Abstract
The norepinephrine transporter critically regulates both neurotransmission and homeostasis of norepinephrine in the nervous system. In this study, we report a previously uncharacterized and common A/T polymorphism at −3081 upstream of the transcription initiation site of the human norepinephrine transporter gene [solute carrier family 6, member 2 (SLC6A2)]. Using both homologous and heterologous promoter-reporter constructs, we found that the −3081(T) allele significantly decreases promoter function compared with the A allele. Interestingly, this T allele creates a new palindromic E2-box motif that interacts with Slug and Scratch, neural-expressed transcriptional repressors binding to the E2-box motif. We also found that both Slug and Scratch repress the SLC6A2 promoter activity only when it contains the T allele. Finally, we observed a significant association between the −3081(A/T) polymorphism and attention-deficit hyperactivity disorder (ADHD), suggesting that anomalous transcription factor-based repression of SLC6A2 may increase risk for the development of attention-deficit hyperactivity disorder and other neuropsychiatric diseases.
Keywords: Snail family, E2-box, Slug, Scratch
Norepinephrine (NE) is a key neurotransmitter in both the central and peripheral nervous systems and regulates many essential functions, including attention, memory, emotion, and autonomic function. The NE transporter (NET) is primarily responsible for reuptake of NE into presynaptic nerve terminals and is a regulator of NE homeostasis (1, 2). Indeed, NET knockout mice exhibited elevated extracellular levels of NE, resulting in altered brain and autonomic function (2, 3). The NET belongs to the SLC6 family of Na+/Cl−-dependent transporters (4). The human NET gene [solute carrier family 6, member 2 (SLC6A2)], spanning ≈45 kb, is located on chromosome 16q12.2 (5). Genomic clones and cDNA encoding SLC6A2 have been isolated and characterized, thus making molecular investigation of regulatory mechanisms of SLC6A2 expression possible (6).
Given the global role of NE in the nervous system, its abnormal regulation/expression may predictably result in various pathological conditions such as neurodegenerative, psychiatric, and cardiovascular disorders (7, 8). In particular, SLC6A2 has been posited as a candidate gene for diverse psychiatric and autonomic disorders. For example, patients suffering from major depression have demonstrated reduced levels of NET, as measured by brain imaging and binding techniques in postmortem brain samples (9). The NET is a primary target of well known tricyclic antidepressant drugs (10). Based on its important role in the maintenance of attention and vigilance, it has been hypothesized that abnormal regulation of NE neurotransmission contributes to attention-deficit hyperactivity disorder (ADHD) (8). Consistent with this hypothesis, an effective medication for ADHD, atomoxetine, selectively targets NET (11). Thus, it is of great interest to test whether functional variants in SLC6A2 may be associated with ADHD.
Several polymorphisms have been identified in the coding and noncoding regions of SLC6A2. A rare missense mutation (Ala457Pro) recently was found in the highly conserved transmembrane domain 9 of SLC6A2 from a patient suffering from orthostatic intolerance and tachycardia (12). Further, a polymorphism in the noncoding region was reported to be associated with anorexia nervosa (13). Nonetheless, there is no direct evidence that these SLC6A2 variants are related to psychiatric disorders. Several recent studies have examined the association between SLC6A2 single-nucleotide polymorphisms (SNPs) and ADHD and have yielded mixed results (14–17). Considering that subtle changes in related gene activities may underlie psychiatric disorders, it is tempting to speculate that sequence variations in the noncoding region that lead to altered regulatory function may contribute to certain brain disorders. Indeed, genetic variations in regulatory regions are beginning to be recognized as risk factors for complex inherited disorders. For example, 10 different alleles containing from 3 to 13 copies of a 40-bp tandem repeat have been found in the 3′ UTR of the dopamine transporter gene. Among them, an allele with 10 copies has been found to be associated with ADHD (18, 19).
At present, there has been no report of functional polymorphisms in the promoter region of SLC6A2. Based on our previous study of the regulatory region (20), we sought to identify potentially functional polymorphisms in the promoter of the SLC6A2. Herein, we report a common polymorphism at −3081 that affects SLC6A2 promoter function. Moreover, we provide a mechanistic basis for loss of SLC6A2 promoter function via ectopic interaction with neural-expressed transcription factors Slug and Scratch. Finally, we demonstrate an association of −3081(T) with ADHD in children and adolescents.
Results
The Upstream Promoter Region for the Noradrenergic Cell-Specific Expression of SLC6A2.
