PM1297 (dkxO::kan; DKX−) on TPM agar.
Fig. 5.
Sporulation efficiency of M. xanthus DK1050 (wild type, DKX+) and PM1284 (dkxA::kan; DKX−) under different conditions after 72 h of development. b.d., below detection limit (0.0003% of wild-type level).
Interestingly, no difference in the total number of spores could be observed as determined by spore counting during development (data not shown). Furthermore, the mutants were able to form glycerol-induced spores to the same extent as the wild type. In glycerol-induced sporulation cellular differentiation proceeds without the formation of fruiting bodies and spores are observed after 2 h of induction. It could be shown for several developmental mutants that the glycerol-sporulation pathway is independent of defects in aggregation and developmental sporulation (23, 24).
Moreover, no significant difference between wild type and mutant was observed in swarming on hard and soft agar indicative for social and adventurous motility. Similarly, polysaccharide production as tested in the trypan blue assay (25, 26) indicative for defects in extracellular matrix formation as well as trehalose accumulation (27) in the spores thought to be involved in heat and desiccation resistance showed no difference between wild type and mutant (data not shown).
Antioxidant Activity of DKxanthene 534.
The antioxidative activity (EC50) of DKxanthene-534 in organic solvents was determined by using the DPPH (2,2-diphenyl-1-picrylhydrazyl) test (28, 29) as 500 μM. This value is almost 3-fold lower than that of ascorbic acid, which served as a control (175 μM). A similar test in aqueous solution using the N,N-dimethyl-p-phenylenediamine (DMPD) method (30–32) revealed that inhibition of the DMPD-radical formation by DKxanthene-534 increased constantly up to 1 mM of DKxanthene-534 and reached a plateau at 40% inhibition in comparison to a noninhibited sample.
Discussion
The DKxanthenes received their name in honor of Dale Kaiser and because of their yellow color. They are the first new family of secondary metabolites isolated from M. xanthus DK1622 after the recent description of myxochromides and myxovirescins (10, 11), two well known classes of myxobacterial secondary metabolites, which had been described from other myxobacterial species (17, 33).
DKxanthenes have an amphiphilic structure consisting of a hydrophilic (hydroxy)asparagine residue and a hydrophobic polyene chain that biosynthetically starts with a pyrrole ring (Fig. 1) as deduced by extensive NMR and MS experiments (Fig. 2 and Table 1). Eleven members of this new class of compounds have been identified that differ in chain length, number of methyl branches and hydroxylation pattern (Fig. 3 and Table 2). The iterative usage (34, 35) of a methylmalonate- and/or malonate-incorporating module after the assembly of the oxazoline ring might explain the creation of chemical diversity in DKXs that is currently investigated in our group. Due to their polyene chain comprising of five to eight double bonds, DKXs are yellow pigments with absorption maxima around 400 nm. They can be found in almost all Myxococcus strains (D. Krug, P.M., and R.M., unpublished work) where they often represent the dominant coloring principle. Despite their structural novelty, the involvement of these pigments in the myxobacterial life cycle has been described almost 30 years ago. In 1977, Burchard et al. (36) stated that “a better understanding of the Y phenotype and its expression will come with characterization of P379 and its biosynthetic pathway …” M. xanthus DK1622 is able to undergo a phase variation between a yellow (Y) and a tan phenotype and phase variants were proposed to play specific roles in development. Mutants locked in the tan phenotype were reported to be unable to produce mature phase-bright spores but instead form phase-dark, round forms (12, 13).
