Abstract
The endosomal sorting complex ESCRT-III, which is formed by the structurally related CHMP proteins, is engaged by HIV-1 to promote viral budding. Here we show that progressive truncations into the C-terminal acidic domains of CHMP proteins trigger an increasingly robust anti-HIV budding activity. Together with biochemical evidence for specific intramolecular interactions between the basic and acidic halves of CHMP3 and CHMP4B, these results suggest that the acidic domains are autoinhibitory. The acidic half of CHMP3 also interacts with the endosome-associated ubiquitin isopeptidase AMSH, and the coexpression of AMSH or its CHMP3-binding domain converts wild-type CHMP3 into a potent inhibitor of HIV-1 release. Point mutations in CHMP3 that prevent binding to AMSH abrogate this effect, suggesting that binding to AMSH relieves the autoinhibition of CHMP3. Collectively, our results indicate that CHMP proteins are regulated through an autoinhibitory switch mechanism that allows tight control of ESCRT-III assembly.
Keywords: AMSH, endosomal sorting machinery, viral budding
By budding through a cellular membrane, HIV-1 acquires a lipid envelope, which ultimately must be severed from the host membrane to allow the formation of extracellular virions. This membrane fission event is facilitated by virus-encoded late assembly (L) domains, which in HIV-1 are found in the C-terminal p6 domain of Gag, the viral protein that drives particle assembly (1). The primary L domain of HIV-1 maps to a highly conserved PTAP motif (2, 3), which binds to Tsg101, a component of the endosomal sorting complex required for transport-I (ESCRT-I) (4–9). Additionally, the HIV-1 p6 domain contains a second L domain that binds AIP1/ALIX, which in turn interacts with both ESCRT-I and the later-acting ESCRT-III sorting complex (10–13).
ESCRT-I mediates the sorting of ubiquitinated cargo into cellular vesicles that bud into the lumen of late endosomes or multivesicular bodies (MVB) and thus, like assembling HIV-1 virions, away from the cytosol (14–16). In yeast, the proper functioning of this endosomal budding pathway requires a series of class E vacuolar protein sorting (Vps) proteins, and mutations that disrupt any of these proteins result in enlarged endosome-derived structures called class E compartments (14, 17). In addition to ESCRT-I, class E Vps proteins form a second cytosolic complex termed ESCRT-II, which acts downstream of ESCRT-I (18). Another set of class E Vps proteins that functions immediately downstream of ESCRT-II is monomeric in the cytosol, but oligomerizes into a large membrane-associated complex called ESCRT-III on recruitment to the cytosolic face of late endosomes (19). Finally, ESCRT-III recruits the ATPase Vps4, which disassembles membrane-associated ESCRT complexes and thus allows further rounds of MVB sorting (19, 20). Catalytic site mutants of Vps4 inhibit not only MVB sorting but also the L domain-mediated budding of HIV-1 and several other enveloped viruses (4, 21–26), strongly supporting the notion that L domains function by engaging the MVB sorting machinery.
The components of ESCRT-III, the charged MVB proteins (CHMPs), belong to a family of coiled coil-containing proteins that is conserved throughout eukaryotic evolution and has six members in yeast and ten members in humans (10, 27). Although the different family members are quite divergent in sequence, they appear to be closely related at the structural level and exhibit a similar charge distribution, with N- and C-terminal halves that are highly enriched in basic and acidic residues, respectively (19, 20, 27, 28). In the case of CHMP3, the N-terminal basic domain has been shown to selectively bind to phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2) (29), a lipid that may regulate the sorting of biosynthetic cargo into MVB (30–32). Based on the highly biased charge distribution of the CHMP proteins, it has been proposed that the binding site for PtdIns(3,5)P2 is masked in soluble CHMP3 through an electrostatic interaction between its N- and C-terminal halves (29).
Interestingly, CHMP proteins that are fused to a bulky tag such as GFP induce class E-like phenotypes and become potent inhibitors of HIV-1 budding (9, 12, 13, 27, 33). One possible explanation for this phenomenon is that a large tag interferes with an inhibitory intramolecular interaction that is necessary to keep CHMP proteins in an inactive soluble form. As predicted by this model, we now report that the anti-HIV-1 budding activity of CHMP proteins can be triggered through the removal of inhibitory regions, and that dominant-negative CHMP proteins interact specifically with their cognate inhibitory domains in trans. Alternatively, CHMP3 can be induced to block HIV-1 release by coexpressing AMSH, a cellular binding partner for its inhibitory domain, suggesting that binding to AMSH relieves the autoinhibition of CHMP3. Our results suggest that intramolecular interactions between the differentially charged halves of CHMP proteins regulate the intermolecular interactions required for the assembly of ESCRT-III.
Results
Truncating the Acidic Domains of CHMP Proteins Triggers Anti-HIV-1 Budding Activity.