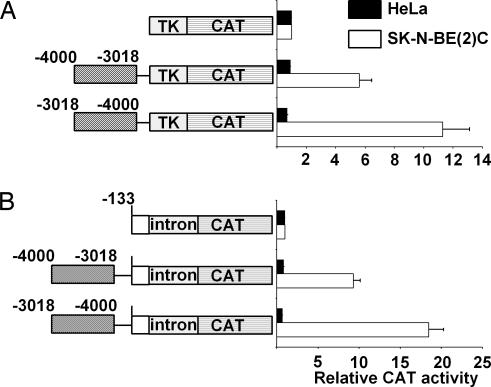
Previously, we have demonstrated that the 4.0-kb SLC6A2 upstream sequences can drive reporter gene expression in a noradrenergic cell-specific manner (20), indicating that these upstream promoter sequences may contain important genetic information about cell-type-specific transcription. In particular, the upstream domain between base pairs −4000 and −3000 may function as a cell-type-specific enhancer because it demonstrates robust activity at a relatively distant position. To further address this finding, we subcloned this region 5′ to the heterologous thymidine kinase (TK) promoter or to the homologous SLC6A2 proximal promoter previously described in ref. 20 [pNET133(i)CAT], either in the sense or antisense orientation. As shown in Fig. 1, this domain prominently increased TK promoter activity in both sense and antisense orientations in NET-expressing SK-N-BE (2)C cells. In addition, this domain was able to increase homologous SLC6A2 proximal promoter activity in both orientations. In contrast, no such effect was observed in NET-negative HeLa cells, thus supporting the notion that this domain has characteristics of a noradrenergic cell-specific enhancer.
Fig. 1.
The sequence between −4000 and −3018 of SLC6A2 promoter contains the noradrenergic cell-specific transcriptional element. The fragment between −4000 and −3018 was subcloned in either sense or antisense orientation 5′ of the pBLCAT2 or 5′ of the pNET133(i)CAT (20). The reporter constructs were transfected into the NET-positive SK-N-BE (2)C cells and NET-negative HeLa cells, and the normalized CAT activity of each construct was expressed relative to the vector pBLCAT2 (A) and pNET133(i)CAT (B), respectively.
Identification of a Previously Uncharacterized Polymorphism in the Upstream Enhancer Domain of SLC6A2.
Based on our preliminary study characterizing potentially important enhancer domains (Fig. 1 and ref. 20), we focused on the upstream enhancer region for the identification of previously uncharacterized polymorphisms instead of random screening of broad genomic regions of SLC6A2. Toward this goal, we amplified the region encompassing from base pair −4000 to −3018 with genomic DNA samples from 88 healthy individuals used for our previous study (21) and analyzed them by DNA sequencing. This analysis revealed one common polymorphism at −3081(A/T) [see supporting information (SI) Fig. 6].
Effect of the −3081(A/T) Polymorphism on Promoter Activity.
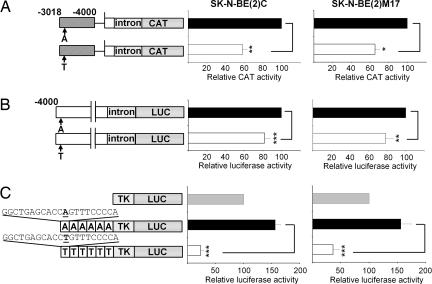
To assess the functional effect of this polymorphism on SLC6A2 promoter activity, we subcloned the enhancer domain from −4000 to −3018 with A or T alleles at nucleotide position −3081 in front of SLC6A2 proximal promoter [designated as pNETB(A)133(i)CAT and pNETB(T)133(i)CAT, respectively] and examined their transcriptional activities in SK-N-BE (2)C and SK-N-BE (2)M17. We found that reporter gene expression driven by the −3081(T) allele construct was reduced by 50% compared with the wild-type construct (P < 0.005) (Fig. 2A). In addition, a 4-kb upstream sequence containing the −3081(A) or −3081(T) alleles was cloned into a luciferase reporter plasmid, and the activity was measured. Using pNET4000T(i)LUC(T) with nucleotide T at −3081, we observed 25% and 28% decreased promoter activity in SK-N-BE (2)C and SK-N-BE (2)M17, respectively, compared with pNET4000A(i)LUC(A) with nucleotide A at −3081 (P < 0.0005) (Fig. 2B). To address more directly the role of the A/T polymorphism at −3081 in transcriptional regulation, we synthesized reporter constructs in which six copies of an oligonucleotide containing base pairs −3092 to −3072 of SLC6A2 promoter with either nucleotide A or T at −3081 were placed in front of the TK promoter. Transient transfection assay demonstrated that the reporter construct with nucleotide A at −3081 slightly stimulated TK promoter activity, whereas the reporter construct with nucleotide T at −3081 decreased TK promoter activity by 85% to 70% in SK-N-BE (2)C and SK-N-BE (2)M17 cells, respectively (P < 0.0005) (Fig. 2C).
Fig. 2.
Basal promoter activities of reporter constructs containing nucleotide A or T at position −3081. (A) The fragment between −4000 and −3018 containing either A or T at base pair −3081 was subcloned in antisense orientation 5′ of the pNET133(i)CAT. The normalized CAT activities driven by an A allele containing construct in each cell line was set to 100 to compare the relative strength of a T allele-containing construct. Significant differences between A and T alleles were evaluated by unpaired t test with two-tailed P values: ∗, P < 0.05; ∗∗, P < 0.005; ∗∗∗, P < 0.0005. (B) A 4-kb upstream sequence and the first intron of SLC6A2 were inserted upstream of the luciferase coding sequence in the pGL3 basic (Promega) vector. These constructs were designed to contain the A or T allele at position −3081 and transiently transfected into cell lines. (C) Six copies of the oligonucleotide containing A or T alleles were coupled to a heterologous TK promoter and a luciferase reporter gene. To compare luciferase activities between two constructs, luciferase activity driven by TK promoter was set to 100.