In this study, we provide evidence that this defect in sporulation is due to the loss of DKX production. The biosynthesis gene cluster for DKxanthene formation was identified by transposon mutagenesis followed by screening for tan mutants and its involvement in DKX biosynthesis was verified by targeted inactivation of the corresponding genes. The biosynthesis gene cluster consists of genes encoding PKSs and NRPSs (data not shown) that are currently analyzed in our group. In contrast to earlier reports about tan phase-locked mutants (12, 13), mutants unable to produce the yellow pigments were still able to build up fruiting bodies and refractile spores (Fig. 4). However, no viable spores can be obtained after 72 h of development, either because they are not truly mature or are not able to germinate (Fig. 5). Because sporulation was observed after 7 d of starvation-induced development to ≈18% of the wild-type level, a much slower maturation of these spores seems to be most likely. To track this effect to the absence of DKX, the mutant strains were supplemented with DKxanthene-534 resulting in restoration of sporulation to ≈25% of the wild-type level. Furthermore, sporulation could be restored by the addition of wild-type cells to mutant cells in a 1:1 ratio leading to a preferential generation of spores derived from mutant cells (ratio of tan:yellow in the spores was 7:3 with overall 1.3% of spores compared with the wild type) as already described for phase-locked mutants (12, 13). The low level of complementation might result from the physical properties and/or cellular localization of the DKXs. With their polar hydroxy asparagine residue and their more lipophilic polyene side chain they very much resemble fatty acids or lipids and therefore are located most likely in the membranes. This is in accordance with the finding that they are not secreted as several other myxobacterial compounds but have been isolated from the cells almost exclusively. However, the exact localization of DKXs remains elusive but assuming a role in sporulation and/or germination a localization to the cytoplasmic membrane seems possible, which would explain the difficulty observed during extracellular complementation.
Structural similarity of the DKxanthenes to polyenes such as the group of carotenoids might indicate their involvement in environmental resistance toward oxidative damage by scavenging free radicals. A similar function has been described for the product of a NRPS biosynthesis gene cluster in Aspergillus fumigatus (37). Disruption of the NRPS encoding gene pes1 resulted in a color change of the conidia from greyish-green to yellow-green and in reduced hydrophobicity and viability of conidia. Furthermore, the Δpes1 conidia were less virulent when injected into insects.
The antioxidative activity of DKX could be shown in two standard test systems. DKXs might also be involved in scavenging oxygen directly in the membrane to protect the cells against fatty acid oxidation or in rigidifying the membranes to maintain the required membrane fluidity (38). However, further experiments are necessary to clarify the function of DKXs during the developmental life cycle of M. xanthus, which will lead to a better understanding of this highly coordinated process and bacterial sporulation in general.
Materials and Methods
Isolation of the Compounds.
M. xanthus DK1050 (39), a stable yellow derivative of M. xanthus FB (40) was cultivated at 30°C in a 300-liter fermenter in MD6 medium (6 g/liter casein-pepton (Marcor)/0.5 g/liter CaCl2·2H2O/2 g/liter MgSO4·7H2O, pH 7.2) with 1% of Amberlite XAD-16 adsorber resin (Fluka). During fermentation the pH was adjusted between 7.0 and 7.4 with 5 M KOH and 50% acetic acid, respectively. Ten grams of behenyl alcohol was added before sterilization and a 1:1 mixture of dodecanol-methanol was pumped into the culture at constant flow-rate (14 ml/d). Cells and XAD were harvested separately after 5 d of cultivation. Analytical extraction of the cells with ethylacetate/methanol (1:4) followed by HPLC-MS analysis revealed that the DKxanthenes reside in the cells rather than being secreted because only traces could be detected in the XAD extract. DKX isolation from cells was performed in the dark whenever possible because early experiments indicated the isomerization or even degradation in the presence of light.
Two hundred grams of the cells was extracted repeatedly with overall 5 liters of ethylacetate/methanol (1:4) until the eluent was colorless. After evaporation of the solvent, the extract was first fractionated by a silica-gel flash chromatography using a stepwise gradient from hexane (fraction 1) to chloroform to methanol to methanol:water 1:1 (fraction 11). DKXs eluted with pure methanol and were further enriched by gel permeation chromatography (Sephadex LH-20 in methanol). Finally, pure compounds were obtained by sequential RP-HPLC (ZORBAX Eclipse XDB-C8; Agilent Technologies; solvent MeOH/H2O 7:3).
Structure Elucidation.
The structures of the main components DKxanthene-534 (10 mg) and DKxanthene-560 (1.2 mg) were elucidated by using 1D (1H, 13C, distortionless enhancement by polarization transfer) and 2D NMR analysis (1H-1H-COSY, heteronuclear sequential quantum correlation, HMBC, NOESY) on Bruker DRX 500 or Bruker Avance 500 spectrometers using different solvents and solvent mixtures ([D6]DMSO, CD3OD, and CD3OD/[D6]benzene). Calculation of theoretical NMR-spectra was performed with ACD/Labs 7.00 software.