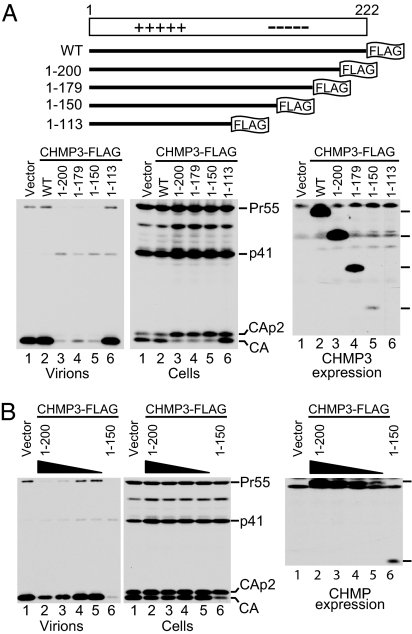
ESCRT-III components such as CHMP3 become potent inhibitors of HIV-1 budding when fused to a bulky tag like GFP or red fluorescent protein (RFP) (10, 12, 13). In contrast, if the relatively short FLAG epitope is fused to CHMP3, the tagged protein has no effect on HIV-1 budding (Fig. 1A). To determine whether FLAG-tagged CHMP3 acquires anti-HIV-1 budding activity once putative autoinhibitory regions are removed, expression vectors for full-length CHMP3 and progressive C-terminal truncation mutants, all tagged at their C-termini with a FLAG epitope, were transfected into 293T cells together with HIV-1 proviral DNA. Although viral particle production was unaffected by full-length CHMP3-FLAG, it was markedly inhibited by variants in which 22, 43, and 72 C-terminal CHMP3 residues were removed (Fig. 1A). The dominant-negative CHMP3 truncation mutants did not affect the overall levels of cell-associated Gag but caused an accumulation of the Gag cleavage intermediates CA-p2 and p41 (Fig. 1A), which is considered a hallmark of late assembly defects (2, 4).
Fig. 1.
Truncating the acidic domain of CHMP3 triggers anti-HIV-1 budding activity. (A) Schematic representation of FLAG-tagged CHMP3 constructs and their effects on HIV-1 particle production. 293T cells were transfected with full-length HIV-1 proviral DNA (1.5 μg) and vectors expressing full-length or truncated CHMP3-FLAG (500 ng). Virion production and Gag expression levels were compared by Western blotting with anti-CA serum, and CHMP3 expression levels were compared by Western blotting with anti-FLAG. (B) CHMP31–200FLAG inhibits HIV-1 particle production less potently than CHMP31–150FLAG when expressed at a comparable level. 293T cells were transfected with proviral DNA (1.5 μg) and expression vectors for CHMP31–200FLAG (100, 50, 25, or 12.5 ng) or CHMP31–150FLAG (500 ng).
In the experiment shown in Fig. 1A, CHMP31–150FLAG inhibited the release of HIV-1 virions as well as CHMP31–200FLAG, but was expressed at much lower levels. Therefore, the vector encoding CHMP31–200FLAG was titrated down to obtain more comparable expression levels (Fig. 1B). Assuming that the FLAG tag was equally accessible on both proteins, this experiment indicated that CHMP31–150FLAG is significantly more potent in inhibiting HIV-1 release than CHMP31–200FLAG.
Because of the strong dominant-negative effect of CHMP31–150FLAG on HIV-1 particle production, we opted to truncate CHMP4B and CHMP2B at an analogous position. Although full-length CHMP4B-FLAG had no effect on HIV-1 virion release or on the processing of Gag, CHMP4B1–153FLAG affected both and inhibited HIV-1 particle production about as much as CHMP31–150FLAG [see supporting information (SI) Fig. 6]. Furthermore, CHMP2B1–153FLAG also inhibited HIV-1 virion production as potently as CHMP31–150FLAG (data not shown). Thus, all three CHMP proteins examined acquired anti-HIV-1 budding activity after truncation of their acidic C-terminal halves.
It has recently been shown that the depletion of CHMP3 retards the degradation of the epidermal growth factor receptor (EGFR) in HeLa cells stimulated with EGF (34). In contrast, we did not observe an evident effect on the ligand-induced degradation of the EGFR in HeLa cells stably transduced with a vector expressing CHMP31–150FLAG in a tetracycline-inducible manner (see SI Fig. 7). However, this does not entirely exclude effects of CHMP31–150FLAG on MVB sorting because, under certain conditions, the EGFR can be efficiently targeted to lysosomes even if it is not sorted into intralumenal MVB vesicles (35).
The Basic N-Terminal Half of CHMP3 Is Sufficient to Inhibit HIV-1 Particle Production.