Specific DNA–Protein Interaction at a Perfect Palindromic Sequence Motif Containing −3081(T).
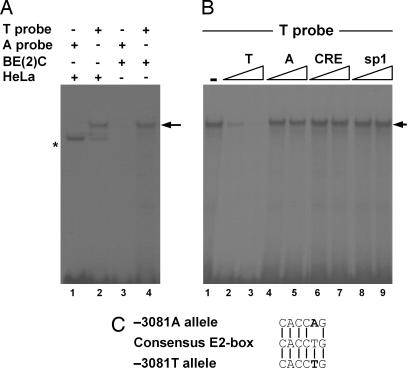
Next, we addressed whether DNA-binding protein(s) interact with promoter sequences encompassing this polymorphism and/or whether the binding activities are altered. When an oligonucleotide probe surrounding the −3081 position was incubated with nuclear extracts from HeLa or SK-N-BE (2)C cells, a single major DNA–protein complex was detected with the −3081(T) allele probe but not with the −3081(A) allele probe, suggesting that the −3081(T) polymorphism creates a new binding site (indicated by an arrow in Fig. 3A). As shown in Fig. 3B, competition assay showed that formation of this DNA–protein complex was diminished by unlabeled −3081(T) oligonucleotide but not by −3081(A) or the unrelated CRE and Sp1 oligonucleotides. Thus, this DNA–protein complex seems to be sequence-specific. To determine whether −3081(T) created a cis-acting element(s), we used the TRANSFAC database to search for known transcription factor-binding motifs (22). Interestingly, the −3081(T) allele, but not the −3081(A) allele, contains a potential E2-box motif (CACCTG) (Fig. 3C), which is known to be a binding site for the basic helix–loop–helix class of transcription factors.
Fig. 3.
Allele-specific binding of protein to oligonucleotide surrounding polymorphic site. (A) EMSA were conducted by using nuclear extracts from SK-N-BE (2)C and HeLa cells with labeled probes for the allele A (lanes 1 and 3) and for the allele T (lanes 2 and 4). An allele-specific complex was indicated by the arrow at right. A nonspecific complex was shown by the asterisk. (B) EMSA were performed by using nuclear extracts from SK-N-BE (2)C cells with 32P-labeled oligonucleotide containing allele T. This DNA–protein complex was competed by unlabeled −3081(T) oligonucleotide but not by either −3081(A) or unrelated CRE and sp1 oligonucleotides. (C) Schematic representation of E2-box DNA sequence created by polymorphism at −3081.
Zinc Finger Transcriptional Repressors Slug and Scratch Interact with the E2-box Element Created by the −3081(T) Polymorphism.
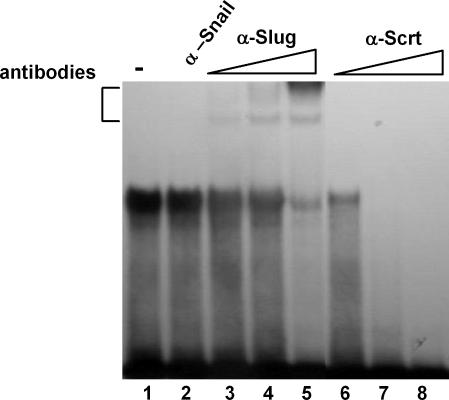
Because the −3081(T) polymorphism specifically repressed promoter activities, we searched for a transcriptional repressor(s) that can bind the consensus E2-box element. Transcription regulators called δEF1/ZEB and Snail superfamily (Snail, Slug, and Scratch) have been reported to bind to a consensus E2-box (CACCTG) and function as potential transcriptional repressors (23, 24). These zinc finger transcription factors are implicated in neural crest development and/or promoting neuronal differentiation. To investigate whether these transcriptional repressors may be involved in DNA–protein interaction with the oligonucleotide probe containing the T allele, we performed EMSA by using in vitro-translated proteins. Incubation of recombinant δEF1/ZEB and Snail produced no complexes with the oligonucleotide probe containing either the A or T allele. In contrast, recombinant Slug and Scratch generated a single complex with the oligonucleotide probe containing the T allele but not the A allele (see SI Fig. 7). Next, we carried out supershift EMSA by using antibodies against Snail, Slug, and Scratch. Incubation of nucleic extracts with anti-Slug and anti-Scratch, but not anti-Snail, retarded the migration of the major DNA–protein complex or diminished formation of complex (Fig. 4), strongly suggesting that Slug and Scratch are components of the major DNA–protein complex formed with the E2-box created by the −3081(T) allele. To investigate whether Slug and Scratch are coexpressed with NET, we performed RT-PCR by using NET-expressing or -nonexpressing cell lines. Slug and Scratch were expressed not only in NET-expressing but also in some NET-nonexpressing cell lines (see SI Fig. 8).
Fig. 4.
