Abstract
Type II protein secretion is critical for Legionella pneumophila infection of amoebae, macrophages, and mice. Previously, we found several enzymes to be secreted by this (Lsp) secretory pathway. To better define the L. pneumophila type II secretome, a 2D electrophoresis proteomic approach was used to compare proteins in wild-type and type II mutant supernatants. We identified 20 proteins that are type II-dependent, including aminopeptidases, an RNase, and chitinase, as well as proteins with no homology to known proteins. Because a chitinase had not been previously reported in Legionella, we determined that wild type secretes activity against both p-nitrophenyl triacetyl chitotriose and glycol chitin. An lsp mutant had a 70–75% reduction in activity, confirming the type II dependency of the secreted chitinase. Newly constructed chitinase (chiA) mutants also had ≈75% less activity, and reintroduction of chiA restored the mutants to normal levels of activity. Although chiA mutants were not impaired for in vitro intracellular infection, they were defective upon intratracheal inoculation into the lungs of A/J mice, and antibodies against ChiA were detectable in infected animals. In contrast, mutants lacking a secreted phosphatase, protease, or one of several lipolytic enzymes were not defective in vivo. In sum, this study shows that the output of type II secretion is greater in magnitude than previously appreciated and includes previously undescribed proteins. Our data also indicate that an enzyme with chitinase activity can promote infection of a mammalian host.
Keywords: bacterial protein secretion, bacterial virulence, Legionnaires' disease
Legionella pneumophila is the etiological agent of Legionnaires' disease, a potentially fatal pneumonia that especially affects immunocompromised individuals (1). As an environmental pathogen, this Gram-negative bacterium exists in fresh waters, in protozoan hosts, and as a part of biofilms. It is also present in man-made systems, such as potable water systems and cooling towers. Transmission occurs when aerosolized bacteria are inhaled. L. pneumophila colonizes the respiratory tract, where it invades alveolar macrophages. Bacterial degradative enzymes, host cell lysis, and excessive inflammation combine to promote damage to lung tissue. We have shown that a type II protein secretion system is important in the ecology and pathogenesis of L. pneumophila (2). Present in many but not all gram negatives, including animal and plant pathogens (2), type II secretion is a two-step process in which nascent proteins are first translocated across the inner membrane by the Sec or Tat pathway and then exit the cell through an outer membrane secretin (3). L. pneumophila type II secretion pathway (Lsp) mutants display a growth defect in amoebae, macrophages, and the lungs of mice (4–6). L. pneumophila is the only intracellular pathogen shown to possess a functional type II system. Other phenotypes ascribed to L. pneumophila type II mutants include reduced growth at low temperatures, altered colony morphology, and diminished invasion of host cells (6–9). Thus far, proteins secreted through the L. pneumophila type II system have been first identified based on the detection of enzymatic activities in culture supernatants, including a metalloprotease, acid phosphatases, lipases, phospholipase A, phospholipase C, lysophospholipase A, cholesterol acyltransferase, and ribonuclease (4–6, 10–15). Both the Sec and Tat pathways are implicated in the processing of L. pneumophila type II substrates (16). Because mutants lacking individual type II effectors infect normally (10, 12–14, 17, 18), we hypothesize that more proteins are secreted by Lsp, and that one or more of them are virulence determinants. Although L. pneumophila has been sequenced (19, 20), it is not possible to readily identify effectors by bioinformatics, because there is no motif that specifically defines a protein as a type II substrate (3). Thus, we embarked on a 2D electrophoresis (2DE)-based proteomic approach to more fully define the scope of L. pneumophila type II secretion. This effort has revealed a large set of effectors, including previously undescribed exoproteins and a chitinase, which remarkably promotes bacterial persistence in the lungs.
Results
Analysis of the L. pneumophila Type II Secretome.
To further define the proteins secreted by Lsp, we analyzed proteins in wild-type and lsp mutant supernatants by 2DE and then used mass spectrometry to obtain the identity of the secreted proteins. Twenty-seven proteins present in wild-type strain 130b supernatants but absent or greatly reduced in supernatants of the NU275 lspF mutant were identified [Table 1; 2D gel images appear as supporting information (SI) Fig. 4]. Three of these, the ProA metalloprotease, PlaA lysophospholipase A, and Map acid phosphatase, were defined as type II exoproteins before (Table 1; refs. 4–6, 9, 12, and 21). A fourth protein was identified as a ribonuclease, undoubtedly representing the type II-dependent ribonuclease activity that we had reported before, but for which the gene had been unknown (Table 1; refs. 6 and 11). Of the remaining 23 proteins, 13 were also predicted to contain a signal sequence and likely represent new type II-dependent exoproteins (Table 1). Two of these were annotated in the database as a leucine aminopeptidase and a chitinase. Four others were annotated as hypothetical proteins having similarities to bacterial enzymes and included an amidase, aminopeptidase, cysteine protease, and endoglucanase. Two others had similarities to eukaryotic proteins; i.e., spot 27 had a protein with collagen-like repeats and spot 21 a protein with greatest homology to a eukaryotic zinc proteinase. Five other proteins were annotated as hypothetical with no similarities to any known protein or domain in the database and therefore may represent novel activities. As noted in Table 1, several of the type II exoproteins, including the leucine aminopeptidase, some of the hypothetical proteins, and others (see IcmX and LvrE, below) were in multiple spots. This is common and can be due to protein modifications such as phosphorylation, glycosylation, or limited proteolysis.