CHMP31–113FLAG did not inhibit virus production and was not detectably expressed when 0.5 μg of the expression vector were used (Fig. 1A). However, at 2.5 μg, the CHMP31–113FLAG vector clearly inhibited HIV-1 release (see SI Fig. 8A). Although the effect of CHMP31–113FLAG remained somewhat less pronounced than that of CHMP31–150FLAG, it was also expressed at a lower level (SI Fig. 8A). Furthermore, whereas full-length CHMP3-FLAG was diffusely distributed throughout the cell and CHMP31–150FLAG predominantly associated with the plasma membrane, CHMP31–113FLAG appeared concentrated in the nucleus (SI Fig. 8B). To rule out passive diffusion of CHMP31–113 through the nuclear pore complex, we attached its C terminus to one or two tandem copies of RFP. CHMP31–113RFP and CHMP31–1132RFP were both excluded from the nucleus and instead localized to punctate perinuclear structures (SI Fig. 8B). Although CHMP31–113RFP and CHMP31–1132RFP remained poorly expressed compared with authentic RFP, this low level of expression was sufficient to block HIV-1 particle production (SI Fig. 8C). Given that CHMP31–150FLAG and CHMP31–113RFP both profoundly inhibited HIV-1 particle production, their different subcellular localization is surprising and may indicate that the accumulation at a specific membrane compartment is not crucial for the antiviral effect.
CHMP Basic Domains Interact Specifically with Their Cognate Acidic Domains.
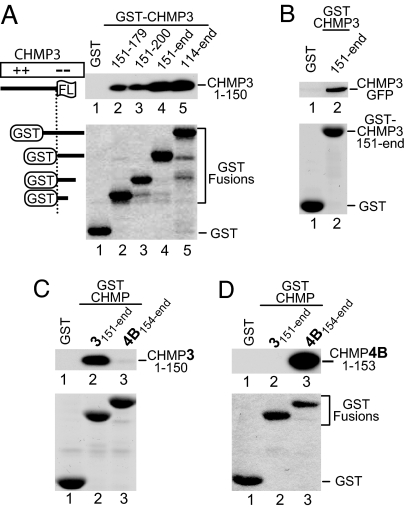
The profound effect of N-terminal CHMP fragments on HIV-1 budding suggested that the truncation mutants might be in a constitutively active state because of the loss of an autoinhibitory region. Because this model predicts a physical interaction between the N-terminal CHMP fragments and their cognate autoinhibitory regions, we fused the acidic C-terminal half of CHMP3 or portions thereof to GST. We observed that GST-CHMP3151–end, but not GST alone, pulled down the dominant-negative proteins CHMP31–150FLAG and CHMP3-GFP (Fig. 2A and B) but not CHMP3-FLAG (see Fig. 3), which does not inhibit HIV-1 budding. Further analysis revealed that even a 29-amino acid peptide from the acidic domain of CHMP3 (residues 151–179) was sufficient to pull down CHMP31–150FLAG, albeit with lower efficiency (Fig. 2A, lane 2). Except for GST-CHMP3114–end, none of the GST fusion proteins tested overlapped with CHMP31–150FLAG (Fig. 2A), providing evidence for an intramolecular interaction within CHMP3.
Fig. 2.
Dominant-negative CHMP truncation mutants specifically bind to nonoverlapping fragments from their cognate acidic domains. (A) Binding of dominant-negative CHMP31–150FLAG to CHMP3 acidic domain fragments. Beads decorated with the indicated GST fusion proteins or GST were incubated with lysates from 293T cells expressing CHMP31–150FLAG. The levels of the GST fusion proteins on the beads were monitored by SDS/PAGE and Coomassie staining. Captured CHMP31–150FLAG was detected by Western blotting with anti-FLAG antibody. (B) GST pull-down showing that full-length CHMP3 fused to GFP binds to the acidic domain fragment CHMP3151–end. (C) GST pull-down showing that dominant-negative CHMP31–150FLAG binds to the nonoverlapping CHMP3151–end fragment but not to the corresponding region of CHMP4B. (D) Conversely, dominant-negative CHMP4B1–153FLAG binds to CHMP4B154–end but not to the corresponding region of CHMP3.
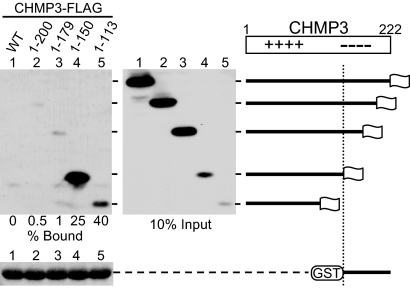
Fig. 3.
Binding of CHMP3 truncation mutants to a C-terminal acidic domain fragment correlates with anti-HIV-1 budding activity. Beads decorated with GST-CHMP3151–end were incubated with lysates from 293T cells expressing full-length CHMP3-FLAG (WT) or the indicated truncation mutants. Captured proteins and total cell lysates representing 10% of the input were analyzed by Western blotting with anti-FLAG antibody, and the amount of GST fusion protein in each of the pull-down samples was compared by Coomassie staining. The percentage of input protein bound is shown.