Slug and Scratch, but not Snail, bind to the E2-box generated by polymorphism at −3081 of SLC6A2. The −3081(T) oligonucleotide radiolabeled probe was incubated with SK-N-BE (2)C nuclear extract in the absence (lane 1) or presence of antibodies (lanes 2–8). Coincubation of nuclear proteins with increasing amounts of Slug- and Scratch-specific antibodies [0.2 μg (lanes 3 and 6), 0.5 μg (lanes 4 and 7), and 1 μg (lanes 5 and 8)] resulted in the generation of a supershifted band in a dose-responsive manner. In addition, formation of DNA–protein complex was significantly diminished. In contrast, coincubation with Snail-specific antibody (1 μg) neither generated the supershifted band nor diminished formation of DNA–protein complex (lane 2).
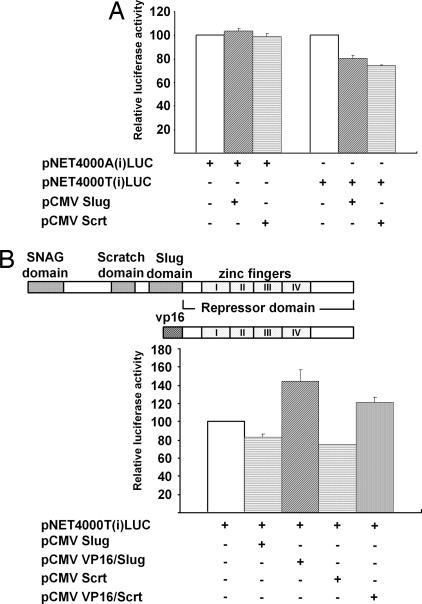
Slug and Scratch Decrease the SLC6A2 Promoter Activity in an Allele-Specific Manner.
To investigate whether Slug and Scratch directly affect the SLC6A2 promoter activity, SK-N-BE (2)C was cotransfected with Slug or Scratch expression vector along with pNET4000(i)LUC(A) or pNET4000(i)LUC(T) reporter constructs. Interestingly, both Slug and Scratch repressed the promoter activity of pNET4000(i)LUC(T), but not that of pNET4000(i)LUC(A), indicating that Slug and Scratch can repress the reporter gene activity in an allele-specific manner (Fig. 5A). To investigate whether Slug and Scratch are responsible for repression observed in the pNET4000(i)LUC(T) constructs, we replaced the region N-terminal to the DNA-binding domain of Slug and Scratch with the transactivation domain from herpes simplex virus VP16, respectively. When cotransfected with the pNET4000(i)LUC(T) construct, Slug or Scratch expression vectors further decreased the reporter gene activity (Fig. 5B). Interestingly, decreases of promoter activity by T allele-derived sequence could be recovered by cotransfection with VP16/Slug expression vector, suggesting the direct involvement of the Slug and Scratch proteins in repression by T allele-derived sequences. Taking these results together, we conclude that Slug and Scratch proteins can bind to the E2-box element created by the −3081(T) polymorphism and result in decreased transcriptional activity.
Fig. 5.
Slug and Scratch transrepress the SLC6A2 promoter in an allele-specific manner. (A) SK-N-BE (2)C cells were transiently cotransfected with reporter constructs and Slug or Scratch expression vectors. To compare the transrepression directly, basal luciferase activity driven by each reporter construct was set to 100%. (B) Diagram of the overall structure of Snail family protein and dominant-negative construct. The activating forms of Slug and Scratch were constructed by replacing the N-terminal repressor domain with the activation domain of VP16 (Upper). The −3081(T) allele of the 4.0-kb SLC6A2 promoter-luciferase reporter constructs were cotransfected with the either Slug or Scratch expression vector and dominant-negative expression plasmids. Here, basal promoter activity driven by the 4.0-kb SLC6A2 promoter-luciferase reporter constructs containing the T allele at −3081 was set to 100%.
The −3081(T) Polymorphism Is Associated with ADHD.
We tested whether the SLC6A2 −3081(A/T) polymorphism was associated with childhood ADHD by genotyping this SNP using PCR-restriction fragment length polymorphism (RFLP) analysis (see SI Fig. 6) in 94 patients who met Diagnostic and Statistical Manual of Mental Disorders, 4th Ed. (DSM-IV) (25) criteria for ADHD and in 60 unaffected controls. We evaluated statistical significance by using nominal P values but also examined significance by using permutation tests given our sample size. The permutation-based significance levels were only slightly higher than standard P values (i.e., ≈5–10%) and thus did not materially alter any of our conclusions regarding the associations between ADHD and the −3081(A/T) SNP. We first established that this SNP was in Hardy–Weinberg equilibrium by using a simulation-based method (26) in the full sample (χ2 = 0.59, P = 0.566), as well as separately in cases (χ2 = 0.35, P = 0.711) and controls (χ2 = 0.001, P = 1.000). The frequency of the −3081(T) allele in ADHD cases was 0.37 compared with 0.22 in controls, a statistically significant difference [χ2 = 7.07, P = 0.008, odds ratio (OR) = 2.00, 95% confidence interval (CI) = 1.19 − 3.37]. Differences in the genotypes of ADHD cases and controls also were significant (logistic regression results for the linear trend contrasting the number of T alleles: χ2 = 6.49, P = 0.011, R2 = 0.06). Similar to the allele-wise results, the AT and TT genotypes were overrepresented in ADHD cases, whereas the AA genotype was overrepresented in controls (ADHD cases: AA = 0.42, AT = 0.44, TT = 0.15; controls: AA = 0.60, AT = 0.35, TT = 0.05).