Table 1.
Type II-dependent exoproteins of L. pneumophila identified by 2DE analysis
| Spot no.* | PubMed accession no. | Score | Sequence covered, % | Identity (as defined in the annotated genome) | Gene designation† |
|||
|---|---|---|---|---|---|---|---|---|
| Philadelphia | Paris | Lens | SignalP | |||||
| 1, 42 | 2110146 | 108 | 16 | Zinc metalloprotease | lpg0467 | lpp0532 | lpl0508 | Y |
| 2 | 11119504 | 209 | 18 | Major acid phosphatase | lpg1119 | lpp1120 | lpl1124 | Y |
| 3 | 15448271 | 146 | 11 | Ribonuclease, T2 family | lpg2848 | lpp2906 | lpl2760 | Y |
| 4 | 12379686 | 229 | 15 | Lysophospholipase A | lpg2343 | lpp2291 | lpl2264 | Y |
| 5, 11, 12, 37 | 15448271 | 50 | 29 | Leucine aminopeptidase | lpg2814 | lpp2866 | lpl2729 | Y |
| 6–10, 34 | 11923356 | 261 | 32 | LvrE | lpg1244 | lpp0181 | lpl0163 | Y |
| 13 | 15448271 | 174 | 7 | Chitinase | lpg1116 | lpp1117 | lpl1121 | Y |
| 14, 15, 17 | 15003709 | 448 | 36 | IcmX | lpg2689 | lpp2743 | lpl2616 | Y |
| 16 | 15448271 | 383 | 26 | Hypothetical protein | lpg0189 | lpp0250 | lpl0249 | Y |
| 18, 35, 41 | 15448271 | 92 | 9 | Hypothetical protein | lpg1809 | lpp1772 | lpl1773 | Y |
| 19, 41 | 15467720 | 345 | 46 | Hypothetical protein - has VirK domain | lpg1832 | lpp1795 | lpl1796 | Y |
| 20 | 15467720 | 319 | 46 | Hypothetical protein - weak similarity to amidase | lpg0264 | lpp0335 | lpl0316 | Y |
| 21 | 15448271 | 415 | 39 | Similar to eukaryotic zinc metalloproteinase | lpg2999 | lpp3071 | lpl2927 | Y |
| 22, 25, 26 | 15467720 | 583 | 41 | Hypothetical protein - weakly similar to endoglucanase | lpg1918 | lpp1893 | lpl1882 | Y |
| 23, 24, 38 | 15448271 | 357 | 23 | Hypothetical protein | lpg0956 | lpp1018 | lpl0985 | Y |
| 27, 36, 39 | 15448271 | 363 | 27 | Tail fiber protein (collagen-like protein SclB) | lpg2644 | lpp2697 | lpl2569 | Y |
| 28, 40, 42 | 15448271 | 227 | 33 | Hypothetical protein | lpg0873 | lpp0936 | lpl0906 | Y |
| 29 | 15467720 | 431 | 12 | Aconitate hydratase | lpg1690 | lpp1659 | lpl1653 | N |
| 30 | 9332363 | 506 | 12 | Chaperone protein dnaK | lpg2025 | lpp2007 | lpl2002 | N |
| 31 | 7790062 | 590 | 46 | Flagellin | lpg1340 | lpp1294 | lpl1293 | N |
| 32, 33 | 15467720 | 92 | 25 | Hypothetical protein | – | lpp1177 | lpl1183 | N |
| 34 | 15448271 | 102 | 11 | Hypothetical protein - weakly similar to cysteine protease | lpg2622 | lpp2675 | lpl2547 | Y |
| 35 | 15448271 | 105 | 25 | Hypothetical protein - carboxymuconolactone decarboxylase | lpg0406 | lpp0472 | lpl0048 | N |
| 36 | 15467720 | 415 | 24 | Hypothetical protein - some similarity to nucleotidase | lpg2526 | lpp2591 | lpl2446 | N |
| 37 | 15467720 | 489 | 25 | Hypothetical protein - similar to aminopeptidase | lpg0032 | lpp0031 | lpl0032 | Y |
| 38 | 15467720 | 146 | 19 | Hypothetical protein | lpg1954 | lpp1936 | lpl1923 | N |
| 40 | 15448271 | 153 | 16 | Hypothetical protein | lpg1385 | lpp1340 | lpl1336 | Y |
*As is commonly seen, some proteins were represented in more than one spot, and some spots contained multiple proteins. For those proteins found in multiple spots, comparable scores and sequence coverage were obtained from the different spots.
Three more proteins identified in wild-type but not mutant supernatants had signal sequences (Table 1), but other information makes a simple connection to type II secretion less immediate. Two of these, IcmX and LvrE, are linked to type IV secretion systems of L. pneumophila. IcmX is linked to Dot/Icm and is required for the biogenesis of the intracellular phagosome, although its precise function is unknown (22, 23). A previous study also found IcmX in supernatants, in addition to being in the periplasm, but that secretion depended on Dot/Icm (23). The gene for LvrE is linked to those encoding the Lvh system, but it is still not clear whether LvrE has a role in type IV secretion (24, 25). LvrE has also been observed in supernatants (26). The third protein, spot 19, was annotated as hypothetical but with homology to VirK, an Agrobacterium tumefaciens protein encoded within the region of the VirB/D4 type IV secretion system but not required for Ti-plasmid-DNA transfer (27, 28). These data raise the possibility of a mechanistic connection between types II and IV secretion. Alternately, these data indicate that IcmX, LvrE, and the VirK-like protein are merely genetically linked to type IV secretion, and that their secretion is in fact by the type II system.