To examine the specificity of the interaction, we compared the ability of dominant-negative CHMP31–150FLAG to bind to its cognate acidic domain versus that of CHMP4B. As shown in Fig. 2C, dominant-negative CHMP31–150FLAG bound far better to GST-CHMP3151–end than to GST-CHMP4B154–end. Conversely, dominant-negative CHMP4B1–153FLAG was almost quantitatively recovered by GST-CHMP4B154–end but did not bind to GST-CHMP3151–end (Fig. 2D). These results indicate that the N-terminal basic domains of CHMP3 and CHMP4B interact preferentially with their cognate acidic domains.
Accessibility of the Binding Site for the Acidic Domain Correlates with Antiviral Activity of CHMP3 Mutants.
As a measure for the relative exposure of the binding site for the acidic domain, we directly compared the ability of full-length CHMP3 and of different C-terminal truncation mutants to bind GST-CHMP3151–end. Full-length CHMP3-FLAG was not detectably pulled down by GST-CHMP3151–end (Fig. 3, lane 1), even though CHMP3-FLAG was expressed at very high levels compared with CHMP31–150FLAG or CHMP31–113FLAG. This result suggests that the binding site in wild-type (WT) CHMP3 is fully occupied by its autoinhibitory domain, which prevents CHMP3151–end from binding in trans.
CHMP31–200FLAG and CHMP31–179FLAG, which exhibited lower specific activity in inhibiting HIV-1 release than CHMP31–150FLAG (Fig. 1), both bound only poorly to CHMP3151–end (Fig. 3, lanes 2 and 3). Under the same conditions, GST-CHMP3151–end bound significantly better to the strongly dominant-negative CHMP31–150FLAG mutant (Fig. 3, lane 4). Indeed, between 25 and 47% of the input CHMP31–150FLAG was pulled down in independent experiments. The isolated basic domain (CHMP31–113FLAG) was pulled down even more efficiently, with a recovery rate between 40 and 80% (Fig. 3 and data not shown). These results suggest that the strongly dominant-negative CHMP3 truncation mutants assumed a relatively open conformation that allowed efficient binding to the acidic inhibitory domain in trans.
Full-Length CHMP3 Blocks HIV-1 Particle Production When Overexpressed Together with its Binding Partner AMSH.
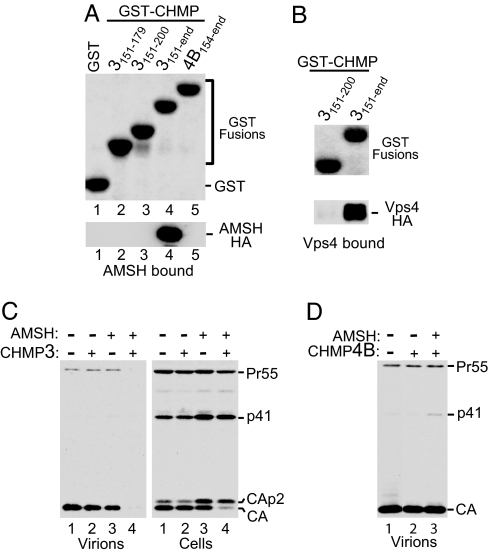
A large-scale two-hybrid screen for protein interactions in Drosophila suggested that CHMP3 interacts with AMSH (36), an endosome-associated deubiquitinating enzyme (37). Based on these data, we performed GST pull-down assays with the human proteins and found that AMSH binds strongly to the C-terminal 72 residues of CHMP3 but not to the corresponding region of CHMP4B (Fig. 4A, lanes 4 and 5). Consistent with this finding, it was recently reported that CHMP3 interacts both with full-length AMSH and with an N-terminal fragment lacking most of the JAMM (Jab1/MPN/Mov34) domain involved in deubiquitination (38).
Fig. 4.
AMSH binds to the acidic domain of CHMP3 and blocks HIV-1 release when coexpressed with WT CHMP3. (A) GST pull-down showing that AMSH-HA binds to a C-terminal region of CHMP3 but not to the corresponding region of CHMP4B. (B) GST pull-down showing that Vps4A binds to a C-terminal region of the CHMP3 acidic domain. (C) GFP-AMSH(D348A) blocks HIV-1 release together with WT CHMP3 but not if expressed alone. 293T cells were cotransfected with 1.5 μg of HIV-1 proviral DNA and expression vectors (500 ng each) for native CHMP3, GFP-AMSH(D348A), or both, along with empty vector controls as appropriate. Virus pellets and cell lysates were analyzed by Western blotting with anti-CA serum. (D) GFP-AMSH(D348A) does not block HIV-1 release when coexpressed with CHMP4B. The experiment was performed as in C.