A potential confound in case-control studies of association is population stratification biases, which most commonly are attributable to the sampling of individuals from different ethnic backgrounds that vary in both allele frequencies at the tested marker and in rates of the disorder being studied (27). A search of the HapMap database (28) revealed substantial allele frequency differences between the European-American and African-American samples for many of the SNPs in SLC6A2, suggesting the need to control for ethnic background in testing for association between SNPs in this gene and ADHD. Thus, we first examined ethnic differences in allele frequencies at the −3081(A/T) SNP (see SI Table 1). Our analyses showed considerable differences in allele frequency between individuals of African-American background and those of European-American, Hispanic, or Asian ethnicity (χ2 = 15.50, P < 0.001, OR = 4.85, 95% CI = 2.08 − 11.36), such that the frequency of the T allele was 0.65 in individuals of African-American background as compared with 0.28 in individuals of European-American, Hispanic, or Asian ethnicity. Similar differences were observed for the comparison of genotype frequencies between individuals of African-American and the other ethnic backgrounds (χ2 = 19.11, P < 0.001, φ = 0.35). In contrast, no such differences were observed among the individuals of European-American, Hispanic, or Asian ethnicity (χ2 = 2.21, P = 0.696, φ = 0.12), suggesting that these groups could be combined in the subsequent case-control analyses in which ethnicity was controlled (also see SI Table 1).
Because our clinic samples were collected at two independent sites, it also was important to control for any possible heterogeneity across site (see SI Table 2). After removing individuals of African-American ethnic background from the sample, we thus tested for and found genotype differences between the cases sampled from Vanderbilt University Medical Center and McLean Hospital (χ2 = 9.88, P = 0.007, φ = 0.35). (Similar analyses using logistic regression in which ethnicity was controlled yielded virtually identical results).
Given the heterogeneity we observed because of both ethnic background and clinic site, we next reanalyzed the association presented above between ADHD and the −3081(A/T) SNP after removal of the 13 individuals of African-American ethnic background and controlling for clinic site. (We also statistically controlled for African-American versus European-American, Hispanic, or Asian ethnicity and obtained virtually identical results.) Differences in the genotypes of ADHD cases and controls remained significant after removal of the individuals of African-American ethnic background and when clinical site was controlled via its inclusion as a covariate in the analyses (logistic regression results for the linear trend contrasting the number of T alleles: χ2 = 4.79, P = 0.029, OR = 3.38, 95% CI = 1.14 − 10.10, R2 = 0.06). Similar to the results above, the AT and TT genotypes were still overrepresented in ADHD cases, whereas the AA genotype was overrepresented in controls.
Discussion
The NET critically regulates NE neurotransmission and homeostasis (2) and is a prime target for the treatment of psychiatric disorders such as depression and ADHD. Accordingly, genetic variations of SLC6A2 are considered to be promising candidates for NE-related disorders. For instance, several polymorphisms have been identified in the coding regions of SLC6A2 (12, 29) including a functional missense mutation (Ala457Pro) that is linked to orthostatic intolerance. In contrast, similar analyses of SLC6A2 variants with various psychiatric disorders, including major depression, Tourette syndrome, bipolar disorder, schizophrenia, and alcoholism have failed to reveal significant association (29). Studies of the association between SLC6A2 SNPs and ADHD have yielded mixed results, with several studies finding no evidence for association (14, 15) and more recent studies providing some evidence for association (16, 17). It is likely that future association studies will show greater yield if they focus on polymorphisms known to be functional, such as the promoter SNP that we have reported on herein.
We focused on the identification of functional polymorphisms in the SLC6A2 promoter sequence because a more common, regulatory variant may be more likely to support vulnerability to the development of common psychiatric disorders than the known rare variants found within the transporter's coding region (30). In the present study, we have screened and identified a common polymorphism at the base pair −3081 promoter of SLC6A2 (20). Several lines of evidence suggest that this −3081(A/T) variant is a functional promoter polymorphism. First, the T allele significantly down-regulated the proximal promoter function of SLC6A2 by ≈50% in NET-expressing cell lines (P < 0.005) (Fig. 2A). Second, the T allele again down-regulated the intact SLC6A2 promoter construct containing the 4-kb upstream sequence as well as the first untranslated intron region, by 25% to 28% (P < 0.0005) (Fig. 2B). Third, a synthetic promoter containing the −3081(T) allele exhibited dramatically decreased promoter function compared with the wild-type allele (P < 0.0005) (Fig. 2C). Together, our results strongly suggest that this A to T variation represents a functional polymorphism that significantly down-regulates the SLC6A2 promoter activity.