The seven remaining proteins that were differentially observed in supernatants were not predicted to have a signal peptide and thus are unlikely to be Lsp substrates (Table 1). The presence of flagellin (29) in supernatants may not be surprising, because the release of flagellar proteins into the extracellular medium is commonly seen. The detection of aconitase and DnaK, cytosolic proteins of L. pneumophila (30, 31), might be the result of some cell lysis. The remaining four proteins were not predicted to be cytoplasmic by PSORTb. Beyond lytic or flagellar release, it is conceivable that one or more of these proteins exit the cell by type IV or I systems (32). It is not apparent why type II mutant supernatants should lack these proteins, but perhaps there is a feedback loop triggered in the absence of type II secretion that decreases expression of these factors.
ChiA Encodes the Type II-Secreted Chitinase ofL. pneumophila.
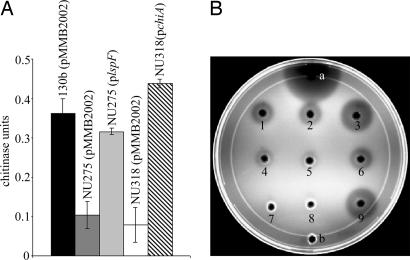
One of the newly identified type II dependent proteins was annotated as a chitinase. In all three sequenced L. pneumophila strains (19, 20), the gene encoding the protein is monocistronic, with the gene on either side of it being transcribed in the opposite direction. The C terminus of the protein has a glycosyl hydrolase domain and a conserved active site characteristic of the family 18 of glycosyl hydrolases, the group that contains most of the known bacterial chitinases (33, 34). The protein did not contain the chitin-binding or fibronectin type III-like domains that are sometimes present in chitinases (35–38). For reasons below, we designated this protein and its gene as ChiA and chiA, respectively. Because Legionella had not been previously investigated for chitinases, we next assayed supernatants from wild-type 130b for activity against known substrates of chitinases. The supernatants had activity against p-nitrophenyl β-d-N,N′,N″ triacetyl chitotriose (pNP-[GlcNAc]3), indicating that L. pneumophila secretes an endochitinase (Fig. 1A; ref. 39). No activity was seen against exochitinases substrates, p-NP-N-acetyl-β-d-glucosamine and p-NP β-d-N,N′-diacetylchitobiose (data not shown; refs. 39 and 40). 130b supernatants also showed clearance on chitin-containing plates (Fig. 1B). When assayed with pNP-[GlcNAc]3, the type II mutant NU275 showed a 70–75% reduction in secreted chitinase activity and minimal clearance on chitin plates (Fig. 1). Both activities were restored to wild-type levels in NU275 containing a plasmid copy of lspF (ref. 6; Fig. 1). Together, these data confirm that L. pneumophila secretes a chitinase secreted by the type II system.
Fig. 1.
Secreted chitinase activity of L. pneumophila wild type and lspF and chiA mutants. (A) Culture supernatants of wild-type 130b (pMMB2002; black bar), lspF mutant NU275 (pMMB2002; dark gray bar), NU275 (plspF; light gray bar), chiA mutant NU318 (pMMB2002; white bar), and NU318 (pchiA; crosshatched bar) were assayed for chitinase activity against p-NP-[GlcNAc]3. Data represent the mean and standard deviation for triplicate cultures for each strain. The reductions in enzymatic activity for NU275 (pMMB2002) and NU318 (pMMB2002) were significant (Student's t test; P < 0.05). 130b, NU275, and NU318 behaved identically to their corresponding derivatives that contained only the cloning vector pMMB2002 (data not shown). (B) Utilization of glycol chitin by the secreted chitinase in the supernatants of strains 130b (1), 130b (pMMB2002; 2), 130b (pchiA; 3), NU275 (4), NU275 (pMM2002; 5), NU275 (plspF; 6), NU318 (7), NU318 (pMMB2002; 8), and NU318 (pchiA; 9). Twenty microunits of S. griseus chitinase (a) and BYE medium (b) were used as the positive and negative controls, respectively. Similar results for A and B were obtained on at least two other occasions. The independently derived chiA mutant NU319 had a phenotype identical to that of mutant NU318 (data not shown).
To determine whether ChiA is the secreted chitinase activity, two independent chiA mutants of strain 130b were constructed. Mutants NU318 and NU319 showed normal behavior when cultured in buffered yeast extract broth and on buffered charcoal yeast extract agar, indicating that chiA is not required for general growth (data not shown). Mutant supernatants had a ≈75% reduction in activity against pNP-[GlcNAc]3 and no clearance on chitin plates (Fig. 1). That NU318 and NU319 gave similar results, coupled with the fact that chiA is monocistronic, indicated that the observed secretion defect was due to the loss of chiA and not a polar effect or spontaneous second-site mutation. As expected, a plasmid-borne copy of chiA (pchiA) restored the mutant to wild-type levels of activity (Fig. 1). That chiA encodes a chitinase was further confirmed when lysates of chiA-expressing Escherichia coli had activity against pNP-[GlcNAc]3 and chitin (SI Fig. 5). Because the type II and chiA mutants had similar reductions in activity, we believe that ChiA is the only L. pneumophila type II chitinase.