Because ubiquitin has been implicated in retroviral budding (39), we examined whether an active site mutant of AMSH affects HIV-1 release. It has been shown previously that a point mutation (D348A) in the JAMM domain of AMSH ablates its ubiquitin isopeptidase activity in vitro and that the expression of inactive GFP-AMSH(D348A) in human cells induces the accumulation of ubiquitin on endosomes (37). When an expression vector for GFP-AMSH(D348A) was cotransfected with the full-length HIV-1 clone HXBH10, we observed an increase in the Gag processing intermediate CA-p2 relative to mature CA in the transfected cells (Fig. 4C, lane 3). However, this effect was relatively moderate, and HIV-1 virion production was at most slightly reduced (Fig. 4C). In contrast, HIV-1 particle production was almost completely blocked when the vector expressing GFP-AMSH(D348A) was cotransfected with an expression vector for full-length untagged CHMP3 (Fig. 4C, lane 4). As expected, the expression vector for native CHMP3 by itself had no effect on HIV-1 particle production (Fig. 4C, lane 2). Furthermore, HIV-1 particle production was unaffected when GFP-AMSH(D348A) was coexpressed with CHMP4B (Fig. 4D), which does not bind to AMSH (Fig. 4A).
Inhibition of Particle Production by WT CHMP3 Depends on the Ability to Interact with AMSH.
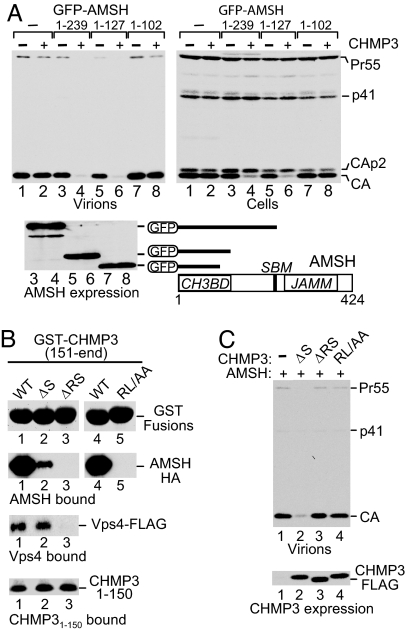
In addition to the C-proximal JAMM domain, AMSH contains a binding site for the class E Vps protein STAM (40). To determine the contribution of these features to the inhibition of HIV-1 budding in combination with CHMP3, we tested the effects of GFP-AMSH truncation mutants. An N-terminal region of AMSH (residues 1–239) that lacked the JAMM motif but contained a functional binding site for STAM (40) retained the ability to profoundly inhibit the release of HIV-1 virions when overexpressed together with CHMP3 (Fig. 5A, lane 4) but, by itself, had no effect on virus production (lane 3). A shorter N-terminal AMSH fragment (residues 1–127) that lacked both the JAMM motif and the STAM binding site inhibited HIV-1 particle production as well as AMSH1–239 but again only when coexpressed with CHMP3 (Fig. 5A, lanes 5 and 6). In contrast, the combination of AMSH1–102 and CHMP3 did not inhibit HIV-1 release (Fig. 5A, lane 8). Furthermore, when expressed together with exogenous CHMP3, AMSH1–239, and AMSH1–127, but not AMSH1–102, caused the inversion of the CA to CA-p2 ratio that is typical for virus release defects (Fig. 5A Right). Interestingly, in GST pull-down assays AMSH1–127 bound CHMP3 nearly as efficiently as full-length AMSH, whereas AMSH1–102 showed no detectable association with CHMP3 (Y. M. Ma and T. Kirchhausen, personal communication). Taken together, these results indicate that the N-terminal CHMP3-binding region of AMSH is both necessary and sufficient to trigger the inhibition of HIV-1 release in conjunction with CHMP3.
Fig. 5.
Inhibition of HIV-1 release by full-length CHMP3 is triggered by the CHMP3-binding domain of AMSH and abrogated by point mutations in the AMSH-binding site of CHMP3. (A) The N-terminal CHMP3-binding domain (CH3BD) of AMSH is both necessary and sufficient to trigger the inhibition of HIV-1 release by WT CHMP3. 293T cells were cotransfected with 1.5 μg of HIV-1 proviral DNA and expression vectors for GFP-AMSH truncation mutants and WT CHMP3, or with the appropriate empty vectors (2.5 μg each). HIV-1 Gag and GFP-AMSH fusions were detected by Western blotting with anti-CA and anti-GFP serum, respectively. SBM, STAM-binding motif. (B) GST pull-down showing that C-terminal CHMP3 residues are essential for binding to AMSH-HA and Vps4A-HA but not to CHMP31–150FLAG. (C) Mutations that abrogate binding to AMSH also abrogate the ability of CHMP3 to inhibit HIV-1 release in the presence of GFP-AMSH(D348A). 293T cells were cotransfected with 1.5 μg of HIV-1 proviral DNA and expression vectors (500 ng each) for GFP-AMSH(D348A) and WT or mutant CHMP3-FLAG. HIV-1 Gag and CHMP3-FLAG mutants were detected by Western blotting with anti-CA serum and anti-FLAG antibody, respectively.
