Abstract
High-throughput screening and optimization experiments are critical to a number of fields, including chemistry and structural and molecular biology. The separation of these two steps may introduce false negatives and a time delay between initial screening and subsequent optimization. Although a hybrid method combining both steps may address these problems, miniaturization is required to minimize sample consumption. This article reports a “hybrid” droplet-based microfluidic approach that combines the steps of screening and optimization into one simple experiment and uses nanoliter-sized plugs to minimize sample consumption. Many distinct reagents were sequentially introduced as ≈140-nl plugs into a microfluidic device and combined with a substrate and a diluting buffer. Tests were conducted in ≈10-nl plugs containing different concentrations of a reagent. Methods were developed to form plugs of controlled concentrations, index concentrations, and incubate thousands of plugs inexpensively and without evaporation. To validate the hybrid method and demonstrate its applicability to challenging problems, crystallization of model membrane proteins and handling of solutions of detergents and viscous precipitants were demonstrated. By using 10 μl of protein solution, ≈1,300 crystallization trials were set up within 20 min by one researcher. This method was compatible with growth, manipulation, and extraction of high-quality crystals of membrane proteins, demonstrated by obtaining high-resolution diffraction images and solving a crystal structure. This robust method requires inexpensive equipment and supplies, should be especially suitable for use in individual laboratories, and could find applications in a number of areas that require chemical, biochemical, and biological screening and optimization.
Keywords: droplets, plugs, protein structure, high-throughput, miniaturization
This work reports a “hybrid” microfluidic approach that uses nanoliter plugs to perform screening and optimization simultaneously in the same experiment. To validate this method using a challenging problem, we demonstrate its compatibility with crystallization of membrane proteins. Small-scale screening and optimization experiments are important for biological assays, chemical screening, and protein crystallization (1–3). Screening and optimization are usually carried out sequentially. In the case of protein crystallization, random sparse matrix screening initially identifies the precipitants that may lead to crystallization. Subsequent gradient optimization establishes concentrations of these precipitants that lead to diffraction-quality crystals (4). Combining screening and optimization steps into a single hybrid experiment would eliminate the need to wait for the outcome of the initial screen before carrying out subsequent optimizations. Furthermore, a hybrid experiment would reduce the false negatives (5) associated with screens performed at a single concentration. The hybrid experiment could also be more conclusive, because a single batch of the sample would be used for both screening and optimization. Simultaneous screening and optimization could increase consumption of samples, so miniaturization is essential for the success of the hybrid method. Such a hybrid method could be especially valuable for the crystallization of membrane proteins.
Determining crystal structures of membrane proteins is the focus of major research efforts, with significant recent progress (6–10). Membrane proteins control a number of cellular signaling pathways and are targets of >50% of pharmaceutical drugs (11, 12). Crystallization of membrane proteins remains challenging (13, 14), because samples are often limited in quantity (15) and may be unstable over time. Also, an extensive range of crystallization conditions must be searched, including screening detergents used to solubilize membrane proteins (15–17). Technologies for miniaturization of crystallization experiments are needed, but handling solutions of membrane proteins is complicated by their low surface tension and high viscosity. To address these challenges, both robotic (18, 19) and microfluidic (20–23) technologies have been developed to screen crystallization conditions on a submicroliter scale for both soluble and membrane proteins, but no universally applicable technology has yet emerged. Given the complexity of the problem of crystallization of membrane proteins, it is unlikely that a single technology would provide a universal solution, but there is a clear need for miniaturized technologies sufficiently robust, simple, and inexpensive to be accessible to individual laboratories.
Results
Developing a Plug-Based Hybrid Screening Method.
To perform simultaneous hybrid screening and optimization, we used plugs [droplets surrounded by fluorinated carrier fluid and transported through microfluidic channels (24)]. Plugs have been used to perform the initial screening in a number of applications, including biochemical assays (25), chemical reactions (26), and crystallization of soluble proteins (22). In separate experiments, plugs have been used for optimization of protein crystallization conditions (23). Implementation of the hybrid method while minimizing sample consumption and labor was enabled by four key developments (Fig. 1):
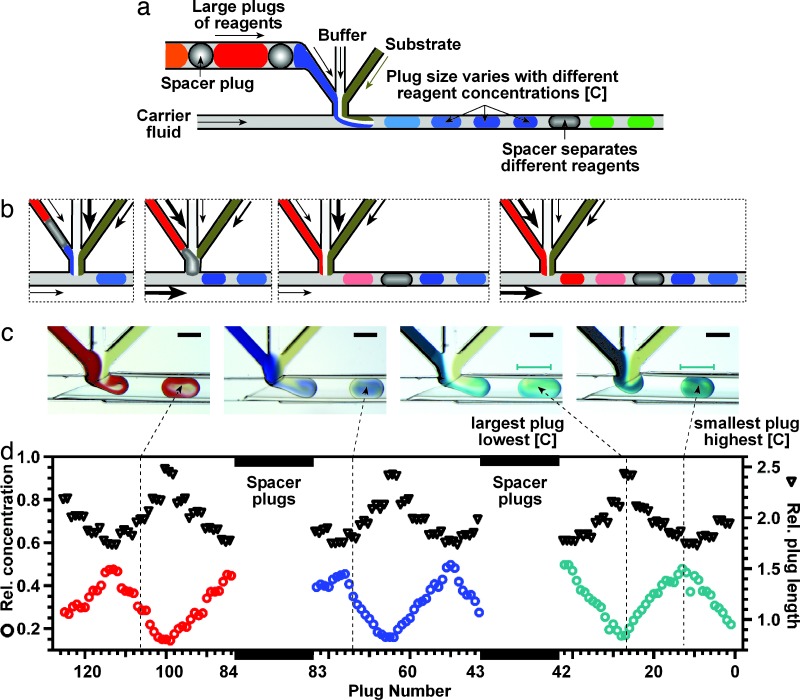
Fig. 1.