ChiA Promotes Persistence in the Lungs.
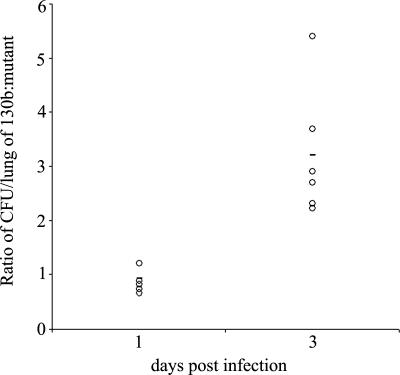
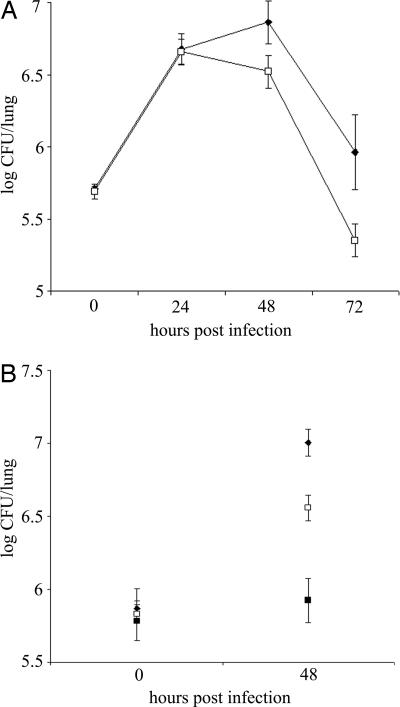
To begin to investigate the role of ChiA in L. pneumophila ecology and infection, we analyzed the capacity of chiA mutants to grow in Hartmannella vermiformis amoebae and macrophage-like U937 cells. The mutants grew similarly to wild type in both cells (data not shown), indicating chiA is not required for intracellular growth. We next analyzed the chiA mutants in the A/J mouse model of Legionnaires' disease, initially doing a competition assay. After introduction into the mouse lung, mutant NU318 exhibited a 3.5-fold deficiency compared with wild type (Fig. 2). That independently derived NU319 had a 4-fold deficiency (SI Fig. 6) indicated that the defects were due to inactivation of monocistronic chiA and not spontaneous second-site mutation. Further confirmation was not possible, because plasmids containing cloned chiA reduced the growth of wild type in the host (data not shown), precluding complementation analysis. At the least, these data indicate that ChiA is indirectly involved in persistence in the lung. To further explore the in vivo defect, we compared the ability of wild type and a chiA mutant to exist in the lungs when injected into separate groups of animals. Once again, NU318 had a 4-fold deficiency (Fig. 3A). In another trial, the reduced recoverability of the chiA mutant was found to be about one-half that of a type II mutant (Fig. 3B). Taken together, these data indicate that ChiA, directly or indirectly, is required for optimal survival of L. pneumophila in the lung. That ChiA is expressed in vivo was confirmed by Western blot by using antisera from animals intratracheally inoculated with wild-type L. pneumophila (SI Fig. 7). ChiA is first type II effector to be implicated in L. pneumophila virulence, because no other effector mutant tested, including strains lacking acid phosphatase (map), lipases A and B (lipAB), lysophospholipase A (plaA), metalloprotease (proA), or phospholipase C (plcA), displayed a defect in the mice (SI Fig. 8). In sum, these data represent an indication that a protein having chitinase activity can promote survival of a pathogen in a mammalian host.
Fig. 2.
In vivo competition of L. pneumophila wild type and a chiA mutant in the lung. An equal mixture of wild-type strain 130b and chiA mutant NU318 was inoculated intratracheally into the lungs of A/J mice. The ratio of the wild type to the mutant was determined at days 1 and 3 after inoculation. Data are representative of actual values obtained per mouse (n = 5), and the solid bar indicates the mean value. The differences in the ratios of 130b and NU318 on day 1 vs. day 3 were significant (Student's t test; P < 0.05).
Fig. 3.
Growth and persistence of wild type and mutant L. pneumophila in the lung. A/J mice were intratracheally inoculated with equal numbers of 130b (♦; A) and the chiA mutant NU318 (□) and 130b (♦; B), NU318 (□) and the type II mutant NU275 (■), and the cfus in the infected lungs were determined at various time points. The data represent the mean and standard deviations of four to six mice for each strain. The differences between the cfus recovered for 130b and NU318 at 48 and 72 h after inoculation (A) and 130b and NU275 and 130b and NU318 at 48 h after inoculation (B) were significant (Student's t test, P < 0.01).