Because the last 22 residues of CHMP3 are required to bind both AMSH (Fig. 4A) and Vps4A (Fig. 4B), we tested the contribution of a heptad repeat motif at the very C terminus of CHMP3 (residues 210–222). Replacing Arg216 and Leu217 in the middle of this motif with alanine residues abolished binding of GST-CHMP3151–end to AMSH-HA (RL/AA; Fig. 5B). The interaction with AMSH was also abolished by removing the last two CHMP3 residues (ΔRS), whereas the removal of only the C-terminal Ser (ΔS) was not sufficient to completely prevent the interaction (Fig. 5B). Similarly, the ΔRS but not the ΔS truncation largely eliminated the interaction of GST-CHMP3151–end with Vps4A (Fig. 5B), shown previously to bind to the C-terminal domains of CHMP proteins (41). In contrast, neither mutation affected binding to the N-terminal CHMP31–150FLAG fragment (Fig. 5B). Furthermore, neither mutation converted CHMP3-FLAG by itself into an inhibitor of HIV-1 budding (data not shown), indicating that the disruption of the C-terminal Vps4 binding site is not sufficient to confer a dominant-negative phenotype. Interestingly, the inhibition of HIV-1 particle production by CHMP3-FLAG in the presence of GFP-AMSH(D348A) was abolished by the RL/AA and ΔRS mutations but not by the ΔS mutation (Fig. 5C). Thus, the inhibitory effect is strictly dependent on the ability of CHMP3 to interact with AMSH, supporting a model in which the binding to AMSH is required to relieve the autoinhibition of CHMP3.
Discussion
The results show that CHMP proteins can be converted into strong dominant-negative inhibitors of HIV-1 budding through deletions into their acidic C-terminal halves. This behavior suggested that the acidic halves function as autoinhibitory modules that repress the WT proteins through intramolecular interactions with their N-terminal basic halves. Indeed, we found that the isolated basic domain of CHMP3 interacts avidly with a fragment from the acidic domain in GST pull-down assays. Further support for an intramolecular interaction is provided by the observation that the acidic domain fragment interacted with dominant-negative CHMP3 truncation mutants but not with full-length CHMP3. This observation suggests that in the native molecule the binding site is fully occupied by the intact autoinhibitory domain and becomes accessible only if at least a portion of the autoinhibitory domain is removed. Of note, the apparent affinity of the CHMP3 truncation mutants for the acidic domain fragment correlated with their potential to act as inhibitors of HIV-1 budding. We infer that the weakly dominant-negative mutants still retained part of the autoinhibitory domain, which prevented efficient binding to autoinhibitory sequences in trans.
Additionally, we find that even native CHMP3 can be converted into a strong inhibitor of HIV-1 budding by coexpressing it with a catalytically inactive mutant of AMSH, an ubiquitin isopeptidase that interacts with CHMP3 and other components of the MVB sorting machinery (37, 38, 42). HIV-1 budding was also inhibited by coexpressing CHMP3 with WT AMSH, albeit somewhat less efficiently (data not shown). We speculate that this is because the in vivo association with CHMP3 is stabilized if AMSH cannot complete its catalytic cycle. Accordingly, the small inhibitory effect of the AMSH active site mutant when expressed alone may be attributable to its relatively stable association with endogenous CHMP3.
AMSH binds tightly to a C-terminal portion of the CHMP3 acidic domain that is also required for Vps4 binding. However, it is unlikely that AMSH inhibited HIV-1 budding simply by displacing Vps4, because a CHMP3 mutant with a disrupted Vps4-binding site had no effect on HIV-1 release. Another possibility is that CHMP3 became sequestered or relocalized on AMSH overexpression, thus preventing its interaction with other components of ESCRT-III. However, this model does not explain why the effect of AMSH overexpression on HIV-1 release was minor when CHMP3 was expressed at endogenous levels and increased considerably when CHMP3 was also overexpressed. We thus favor a model in which the intermolecular interaction with AMSH disrupts an intramolecular interaction that prevents the spontaneous oligomerization of CHMP3 with itself and other members of the CHMP protein family. As predicted by this model, the N-terminal CHMP3 binding domain of AMSH was both necessary and sufficient to inhibit HIV-1 release when coexpressed with native CHMP3 but had no effect when expressed alone. Furthermore, as expected, the inhibition of HIV-1 particle production was abrogated by point mutations at the C terminus of CHMP3 that prevented binding to AMSH. Nevertheless, we cannot exclude that the overexpression of AMSH together with CHMP3 inhibited HIV-1 budding by a different mechanism than the CHMP3 truncation mutants.
In addition to CHMP3, at least two other CHMP family members can be activated to inhibit HIV-1 budding by truncating their C-terminal acidic domains. Because the 10 human CHMP proteins share a similar structural organization (27), it appears likely that all are regulated through autoinhibitory interactions. These interactions do not appear to be driven solely by electrostatic interactions because our results indicate that the interactions between cognate basic and acidic domains are highly specific. One attractive hypothesis is that the autoinhibitory domains bind to and mask residues on the cognate N-terminal basic domains that are involved in CHMP protein oligomerization and ESCRT-III assembly. This situation would be analogous to the regulation of SNARE complex assembly through an inhibitory intramolecular interaction between the N- and C-terminal domains of syntaxins, which competes with the intermolecular interaction between the C-terminal domain and other SNAREs (43, 44).