High-resolution electrospray ionization MS analysis was performed on a Bruker microTOF after HPLC separation (Agilent 1100 Series) using an acetonitrile:water gradient (5–95% acetonitrile with 0.1% formic acid) and calibration with sodium formiate cluster (m/z 300–900) before and after the analysis.
Additional DKxanthenes could be detected by HPLC-MS (for HPLC, Agilent 1100 Series; for MS, Bruker HCT Plus ion trap), and their structures were determined by a combination of feeding experiments with [13C4,15N]threonine and [D6]propionic acid followed by extensive MS/MS-analysis. A total of 30 mg of [13C4,15N]threonine was fed in three portions after 12, 24, and 36 h of fermentation to DK1050 in 100 ml of CTT medium (41), and cells were harvested after 72 h of fermentation and extracted with methanol. The extract was concentrated to 200 μl of methanol and analyzed by HPLC-MS. Similarly, 37 mg of [D6]propionic acid was fed to a 50-ml CTT culture of DK1050 in three portions and analyzed as described above.
Identification of the DKxanthene Biosynthesis Gene Cluster.
Identification of the DKxanthene biosynthesis gene cluster in DK1050 was accomplished by transposon mutagenesis using the transposon pMycoMar (21) and by targeted gene disruption experiments using standard procedures (22). Briefly, internal fragments of genes dkxA (encoding a NRPS adenylation domain) and dkxO (encoding a thioesterase) were amplified by PCR using primers pcrart1284–1 (5′-GCGTCCGAGCTCCACAAC-3′) and pcrart1284–2 (5′-GCCAATGGGCACCGGCTC-3′) for dkxA and primers pcrart1297–1 (5′-CAGGCGTCGCCTCGTCTC-3′) and pcrart1297–2 (5′-GACGGCCTGGACCACCTG-3′) for dkxO. The PCR constructs were cloned into plasmid pCR2.1-TOPO (Invitrogen), and the resulting plasmids were purified from E. coli TOP10 (Invitrogen) and introduced into M. xanthus DK1050 by electroporation as described (22), leading to kanamycin-resistant DKxanthene negative mutants PM1284 (dkxA::kan; DKX−) and PM1297 (dkxO::kan; DKX−), respectively.
Phenotypic Analyses.
For phenotypic comparison between wild type and mutants, M. xanthus strains were grown in CTT medium (41), and trehalose (42) and fibril content (25) was determined as described previously.
Fruiting body formation in submerged culture and on agar plates (43) of M. xanthus DK1050 and mutant strains PM1284 and PM1297 were conducted as described. Mature sporulation was examined by incubating 2 × 108 cells per ml in MC7-buffer in 24-well microtiter plates (43). After 3, 5, 7, 10, or 14 d of development, respectively, the suspensions were sonicated and incubated at 50°C for 2 h, and dilutions of the suspensions were plated on CTT-agar plates with and without kanamycin. Formation of colonies was observed within the following 2 weeks. Complementation of sporulation deficient mutants was tested under the conditions described above applying two different approaches. (i) 1 × 108 mutants cells were developed in presence of 1 × 108 wild-type (DK1050) cells. In this case, cells were plated on CTT-agar with or without kanamycin to differentiate between wild-type and mutant colonies, because only mutants were resistant against kanamycin. (ii) 2 × 108 mutant cells were developed in the presence of 25 μg/ml of DKxanthene-534, which was added at the beginning of developmental cultivation.
Glycerol induced sporulation (44) was examined by adding glycerol to a final concentration of 0.5 M to vegetative CTT-cultures of wild-type and the mutant strains. Spore formation was controlled microscopically, and spores were plated on CTT-agar after 12 h when complete sporulation was observed.
Antioxidant Activity of DKxanthene-534.
Antioxidative capacity of DKxanthene-534 in methanol was determined by using 2,2-diphenyl-1-picrylhydrazyl (DPPH) (28, 29). Briefly, 2 ml of DPPH (25 mg/l in methanol) was mixed with sample solutions of defined concentrations as well as ascorbic acid as a standard. After 30 min the absorbance at 515 nm was measured, the percentage of remaining DPPH was calculated and the EC50 value (concentration of inhibitor at 50% decrease of DPPH) was determined as described (28).