Discussion
Our proteomic analysis identified 20 proteins that are substrates for type II secretion; i.e., they were in wild-type but not mutant supernatants, and their corresponding ORF had a signal sequence. These 20 are not, however, the complete set of type II substrates, for several reasons. First, five enzymes, lipases, phospholipases, and lysophospholipases that were previously defined as type II-dependent were not identified in this study. The reasons that these proteins and perhaps other type II effectors were not seen include low-level expression that precluded detection by Coomassie stain and degradation due to the processing of the sample or to the specific action of a secreted protease. Second, although we examined 42 spots, there are additional spots that we did not submit for MS analysis. Third, our samples were from bacteria grown under a single growth condition. Therefore, we suspect that L. pneumophila type II effectors number >25. Indeed, an in silico screen of the L. pneumophila genome revealed 254 proteins that both contain a signal peptide and are predicted by a combination of programs not to be in the cytoplasm, periplasm, or outer membrane or have a function clearly linked to the cell interior (SI Tables 2 and 3). Among these proteins, 60 were predicted to be extracellular by at least one program and 13 by two programs. Our proteomic analysis identified proteins belonging to this group as well as some proteins whose predicted location is unknown. We do not imagine that the output of the type II system is 254, but it is reasonable to think that the system can process 60 substrates. Even at 25, the experimentally defined catalog of L. pneumophila type II effectors (i.e., proteins/activities that are in wild-type but not mutant supernatants and whose ORF had a signal sequence) is the largest known in bacteria.
Our data increase our appreciation not only for the magnitude of the type II output but also for the types of proteins exported. First, we confirmed that a number of types of enzymes secreted by other bacterial type II systems are elaborated by L. pneumophila, including peptidases, lipolytic enzymes, cellulases, chitinases, and phosphatases (2). Second, we reaffirmed that L. pneumophila is unique in its secretion of a ribonuclease and gained the first indication of a type II-dependent amidase. Third, we documented that the secretome of L. pneumophila includes proteins that show their greatest similarity to eukaryotic proteins. We had previously reported that a secreted acid phosphatase of L. pneumophila has eukaryotic-like properties (12), and now we add a protein with collagen-like repeats and a protein related to a eukaryotic zinc metalloproteinase. Fourth, based on the type II dependency of IcmX and LvrE, we obtained an indication of a possible connection between types II and IV secretion. Our in silico analysis also revealed secreted proteins that are predicted to have known and novel types of activities, homology to eukaryotic factors, or a connection to type IV secretion (SI Tables 2 and 4). Fifth, and perhaps most significantly, we uncovered a number of secreted proteins that do not bear any similarity to known proteins, raising the possibility of there being novel effectors secreted by the type II system. From proteomic and in silico analysis, the type of proteins most represented are proteases/peptidases (Tables 1, 2, and 4), a finding consistent with the fact that amino acids are the main source of carbon and energy for broth-grown L. pneumophila (41). The legionellae may also be able to catabolize carbohydrate derivatives (42), and our list of putative Lsp exoproteins also includes proteins with predicted glycosidase activity (SI Tables 2 and 4), such as a eukaryotic-like glucoamylase. Paralogs of genes from the enhABC locus, including EnhC and LpnE, which promote invasion of macrophages (43–45), were found in our screen (SI Tables 2 and 4). Such a finding is compatible with the fact that, under some growth conditions, Lsp mutants exhibit a defect in amoebae and macrophage invasion (8) (O.R. and N.P.C., unpublished results). Based on both the number of proteins uncovered and the types of proteins detected, our analysis highlights more than ever the impact that type II secretion has on bacterial function.
One of the type II exoproteins was a chitinase. Chitin is an insoluble polymer consisting of alternating β-1,4-linked N-acetylglucosamine residues and is the second most-abundant organic compound in nature after cellulose (46). It is prominently found in the cell walls of mold, fungi, and algae and is the major constituent of cuticles and exoskeletons of worms, mollusks, and arthropods (46). Chitinases, in turn, are present in plants, insects, crustaceans, bacteria (especially marine bacteria), protozoa, fish, animals, and humans (46–49). Bacterial chitinases are often involved in nutrient acquisition (50, 51). However, chitinases may also provide protection/advantage against invading or competing microorganisms that have chitin in their composition (52). Thus, it is quite plausible that ChiA is important for L. pneumophila to succeed in the environment. Although mutating chiA did not reduce replication in H. vermiformis trophozoites, ChiA may play a role in the usage or degradation of chitin in amoebal cysts (53). Beyond obviously acting upon other chitins in the environment, ChiA might act on peptidoglycan fragments that have similarity to chitin (54).
Our most striking observation is the reduced ability of the chitinase mutant to persist in the lung. As such, L. pneumophila ChiA is the first protein with chitinase activity to be implicated in infection of a mammalian host and a type II-dependent virulence factor of L. pneumophila. Because the chiA mutant grew normally in U937 cells, we believe that the role of ChiA is not simply associated with macrophage infection. Because the reduced recoverability of the mutant was not manifest in the first 24 h but rather at 72 h, we further suspect that ChiA most significantly promotes persistence vs. replication. At a molecular level, the infection defect could have several explanations. In one scenario, the effect of ChiA could be due to the protein's direct action on a host target. Because mammals do not have chitin, this leads to the hypothesis that there is a chitin-like substance in the lung whose degradation by ChiA aids bacterial persistence. Compatible with this hypothesis is the recent suggestion that O-GlcNAcylated proteins in eukaryotic cells can be acted on by chitinases/chitinase-like enzymes (55). Alternatively, ChiA might be a novel bifunctional enzyme that has another unidentified substrate, whose loss enhances bacterial survival. In a second scenario, the effect of ChiA is not directly due to a bacterial enzymatic activity but involves the host response to the protein; when ChiA is present, the host response is compromised sufficiently to allow progression to pneumonia, but when ChiA is absent, a more effective response is mounted. That chitinases can modulate host response is recently evident from studies of the asthmatic lung in which overexpression of mammalian chitinases contribute to pathogenesis by the recruitment of inflammatory cells (56). Incidentally, this type of observation may help explain the detrimental effect that overexpression of cloned chiA had on legionellae in the lung. Our observations have implications for the pathogenesis of other bacteria that elaborate chitinases, including E. coli, Listeria monocytogenes, Vibrio cholerae, and Vibrio vulnificus (57–60). The V. cholerae, V. vulnificus, and E. coli chitinases are type II-dependent (59–61), but they have not been assessed for a role in infection.