Another salient feature of this study is that the T allele at −3081 creates a new E2-box consensus motif (5′-CACCTG-3′) and that this new sequence motif, but not the wild-type sequence, interacts with proteins in a sequence-specific manner (Fig. 3). Because the T allele decreases SLC6A2promoter activity, we speculated that the interacting protein might be a transcriptional repressor binding to the E2-box motif. Indeed, our EMSA and antibody supershift assays demonstrated that Slug and Scratch, which belong to Snail superfamily, bind to the E2-box motif created by the −3081(T) polymorphism (Fig. 4). Forced expression of Slug or Scratch differentially down-regulated the T allele-containing SLC6A2 promoter function but not the A allele-containing promoter (Fig. 5A). Finally, conversion of the transcriptional repression domain of Slug and Scratch to the VP16 activation domain not only recovered the decreased promoter activity of the T allele but also up-regulated its function (Fig. 5B). Taking these results together, we conclude that the −3081(A/T) polymorphism creates a new E2-box motif, that it interacts with transcriptional repressors Slug and Scratch, and that it results in down-regulated promoter function of SLC6A2. Slug is involved in the formation of mesoderm and neural crest development (24), whereas Scratch is implicated in promoting neuronal differentiation and is expressed in almost all brain regions (31) (also see the Allen Brain Atlas, www.brainatlas.org). Further studies are necessary to delineate functional roles of Slug and/or Scratch on temporal and spatial regulation of SLC6A2 expression and their association with ADHD.
Given the physiological and clinical importance of NET and NET antagonists, our working hypothesis is that the −3081(T) SNP could be associated with a variety of NE-related human disorders. To initiate such analyses, we performed an association study to contrast allele and genotype frequencies for the −3081(A/T) polymorphism in ADHD cases versus unaffected controls. Interestingly, the frequency of the −3081(T) allele in ADHD patients was significantly higher than in controls (0.32 versus 0.22, respectively, in individuals who were not of African-American background). It is particularly noteworthy that this association remained after controlling for heterogeneity because of both ethnic background and clinic site. Thus, our tests of association suggest that the −3081(T) allele in SLC6A2 may be a “risk-inducing” allele for ADHD and may play a significant role in its etiology. Given that the sample size used herein is relatively small, however, a more extensive investigation is warranted to test conclusively whether the −3081(A/T) SNP is associated with ADHD and its constituent diagnostic subtypes and symptom dimensions. In follow-up studies, we plan to test for differential association between this polymorphism and the three ADHD diagnostic subtypes (Inattentive, Hyperactive-Impulsive, and Combined) as well as with treatment response. We also intend to characterize the pattern and magnitude of linkage disequilibrium between this functional promoter polymorphism and tagging SNPs across the gene and to test whether it accounts for any association observed between these other SNPs and ADHD. Furthermore, our demonstration that transcription factors Slug and Scratch interact with the polymorphism suggests the possibility that certain transcription factors/repressors and their own variations may contribute to the predisposition to certain psychiatric and cognitive disorders.
Materials and Methods
Further details of cell culture and mRNA isolation, RT-PCR, transient transfection assays, and preparation of nuclear extracts can be found in SI Text.
Genomic DNA Analysis.
For analysis of genomic DNA samples, a 1,360-bp region containing nucleotides −4000 to −3018 of SLC6A2 was amplified by PCR with primers NET24S (5′-GAATTCAGGGCAGGTCAGCTG-3′) and NET38A (5′-GAGACAGCAAAGGGAAGGAAACCA-3′). PCR was carried out under the condition used in our previous study (21). The resulting PCR products then were purified by 7% polyacrylamide gel electrophoresis and directly sequenced. To detect polymorphisms in SLC6A2, 88 samples from healthy subjects used for our previous study (21) were initially sequenced. One common SNP was detected (SI Fig. 6), for which we developed PCR-restriction fragment length polymorphism (RFLP) strategies for rapid genotyping. A 148-bp region containing nucleotides −3165 to −3018 of SLC6A2 was amplified by PCR with primers NET137S (5′-CTGTAGTTTTCTTGCCCCTCAAG-3′) and NET38A. The PCR fragment then was digested with 5 units of BsrI at 65°C for 2 h for −3081(A/T). Digestion products were run on 7% polyacrylamide gels.
Plasmid DNA Constructs.
The luciferase plasmid pNET4000(i)LUC(A) was obtained by insertion of a SalI and XhoI fragment of the plasmid pNET4000(i)CAT (20) into pGL3-Basic (Promega, Madison, WI). The fragment between −4000 and −3018 was amplified by PCR, digested with HindIII, and subcloned into pBLCAT2 or pNET133(i)CAT (20). Constructs containing the T nucleotide at −3081 were generated by using the Transformer Mutagenesis kit (Clontech, Mountain View, CA). Double-stranded DNAs containing base pairs −3092 to −3072 were generated by annealing the sense and antisense oligonucleotides containing either A or T nucleotide at the −3081 position. Multimers were blunt-ended with the Klenow fragment and inserted upstream of the phRL-TK (Promega). An 807-bp Slug, a 785-bp Snail, and a 1,047-bp Scratch cDNA were produced by RT-PCR from SK-N-BE (2)C mRNA. The PCR fragments were cloned into the pCDNA3.1 zeo(+) (Invitrogen, Carlsbad, CA) to produce pCMVSlug, pCMVSnail, and pCMVScrt, respectively. The VP16/Slug and VP16/Scrt hybrids were constructed by replacing the N-terminal 118 aa of Slug and the N-terminal 162 aa of Scratch with the transactivation domain of the VP16 protein, respectively. δEF1 expressing plasmid was kindly provided by H. Kondoh (Osaka University, Osaka, Japan). The integrity of all sequences was verified by DNA sequence analyses.