Antioxidant activity in aqueous solution was determined by using N,N-dimethyl-p-phenylenediamine (DMPD). DKxanthene-534 was used in final concentrations ranging from 0.01 mM to 0.2 mM and mixed with a solution of the stable DMPD+ cation (30–32). After mixing and incubation at room temperature for 10 min, the absorbance at 505 nm was determined. Ascorbic acid was used as a control.
The antioxidant activity of increasing concentrations of DKxanthene-534 was calculated as percentage of the uninhibited radical cation solution (100% value) as described (31).
Supplementary Material
Acknowledgments
We thank F. Sasse, B. Hinkelmann, and H. Schüler for performing large-scale fermentation; J. Zapp for NMR measurements; R. Welch, J. Jakobsen, B. Goldman, the Monsanto Company, and the Institute for Genomic Research for providing access to the M. xanthus genome sequence before publication; and the Deutsche Forschungsgemeinschaft (grants to H.B.B. and R.M.) and the Bundesministerium für Bildung und Forschung (grants to R.M.) for financial support.
Abbreviations
- DKX
DKxanthenes
- HMBC
heteronuclear multiple bond correlation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606039103/DC1.
References
- 1.Kaiser D. Nat Rev Microbiol. 2003;1:45–54. doi: 10.1038/nrmicro733. [DOI] [PubMed] [Google Scholar]
- 2.Dworkin M, Kaiser D. Science. 1985;230:18–24. doi: 10.1126/science.3929384. [DOI] [PubMed] [Google Scholar]
- 3.Reichenbach H, Dworkin M. Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H. The Procaryotes. New York: Springer; 1992. pp. 3416–3487. [Google Scholar]
- 4.Kaiser D. Annu Rev Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 5.Bode HB, Müller R. J Ind Microbiol Biotechnol. 2006;33:577–588. doi: 10.1007/s10295-006-0082-7. [DOI] [PubMed] [Google Scholar]
- 6.Gerth K, Pradella S, Perlova O, Beyer S, Müller R. J Biotechnol. 2003;106:233–253. doi: 10.1016/j.jbiotec.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen J, Ronning CM, Barbazuk WB, Blanchard M, Field C, et al. Proc Natl Acad Sci USA. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 10.Wenzel SC, Meiser P, Binz T, Mahmud T, Müller R. Angew Chem Int Ed. 2006;45:2296–2301. doi: 10.1002/anie.200503737. [DOI] [PubMed] [Google Scholar]
- 11.Simunovic V, Zapp J, Rachid S, Krug D, Meiser P, Müller R. ChemBioChem. 2006;7:1206–1220. doi: 10.1002/cbic.200600075. [DOI] [PubMed] [Google Scholar]
- 12.Laue BE, Gill RE. J Bacteriol. 1994;176:5341–5349. doi: 10.1128/jb.176.17.5341-5349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laue BE, Gill RE. J Bacteriol. 1995;177:4089–4096. doi: 10.1128/jb.177.14.4089-4096.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Vazquez R, Murillo FJ. J Bacteriol. 1984;160:818–821. doi: 10.1128/jb.160.2.818-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths GL, Sigel SP, Payne SM, Neilands JB. J Biol Chem. 1984;259:383–385. [PubMed] [Google Scholar]
- 16.Ong SA, Peterson T, Neilands JB. J Biol Chem. 1979;254:1860–1865. [PubMed] [Google Scholar]
- 17.Trowitzsch Kienast W, Gerth K, Reichenbach H, Höfle G. Liebigs Ann Chem. 1993:1233–1237. [Google Scholar]
- 18.Silakowski B, Nordsiek G, Kunze B, Blöcker H, Müller R. Chem Biol. 2001;8:59–69. doi: 10.1016/s1074-5521(00)00056-9. [DOI] [PubMed] [Google Scholar]
- 19.Steinmetz H, Forche E, Reichenbach H, Höfle G. Tetrahedron. 2000;56:1681–1684. [Google Scholar]