Another major implication of this study is that factors such as chitinases that are traditionally viewed as being relevant only in the environment may in fact have great importance in human and animal infection. In support of this view, a chitin-binding protein of V. cholerae was recently shown to promote attachment to intestinal epithelia (62). Thus, future investigations into pathogenic processes may well benefit from the inclusion of “environmental” enzymes such as chitinases and cellulases.
Materials and Methods
Strains, Growth Media, and Chemicals.
L. pneumophila 130b (ATCC BAA-74) was used as the wild type in this study. Its derivatives NU275, NU255, NU267, NU270, NU268, and AA200, which contain mutations in lspF, map, lipAB, plaA, plcA, and proA, respectively, were described before (6, 12–14, 17, 63). Legionellae were cultured at 37°C in buffered yeast extract broth or on buffered charcoal yeast extract agar (6). E. coli DH5α (Invitrogen, Carlsbad, CA) was grown as before (64). Chemicals, unless otherwise noted, were from Sigma (St. Louis, MO.).
2DE Analysis and Protein Identification.
L. pneumophila were grown in 50 ml of broth to late-log phase, and supernatants were concentrated 100-fold as described (15). Samples were treated with the Ready Prep 2-D Cleanup Kit (BioRad, Hercules, CA) and subjected to 2DE (details are published as SI Supporting Text). Protein spots were excised and submitted for identification to Stanford Mass Spectrometry Services (Stanford University, Stanford, CA). The MS/MS spectra were searched by MASCOT (Matrix Science, Boston, MA) by using the bacterial database NCBI-nr. Peptide mass tolerance was set at 2 Da, and fragment mass tolerance was set at 0.8 Da. Unambiguous matches were established by considering the number of peptides matched, the percentage of the ORF covered, and the agreement between the experimental and predicted masses and isoelectric points for the protein. Proteins containing signal peptides were identified by using SignalP (65). Further predictions concerning the cellular location of proteins were determined by using PSORTb (66, 67).
Genetic Techniques.
To construct and complement chitinase mutants, we used our standard cloning, allelic exchange, and transformation procedures (details are published as SI Supporting Text). NU318 and NU319 are independent mutants, because they were derived from separate transformations. For complementation, chiA was cloned into pMMB2002 (6), yielding pchiA.
Detection of Chitinase Activity.
L. pneumophila supernatants and E. coli lysates were assayed for chitinase by monitoring the release of p-nitrophenol from the chromogenic substrates, p-nitrophenyl-N-acetyl-β-d-glucosamine, p-nitrophenyl β-d-N,N′-diacetylchitobiose, and pNP-[GlcNAc]3, as described previously (39, 68). Briefly, filtered supernatants from late-log broth cultures of L. pneumophila and lysates from overnight cultures of E. coli were prepared as described previously (11). Then, 200 μl of supernatant or lysate was incubated in 600 μl of sodium phosphate buffer (pH 6.5) containing 10 μg of substrate for 4 h at 37°C. The reaction was ended by the addition of 200 μl of sodium carbonate, and absorbance was measured at 410 nm. The ability of 50-μl samples of supernatants or lysates to degrade glycol chitin was measured on agarose plates containing 0.1% glycol chitin, as described (69, 70). Immunoblot detection of chitinase in recombinant E. coli was done as before, by using mouse sera from animals intratracheally inoculated with wild-type 130b (6).
Infection Assays.
H. vermiformis and U937 cells were infected with L. pneumophila, as described (4, 6). To assess in vivo growth, 6- to 8-week-old A/J mice (The Jackson Laboratory, Bar Harbor, ME) were inoculated intratracheally, and then the numbers of legionellae in lung homogenates were determined by plating (6). Animal experiments were approved by the Animal Care and Use Committee of Northwestern University.
Supplementary Material
Acknowledgments
We thank Cianciotto laboratory members as well as Mark Strom for helpful discussions. This work was supported by National Institutes of Health Grant AI43987 (to N.P.C.).
Abbreviations
- Lsp
Legionella secretion pathway
- pNP-[GlcNAc]3
p-nitrophenyl β-d-N,N′,N″ triacetyl chitotriose
- 2DE
2D electrophoresis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608279103/DC1.