Although HIV-1 utilizes components of the same cellular machinery that mediates the budding of cellular vesicles into MVB, HIV-1 budding may not be absolutely dependent on an intact MVB sorting pathway. Hanson and coworkers (33) recently reported that both full-length CHMP4B and its N-terminal basic half impaired MVB function and induced the formation of enlarged vacuolar structures. In agreement with this study, we observed that FLAG-tagged full-length CHMP4B induced an accumulation of enlarged perinuclear vacuoles reminiscent of the class E compartment induced by dominant-negative Vps4 (data not shown). However, full-length CHMP4B had no inhibitory effect on HIV-1 release. These observations imply that the profound inhibition of HIV-1 particle production by CHMP proteins with defective autoinhibitory domains is not simply a consequence of a dysfunctional MVB sorting pathway, and they provide a framework for further studies aimed at understanding how these proteins affect HIV-1 budding.
Materials and Methods
Constructs.
DNAs encoding full-length human CHMP3 or residues 1–200, 1–179, 1–150, or 1–113 with a C-terminal FLAG epitope were amplified by using EST clone BC004419 as a template and inserted into the expression vector pBJ5. CHMP4B-FLAG in pBJ5 has been described (13). DNAs encoding CHMP4B residues 1–153, full-length CHMP2B, and CHMP2B residues 1–153 with a C-terminal FLAG tag were amplified from BC011429 and BC001553 (Open Biosystems, Huntsville, AL) and inserted into pBJ5. The DNA encoding CHMP3 residues 1–113 was also inserted into pDsRed2-N1 (Clontech, Palo Alto, CA) to generate CHMP31–113RFP. The coding sequence for CHMP31–113RFP was then amplified and inserted in frame between the XhoI and AgeI sites of pDsRed2-N1, yielding CHMP31–1132RFP. The coding sequence for AMSH with a C-terminal HA tag was amplified from BC007682 and cloned into pcDNA3.1. The GFP-AMSH(D348A) mutant was made by using the QuikChange mutagenesis strategy (Stratagene, La Jolla, CA). The upstream primer for each PCR product also provided a Kozak sequence. For GST fusions, DNAs encoding CHMP3 residues 1–222 (full-length), 114–end, 151–end, 151–221 (ΔS), 151–220 (ΔRS), 151–200, 151–179, and CHMP4B residues 154–end were subcloned into pGEX-4T-1 (Amersham Biosciences, Piscataway, NJ).
Analysis of Viral Particle Production.
293T cells (1.2 × 106) were seeded into T25 flasks and transfected 24 h later by using a calcium phosphate precipitation technique. The cultures were transfected with 1.5 μg of HXBH10, which encodes WT HIV-1, together with expression vectors for WT and mutant CHMP proteins and/or AMSH, or the appropriate empty vectors (0.5 μg each, unless indicated otherwise). The total amount of transfected DNA was brought to 8 μg with carrier DNA (pBluescript; Stratagene). Twenty-four hours after transfection, the cells were lysed by boiling in SDS/PAGE sample buffer, and the culture supernatants were clarified by low-speed centrifugation and passaged through 0.45-μm filters. Virions released into the medium were then pelleted through 20% sucrose cushions and analyzed as described (45) by SDS/PAGE and Western blotting with a rabbit anti-HIV CA serum (Advanced Biotechnologies, Columbia, MD). Proteins in the cell lysates were detected by Western blotting with rabbit anti-HIV CA serum, anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO), rabbit anti-DsRed serum (Clontech), or rabbit anti-GFP serum (Molecular Probes, Carlsbad, CA).
GST Pull-Down Assay.
GST fusion proteins were expressed in Escherichia coli strain BL21 and immobilized on glutathione-Sepharose beads (Amersham Biosciences). The beads were then incubated for 2 h at 4°C with hypotonic lysates of 293T cells transiently expressing epitope-tagged proteins, followed by extensive washing in PBS. Bound proteins were eluted by boiling in SDS/PAGE sample buffer and resolved by SDS/PAGE. GST fusion proteins were visualized with colloidal Coomassie Brilliant Blue G-250, and tagged proteins were detected by Western blotting.
Immunofluorescence Microscopy.
HeLa cells were seeded into glass-bottom dishes and transfected by a calcium phosphate precipitation technique with 1 μg of DNA. Twenty-four hours after transfection the cells were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 5% normal goat serum, and incubated with anti-FLAG M2 followed by FITC-conjugated secondary antibody. Images were acquired on a Nikon (Melville, NY) TE300 inverted microscope equipped with a cooled CCD camera. RFP fusion proteins were visualized directly with a Leica (Deerfield, IL) TCS SP2 laser-scanning confocal microscope.