- 20.Staunton J, Weissman KJ. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 21.Sandmann A, Sasse F, Müller R. Chem Biol. 2004;11:1071–1079. doi: 10.1016/j.chembiol.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Bode HB, Ring MW, Schwär G, Kroppenstedt RM, Kaiser D, Müller R. J Bacteriol. 2006;188:6524–6528. doi: 10.1128/JB.00825-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagen DC, Bretscher AP, Kaiser D. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 24.LaRossa R, Kuner J, Hagen D, Manoil C, Kaiser D. J Bacteriol. 1983;153:1394–1404. doi: 10.1128/jb.153.3.1394-1404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black WP, Yang Z. J Bacteriol. 2004;186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold JW, Shimkets LJ. J Bacteriol. 1988;170:5771–5777. doi: 10.1128/jb.170.12.5771-5777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride MJ, Zusman DR. J Bacteriol. 1989;171:6383–6386. doi: 10.1128/jb.171.11.6383-6386.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D, Ou B, Prior RL. J Agric Food Chem. 2005;53:1841–1856. doi: 10.1021/jf030723c. [DOI] [PubMed] [Google Scholar]
- 29.Xie C, Koshino H, Esumi Y, Takahashi S, Yoshikawa K, Abe N. Biosci Biotechnol Biochem. 2005;69:2326–2332. doi: 10.1271/bbb.69.2326. [DOI] [PubMed] [Google Scholar]
- 30.Fiore A, La Fauci L, Cervellati R, Guerra MC, Speroni E, Costa S, Galvano G, De Lorenzo A, Bacchelli V, Fogliano V, Galvano F. Mol Nutr Food Res. 2005;49:1129–1135. doi: 10.1002/mnfr.200500139. [DOI] [PubMed] [Google Scholar]
- 31.Fogliano V, Verde V, Randazzo G, Ritieni A. J Agric Food Chem. 1999;47:1035–1040. doi: 10.1021/jf980496s. [DOI] [PubMed] [Google Scholar]
- 32.Verde V, Fogliano V, Ritieni A, Maiani G, Morisco F, Caporaso N. Free Radical Res. 2002;36:869–873. doi: 10.1080/1071576021000005302. [DOI] [PubMed] [Google Scholar]
- 33.Trowitzsch W, Wray V, Gerth K, Höfle G. Chem Commun. 1982:1340–1341. [Google Scholar]
- 34.Gaisser S, Trefzer A, Stockert S, Kirschning A, Bechthold A. J Bacteriol. 1997;179:6271–6278. doi: 10.1128/jb.179.20.6271-6278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wenzel SC, Kunze B, Höfle G, Silakowski B, Scharfe M, Blöcker H, Müller R. ChemBioChem. 2005;6:375–385. doi: 10.1002/cbic.200400282. [DOI] [PubMed] [Google Scholar]
- 36.Burchard RP, Burchard AC, Parish JH. Can J Microbiol. 1977;23:1657–1662. doi: 10.1139/m77-238. [DOI] [PubMed] [Google Scholar]
- 37.Reeves EP, Reiber K, Neville C, Scheibner O, Kavanagh K, Doyle S. FEBS J. 2006;273:3038–3053. doi: 10.1111/j.1742-4658.2006.05315.x. [DOI] [PubMed] [Google Scholar]
- 38.Gruszecki WI, Strzalka K. Biochim Biophys Acta. 2005;1740:108–115. doi: 10.1016/j.bbadis.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 39.Ruiz-Vazquez R, Murillo FJ. J Bacteriol. 1984;160:818–821. doi: 10.1128/jb.160.2.818-821.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dworkin M. J Bacteriol. 1962;84:250–257. doi: 10.1128/jb.84.2.250-257.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kroos L, Kuspa A, Kaiser D. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 42.Barsch A, Patschkowski T, Niehaus K. Funct Integr Genomics. 2004;4:219–230. doi: 10.1007/s10142-004-0117-y. [DOI] [PubMed] [Google Scholar]
- 43.Jakobsen JS, Jelsbak L, Welch RD, Cummings C, Goldman B, Stark E, Slater S, Kaiser D. J Bacteriol. 2004;186:4361–4368. doi: 10.1128/JB.186.13.4361-4368.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dworkin M, Gibson SM. Science. 1964;146:243–244. doi: 10.1126/science.146.3641.243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.