References
- 1.Fields BS, Benson RF, Besser RE. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cianciotto NP. Trends Microbiol. 2005;13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Johnson TL, Abendroth J, Hol WG, Sandkvist M. FEMS Microbiol Lett. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- 4.Liles MR, Edelstein PH, Cianciotto NP. Mol Microbiol. 1999;31:959–970. doi: 10.1046/j.1365-2958.1999.01239.x. [DOI] [PubMed] [Google Scholar]
- 5.Hales LM, Shuman HA. Infect Immun. 1999;67:3662–3666. doi: 10.1128/iai.67.7.3662-3666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossier O, Starkenburg S, Cianciotto NP. Infect Immun. 2004;72:310–321. doi: 10.1128/IAI.72.1.310-321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Söderberg MA, Rossier O, Cianciotto NP. J Bacteriol. 2004;186:3712–3720. doi: 10.1128/JB.186.12.3712-3720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polesky AH, Ross JT, Falkow S, Tompkins LS. Infect Immun. 2001;69:977–987. doi: 10.1128/IAI.69.2.977-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossier O, Cianciotto NP. Infect Immun. 2001;69:2092–2098. doi: 10.1128/IAI.69.4.2092-2098.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerji S, Bewersdorff M, Hermes B, Cianciotto NP, Flieger A. Infect Immun. 2005;73:2899–2909. doi: 10.1128/IAI.73.5.2899-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aragon V, Kurtz S, Flieger A, Neumeister B, Cianciotto NP. Infect Immun. 2000;68:1855–1863. doi: 10.1128/iai.68.4.1855-1863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aragon V, Kurtz S, Cianciotto NP. Infect Immun. 2001;69:177–185. doi: 10.1128/IAI.69.1.177-185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aragon V, Rossier O, Cianciotto NP. Microbiology. 2002;148:2223–2231. doi: 10.1099/00221287-148-7-2223. [DOI] [PubMed] [Google Scholar]
- 14.Flieger A, Neumeister B, Cianciotto NP. Infect Immun. 2002;70:6094–6106. doi: 10.1128/IAI.70.11.6094-6106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debroy S, Aragon V, Kurtz S, Cianciotto NP. Infect Immun. 2006;74:5152–5160. doi: 10.1128/IAI.00484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossier O, Cianciotto NP. Infect Immun. 2005;73:2020–2032. doi: 10.1128/IAI.73.4.2020-2032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moffat JF, Edelstein PH, Regula DP, Jr, Cirillo JD, Tompkins LS. Mol Microbiol. 1994;12:693–705. doi: 10.1111/j.1365-2958.1994.tb01057.x. [DOI] [PubMed] [Google Scholar]
- 18.Szeto L, Shuman HA. Infect Immun. 1990;58:2585–2592. doi: 10.1128/iai.58.8.2585-2592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, et al. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 20.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, et al. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 21.Flieger A, Gong S, Faigle M, Stevanovic S, Cianciotto NP, Neumeister B. J Bacteriol. 2001;183:2121–2124. doi: 10.1128/JB.183.6.2121-2124.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand BC, Sadosky AB, Shuman HA. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 23.Matthews M, Roy CR. Infect Immun. 2000;68:3971–3982. doi: 10.1128/iai.68.7.3971-3982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridenour DA, Cirillo SL, Feng S, Samrakandi MM, Cirillo JD. Infect Immun. 2003;71:6256–6263. doi: 10.1128/IAI.71.11.6256-6263.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segal G, Russo JJ, Shuman HA. Mol Microbiol. 1999;34:799–809. doi: 10.1046/j.1365-2958.1999.01642.x. [DOI] [PubMed] [Google Scholar]
- 26.De Buck E, Maes L, Meyen E, Van Mellaert L, Geukens N, Anne J, Lammertyn E. Biochem Biophys Res Commun. 2005;331:1413–1420. doi: 10.1016/j.bbrc.2005.04.060. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Wolf SG, Elbaum M, Tzfira T. Trends Microbiol. 2005;13:295–298. doi: 10.1016/j.tim.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Kalogeraki VS, Winans SC. J Bacteriol. 1998;180:5660–5667. doi: 10.1128/jb.180.21.5660-5667.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuner K, Bender-Beck L, Brand BC, Luck PC, Mann KH, Marre R, Ott M, Hacker J. Infect Immun. 1995;63:2499–2507. doi: 10.1128/iai.63.7.2499-2507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mengaud JM, Horwitz MA. J Bacteriol. 1993;175:5666–5676. doi: 10.1128/jb.175.17.5666-5676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amemura-Maekawa J, Watanabe H. Gene. 1997;197:165–168. doi: 10.1016/s0378-1119(97)00257-6. [DOI] [PubMed] [Google Scholar]
- 32.Jacobi S, Heuner K. Int J Med Microbiol. 2003;293:349–358. doi: 10.1078/1438-4221-00276. [DOI] [PubMed] [Google Scholar]
- 33.Orikoshi H, Baba N, Nakayama S, Kashu H, Miyamoto K, Yasuda M, Inamori Y, Tsujibo H. J Bacteriol. 2003;185:1153–1160. doi: 10.1128/JB.185.4.1153-1160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henrissat B, Bairoch A. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T, Fujiwara S, Nishikori S, Fukui T, Takagi M, Imanaka T. Appl Environ Microbiol. 1999;65:5338–5344. doi: 10.1128/aem.65.12.5338-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikegami T, Okada T, Hashimoto M, Seino S, Watanabe T, Shirakawa M. J Biol Chem. 2000;275:13654–13661. doi: 10.1074/jbc.275.18.13654. [DOI] [PubMed] [Google Scholar]