Supplementary Material
Acknowledgments
We thank Drs. Yu May Ma and Tomas Kirchhausen (Harvard Medical School, Boston, MA) for providing GFP-AMSH constructs, Tsuyoshi Akagi (Osaka Bioscience Institute, Osaka, Japan) for pCXneo/TR-2 and pC4bsrR(TO), and Winfried Weissenhorn for helpful discussions. A.Z. and G.P. were supported by the Ministero dell'Università e della Ricerca Scientifica e Tecnologia. This work was supported by National Institutes of Health Grants AI29873 and AI50466 (to H.G.).
Abbreviations
- ESCRT
endosomal sorting complex required for transport
- L
late assembly
- MVB
multivesicular bodies
- RFP
red fluorescent protein
- Vps
vacuolar protein sorting
- WT
wild-type.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0603788103/DC1.
References
- 1.Freed EO. J Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang M, Orenstein JM, Martin MA, Freed EO. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 5.Demirov DG, Ono A, Orenstein JM, Freed EO. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Serrano J, Zang T, Bieniasz PD. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 7.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Serrano J, Zang T, Bieniasz PD. J Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stuchell MD, Garrus JE, Muller B, Stray KM, Ghaffarian S, McKinnon R, Krausslich HG, Morham SG, Sundquist WI. J Biol Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 10.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Serrano J, Bieniasz PD. J Virol. 2003;77:12373–12377. doi: 10.1128/JVI.77.22.12373-12377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Proc Natl Acad Sci USA. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 14.Katzmann DJ, Odorizzi G, Emr SD. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 15.Gruenberg J, Stenmark H. Nat Rev Mol Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 16.Babst M. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 17.Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 19.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 20.Babst M, Wendland B, Estepa EJ, Emr SD. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottwein E, Bodem J, Muller B, Schmechel A, Zentgraf H, Krausslich HG. J Virol. 2003;77:9474–9485. doi: 10.1128/JVI.77.17.9474-9485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shehu-Xhilaga M, Ablan S, Demirov DG, Chen C, Montelaro RC, Freed EO. J Virol. 2004;78:724–732. doi: 10.1128/JVI.78.2.724-732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop N, Woodman P. Mol Biol Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakaguchi T, Kato A, Sugahara F, Shimazu Y, Inoue M, Kiyotani K, Nagai Y, Yoshida T. J Virol. 2005;79:8933–8941. doi: 10.1128/JVI.79.14.8933-8941.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. J Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. J Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard TL, Stauffer DR, Degnin CR, Hollenberg SM. J Cell Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- 28.Amerik AY, Nowak J, Swaminathan S, Hochstrasser M. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitley P, Reaves BJ, Hashimoto M, Riley AM, Potter BV, Holman GD. J Biol Chem. 2003;278:38786–38795. doi: 10.1074/jbc.M306864200. [DOI] [PubMed] [Google Scholar]
- 30.Odorizzi G, Babst M, Emr SD. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 31.Efe JA, Botelho RJ, Emr SD. Curr Opin Cell Biol. 2005;17:402–408. doi: 10.1016/j.ceb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Ikonomov OC, Sbrissa D, Mlak K, Kanzaki M, Pessin J, Shisheva A. J Biol Chem. 2002;277:9206–9211. doi: 10.1074/jbc.M108750200. [DOI] [PubMed] [Google Scholar]
- 33.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 34.Bache KG, Stuffers S, Malerod L, Slagsvold T, Raiborg C, Lechardeur D, Walchli S, Lukacs GL, Brech A, Stenmark H. Mol Biol Cell. 2006;17:2513–2523. doi: 10.1091/mbc.E05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Futter CE, Collinson LM, Backer JM, Hopkins CR. J Cell Biol. 2001;155:1251–1264. doi: 10.1083/jcb.200108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 37.McCullough J, Clague MJ, Urbe S. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCullough J, Row PE, Lorenzo O, Doherty M, Beynon R, Clague MJ, Urbe S. Curr Biol. 2006;16:160–165. doi: 10.1016/j.cub.2005.11.073. [DOI] [PubMed] [Google Scholar]
- 39.Vogt VM. Proc Natl Acad Sci USA. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato M, Miyazawa K, Kitamura N. J Biol Chem. 2000;275:37481–37487. doi: 10.1074/jbc.M007251200. [DOI] [PubMed] [Google Scholar]
- 41.Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI. Proc Natl Acad Sci USA. 2005;102:13813–13818. doi: 10.1073/pnas.0502165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka N, Kaneko K, Asao H, Kasai H, Endo Y, Fujita T, Takeshita T, Sugamura K. J Biol Chem. 1999;274:19129–19135. doi: 10.1074/jbc.274.27.19129. [DOI] [PubMed] [Google Scholar]
- 43.Munson M, Chen X, Cocina AE, Schultz SM, Hughson FM. Nat Struct Biol. 2000;7:894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- 44.Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Nat Struct Biol. 1998;5:793–802. doi: 10.1038/1834. [DOI] [PubMed] [Google Scholar]
- 45.Accola MA, Strack B, Göttlinger HG. J Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.