EMSA.
The −3081(A/T) oligonucleotides were synthesized with either the A or T variant centrally located and annealed to produce double-stranded DNA (5′-GGCTGAGCACCA/TGTTTCCCCA-3′) and 32P-labeled by T4 DNA kinase. Competition-binding assays were performed by adding nonradioactive oligonucleotides in molar excess before adding 32P-labeled oligonucleotides. For supershift assays, antibodies were coincubated with the nuclear extract for 30 min at 4°C before adding the radiolabeled probe. Antibodies against Slug and Snail were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Scratch was purchased from Affinity Bioreagents (Golden, CO).
Subjects.
All procedures involving human subject participation were reviewed and approved by the Vanderbilt University Medical Center, Emory University, and McLean Hospital Institutional Review Boards. Buccal cell samples were collected from 68 subjects between the ages of 6 and 17 years. These data on the 68 ADHD patients were gathered under the auspices of the Center for Child Development and Clinical Trials Center at the Vanderbilt University Medical Center, whereas data on the 60 nondisordered comparison children were sampled in Atlanta, GA. At the Vanderbilt site, the Kiddie-SADS-Present and Lifetime Version (32) was used to determine whether DSM-IV criteria for ADHD were met (33), whereas at the Emory site, a DSM-IV symptom questionnaire was used to rule out ADHD in the nondisordered controls. At the McLean site, ADHD subjects between the ages of 6 and 17 years were diagnosed when they met DSM-IV criteria.
Statistical Analysis.
Crosstabs and logistic regression analyses with two-tailed P values were conducted by using SPSS for Windows (SPSS, Chicago, IL) version 14 and were used to compare allele and genotype frequencies between ADHD cases and nondisordered controls. In cell studies, unpaired t tests with two-tailed P values were performed to compare transcriptional activities of the A versus T alleles and were performed by using the GraphPad Prism software (GraphPad Software, San Diego, CA).
Supplementary Material
Acknowledgments
We thank R. Steven Couch and Richard C. Shelton for assistance with collection of Pediatric ADHD subjects. We also wish to recognize the contribution of David C. Rowe, who played an integral role in beginning the candidate gene study of ADHD at Emory University. This work was supported by National Institutes of Health Grants MH48866 and DC006501 and a NARSAD Independent award (to K.-S.K.), by National Institutes of Health Grant HL56693 (to R.D.B.), and by National Institute of Mental Health Grants K01-MH01818 (to I.D.W.) and F31-MH072083 (to I.G.).
Abbreviations
- NE
norepinephrine
- NET
norepinephrine transporter
- ADHD
attention-deficit hyperactivity disorder
- TK
thymidine kinase
- DSM-IV
Diagnostic and Statistical Manual of Mental Disorders, 4th Ed.
- OR
odds ratio
- CI
confidence interval.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0510836103/DC1.
References
- 1.Iversen LL. Br J Pharmacol Chemother. 1963;21:523–537. doi: 10.1111/j.1476-5381.1963.tb02020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu F, Gainetdinov RR, Wetsel WC, Jones SR, Bohn LM, Miller GW, Wang YM, Caron MG. Nat Neurosci. 2000;3:465–471. doi: 10.1038/74839. [DOI] [PubMed] [Google Scholar]
- 3.Keller NR, Diedrich A, Appalsamy M, Tuntrakool S, Lonce S, Finney C, Caron MG, Robertson D. Circulation. 2004;110:1191–1196. doi: 10.1161/01.CIR.0000141804.90845.E6. [DOI] [PubMed] [Google Scholar]
- 4.Amara SG, Kuhar MJ. Annu Rev Neurosci. 1993;16:73–93. doi: 10.1146/annurev.ne.16.030193.000445. [DOI] [PubMed] [Google Scholar]
- 5.Bruss M, Kunz J, Lingen B, Bonisch H. Hum Genet. 1993;91:278–280. doi: 10.1007/BF00218272. [DOI] [PubMed] [Google Scholar]
- 6.Pacholczyk T, Blakely RD, Amara SG. Nature. 1991;350:350–354. doi: 10.1038/350350a0. [DOI] [PubMed] [Google Scholar]