- 37.Tsujibo H, Orikoshi H, Shiotani K, Hayashi M, Umeda J, Miyamoto K, Imada C, Okami Y, Inamori Y. Appl Environ Microbiol. 1998;64:472–478. doi: 10.1128/aem.64.2.472-478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe T, Suzuki K, Oyanagi W, Ohnishi K, Tanaka H. J Biol Chem. 1990;265:15659–15665. [PubMed] [Google Scholar]
- 39.Folders J, Algra J, Roelofs MS, van Loon LC, Tommassen J, Bitter W. J Bacteriol. 2001;183:7044–7052. doi: 10.1128/JB.183.24.7044-7052.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tronsmo A, Harman GE. Anal Biochem. 1993;208:74–79. doi: 10.1006/abio.1993.1010. [DOI] [PubMed] [Google Scholar]
- 41.George JR, Pine L, Reeves MW, Harrell WK. J Clin Microbiol. 1980;11:286–291. doi: 10.1128/jcm.11.3.286-291.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, Buchrieser C. Cell Microbiol. 2006;8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 43.Bandyopadhyay P, Xiao H, Coleman HA, Price-Whelan A, Steinman HM. Infect Immun. 2004;72:4541–4551. doi: 10.1128/IAI.72.8.4541-4551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cirillo SLG, Lum J, Cirillo JD. Microbiology. 2000;146:1345–1359. doi: 10.1099/00221287-146-6-1345. [DOI] [PubMed] [Google Scholar]
- 45.Newton HJ, Sansom FM, Bennett-Wood V, Hartland EL. Infect Immun. 2006;74:1683–1691. doi: 10.1128/IAI.74.3.1683-1691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dahiya N, Tewari R, Hoondal GS. Appl Microbiol Biotechnol. 2006;71:773–782. doi: 10.1007/s00253-005-0183-7. [DOI] [PubMed] [Google Scholar]
- 47.Flach J, Pilet P-E, Jolles P. Experimentia. 1992;48:701–716. doi: 10.1007/BF02124285. [DOI] [PubMed] [Google Scholar]
- 48.Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, Place A, Aerts JM. J Biol Chem. 2001;276:6770–6778. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 49.Zheng T, Rabach M, Chen NY, Rabach L, Hu X, Elias JA, Zhu Z. Gene. 2005;357:37–46. doi: 10.1016/j.gene.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 50.Gooday GW. Adv Microb Ecol. 1990;11:387–430. [Google Scholar]
- 51.Roberts WK, Selitrennikoff CP. J Gen Microbiol. 1988;134:169–176. [Google Scholar]
- 52.Metraux JP, Burkhart W, Moyer M, Dincher S, Middlesteadt W, Williams S, Payne G, Carnes M, Ryals J. Proc Natl Acad Sci USA. 1989;86:896–900. doi: 10.1073/pnas.86.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linder M, Winiecka-Krusnell J, Linder E. Appl Environ Microbiol. 2002;68:2503–2508. doi: 10.1128/AEM.68.5.2503-2508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keyhani NO, Roseman S. Biochim Biophys Acta. 1999;1473:108–122. doi: 10.1016/s0304-4165(99)00172-5. [DOI] [PubMed] [Google Scholar]
- 55.Rao FV, Dorfmueller HC, Villa F, Allwood M, Eggleston IM, van Aalten DM. EMBO J. 2006;25:1569–1978. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuttelkopf AW, Andersen OA, Rao FV, Allwood M, Lloyd C, Eggleston IM, van Aalten DM. J Biol Chem. 2006;281:27278–27285. doi: 10.1074/jbc.M604048200. [DOI] [PubMed] [Google Scholar]
- 57.Wortman AT, Somerville CC, Colwell RR. Appl Environ Microbiol. 1986;52:142–145. doi: 10.1128/aem.52.1.142-145.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Somerville CC, Colwell RR. Proc Natl Acad Sci USA. 1993;90:6751–6755. doi: 10.1073/pnas.90.14.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paranjpye RN, Lara JC, Pepe JC, Pepe CM, Strom MS. Infect Immun. 1998;66:5659–5668. doi: 10.1128/iai.66.12.5659-5668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Francetic O, Belin D, Badaut C, Pugsley AP. EMBO J. 2000;19:6697–6703. doi: 10.1093/emboj/19.24.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Connell TD, Metzger DJ, Lynch J, Folster JP. J Bacteriol. 1998;180:5591–5600. doi: 10.1128/jb.180.21.5591-5600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirn TJ, Jude BA, Taylor RK. Nature. 2005;438:863–866. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 63.Aragon V, Kurtz S, McClain M, Engleberg NC, Cianciotto NP. In: Legionella. Marre R, Abu Kwaik Y, Bartlett C, Cianciotto NP, Fields BS, Frosch M, Hacker J, Luck PC, editors. Washington, DC: ASM Press; 2002. pp. 18–21. [Google Scholar]
- 64.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Plainview, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 65.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 66.Gardy JL, Spencer C, Wang K, Ester M, Tusnady GE, Simon I, Hua S, deFays K, Lambert C, Nakai K, Brinkman FS. Nucleic Acids Res. 2003;31:3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 68.Thompson SE, Smith M, Wilkinson MC, Peek K. Appl Environ Microbiol. 2001;67:4001–4008. doi: 10.1128/AEM.67.9.4001-4008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Velasquez L, Hammerschmidt R. J Microbiol Methods. 2004;59:7–14. doi: 10.1016/j.mimet.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 70.Trudel J, Asselin A. Anal Biochem. 1990;189:249–253. doi: 10.1016/0003-2697(90)90116-q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.