- 7.Marien MR, Colpaert FC, Rosenquist AC. Brain Res Brain Res Rev. 2004;45:38–78. doi: 10.1016/j.brainresrev.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Beane M, Marrocco RT. Prog Neurobiol. 2004;74:167–181. doi: 10.1016/j.pneurobio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Klimek V, Stockmeier C, Overholser J, Meltzer HY, Kalka S, Dilley G, Ordway GA. J Neurosci. 1997;17:8451–8458. doi: 10.1523/JNEUROSCI.17-21-08451.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owens MJ, Morgan WN, Plott SJ, Nemeroff CB. J Pharmacol Exp Ther. 1997;283:1305–1322. [PubMed] [Google Scholar]
- 11.Biederman J, Heiligenstein JH, Faries DE, Galil N, Dittmann R, Emslie GJ, Kratochvil CJ, Laws HF, Schuh KJ. Pediatrics. 2002;110:e75. doi: 10.1542/peds.110.6.e75. [DOI] [PubMed] [Google Scholar]
- 12.Shannon JR, Flattem NL, Jordan J, Jacob G, Black BK, Biaggioni I, Blakely RD, Robertson D. N Engl J Med. 2000;342:541–549. doi: 10.1056/NEJM200002243420803. [DOI] [PubMed] [Google Scholar]
- 13.Urwin RE, Bennetts B, Wilcken B, Lampropoulos B, Beumont P, Clarke S, Russell J, Tanner S, Nunn KP. Mol Psychiatry. 2002;7:652–657. doi: 10.1038/sj.mp.4001080. [DOI] [PubMed] [Google Scholar]
- 14.De Luca V, Muglia P, Jain U, Kennedy JL. Am J Med Genet B Neuropsychiatr Genet. 2004;124:38–40. doi: 10.1002/ajmg.b.20075. [DOI] [PubMed] [Google Scholar]
- 15.McEvoy B, Hawi Z, Fitzgerald M, Gill M. Am J Med Genet. 2002;114:665–666. doi: 10.1002/ajmg.10416. [DOI] [PubMed] [Google Scholar]
- 16.Bobb AJ, Addington AM, Sidransky E, Gornick MC, Lerch JP, Greenstein DK, Clasen LS, Sharp WS, Inoff-Germain G, Wavrant-De Vrieze F, et al. Am J Med Genet B Neuropsychiatr Genet. 2005;134:67–72. doi: 10.1002/ajmg.b.30142. [DOI] [PubMed] [Google Scholar]
- 17.Xu X, Knight J, Brookes K, Mill J, Sham P, Craig I, Taylor E, Asherson P. Am J Med Genet B Neuropsychiatr Genet. 2005;134:115–118. doi: 10.1002/ajmg.b.30160. [DOI] [PubMed] [Google Scholar]
- 18.Cook EH, Jr, Stein MA, Krasowski MD, Cox NJ, Olkon DM, Kieffer JE, Leventhal BL. Am J Hum Genet. 1995;56:993–998. [PMC free article] [PubMed] [Google Scholar]
- 19.Waldman ID, Rowe DC, Abramowitz A, Kozel ST, Mohr JH, Sherman SL, Cleveland HH, Sanders ML, Gard JM, Stever C. Am J Hum Genet. 1998;63:1767–1776. doi: 10.1086/302132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim CH, Hwang DY, Park JJ, Kim KS. J Neurosci. 2002;22:2579–2589. doi: 10.1523/JNEUROSCI.22-07-02579.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim CH, Zabetian CP, Cubells JF, Cho S, Biaggioni I, Cohen BM, Robertson D, Kim KS. Am J Med Genet. 2002;108:140–147. [PubMed] [Google Scholar]
- 22.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Development (Cambridge, UK) 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 24.Nieto MA. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 25.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th Ed. Am Psychiatric Assoc: Washington, DC; 1994. [Google Scholar]
- 26.Cubells JF, Kobayashi K, Nagatsu T, Kidd KK, Kidd JR, Calafell F, Kranzler HR, Ichinose H, Gelernter J. Am J Med Genet. 1997;74:374–379. doi: 10.1002/(sici)1096-8628(19970725)74:4<374::aid-ajmg7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 27.Lander ES, Schork NJ. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 28.International HapMap Consortium. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stober G, Hebebrand J, Cichon S, Bruss M, Bonisch H, Lehmkuhl G, Poustka F, Schmidt M, Remschmidt H, Propping P, Nothen MM. Am J Med Genet. 1999;88:158–163. doi: 10.1002/(sici)1096-8628(19990416)88:2<158::aid-ajmg12>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 30.Hahn MK, Blakely RD. Pharmacogenomics J. 2002;2:217–235. doi: 10.1038/sj.tpj.6500106. [DOI] [PubMed] [Google Scholar]
- 31.Marin F, Nieto MA. Dev Dyn. 2006;235:2586–2591. doi: 10.1002/dvdy.20869. [DOI] [PubMed] [Google Scholar]
- 32.Ambrosini PJ. J Am Acad Child Adolesc Psychiatry. 2000;39:49–58. doi: 10.1097/00004583-200001000-00016. [DOI] [PubMed] [Google Scholar]
- 33.Kaufman J, Birmaher B, Brent D, Rao U, Flynn C, Moreci P, Williamson D, Ryan N. J Am Acad Child Adolesc Psychiatry. 1997;36:980–988. doi: 10.1097/00004583-199707000-00021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.