Microfluidic approach to perform hybrid screening. (a) Schematic illustration of the approach. A preformed array of ≈140-nl reagent plugs separated by ≈40-nl spacers is flowed into the microfluidic channel. The stream is combined with streams of buffer and substrate and flowed into a stream of a fluorinated carrier fluid. For each reagent, ≈50 smaller (≈10–15 nl) plugs are formed, each potentially containing a different concentration of the reagent. This concentration may be deduced from the size of the plug. (b) To form plugs containing different concentrations of reagents, the relative flow rates of the three streams are constantly changed to combine streams in several ratios (black arrows). At the same time, to index these concentrations, the sizes of plugs are changed by changing the flow rate of the carrier fluid (black arrows). Higher concentrations correspond to smaller plugs. (c) Microphotographs of plugs forming in hybrid screens, illustrated with plugs colored with dyes. Viscosities of solutions: 36 mPa s (red), 6 mPa s (blue), 1 mPa s (green). (d) A plot quantifying a hybrid screen, performed in a separate experiment. Precipitants were the same as in c but marked with fluorescent dyes instead of absorption dyes. Relative (Rel.) concentrations inside the plugs and sizes of plugs were measured from quantitative fluorescent images. The flow rate of the substrate was kept constant at 10 nl/s. The flow rates of the reagent and buffer streams were cycled with opposite phases from 10 nl/s to 3.3 nl/s in 1.7-nl/s steps, with each step taking 1.5 s. At the same time, the flow rate of the carrier fluid was cycled between 23.3 nl/s to 50 nl/s, in 6.7-nl/s steps, in phase with the reagent stream.
To deliver many distinct reagents in the same experiment, as required for the initial screen, we used a preformed array of reagent plugs, each plug ≈120–140 nl in volume. Each pair of plugs in the array was separated by ≈40-nl spacer to ensure reliable transport of reagent plugs of different viscosities and surface tensions (22, 27). As this array was flowed from the cartridge into the microfluidic device, each plug formed a long segment that could be manipulated as a continuous stream (Fig. 1a). This stream was combined with the streams of buffer (24) and substrate at a microfluidic junction. The three streams flowed continuously and laminarly into a flowing stream of fluorinated carrier fluid, forming a series of ≈50 small plugs (10–15 nl) as each 140-nl plug of reagent combined with buffer and substrate. An array of 20 reagents produced ≈1,000 small plugs.
For each reagent, to accomplish an optimization simultaneously with the initial screen, each of ≈50 small plugs contained a different concentration of the reagent. We used a LabView subroutine for computer-controlled pumps to vary the relative flow rates of the three streams, producing plugs containing different ratios (and different concentrations) of the reagent, buffer, and substrate (Fig. 1b). When the flow rates were changed in steps, each step produced a group of plugs with similar concentration. Higher flow rate of a stream corresponded to a proportionally higher concentration of that stream in the plug (Fig. 1c). Even with simple syringe pumps, concentrations may be controlled reliably over a 16-fold range (28).
To ensure that the optimization covers a full range of concentrations of each reagent, two approaches are possible. In the first, the arrival of each reagent plug is synchronized with the start of a computer subroutine that gradually changes the flow rate. Although such synchronization is possible when the volumes of reagents and spacers are known precisely, it complicates the procedure significantly and is not robust. We used the second approach, in which the subroutine gradually changes the flow rate of the reagent up and down and then repeats the cycle [see supporting information (SI) Fig. 5]. No synchronization was required in this approach as long as each reagent produced small plugs over a period longer than one up/down cycle, regardless of where the cycle started; each reagent produced at least one small plug of each concentration (Fig. 1d). A cycle with a gradual increase and an abrupt decrease is less redundant and saves substrate, but we found that abrupt changes of flow rate were more difficult to control precisely, possibly because of elasticity of poly(dimethlysiloxane) (PDMS). In addition, ≈2-fold redundancy (Fig. 1d) of this method provided an internal check of reproducibility.
To index the concentration of reagents in hundreds of plugs generated by this method, we relied on sensitivity of the size of the plug to the “water fraction” (29), the ratio of the combined volumetric flow rates of the aqueous streams to the total flow rate of the carrier fluid and the aqueous streams. At higher water fractions, larger plugs form (29). As the computer subroutine increased the relative flow rate of the reagent stream, it also decreased the flow rate of the buffer to keep the combined volumetric flow rates of the three aqueous streams constant. At the same time, the subroutine increased the flow rate of the carrier fluid, decreasing the water fraction and decreasing the size of the plugs. Within a series of ≈40 plugs containing the same reagent, the concentration of a reagent in each plug was inferred by measuring the size of the plug relative to the size of plugs in the beginning and the middle of the up/down cycle (Fig. 1 c and d). The size of plugs changes approximately linearly with water fraction, and we found the dependence to be slightly sensitive to viscosity (SI Fig. 6). Calibration would be required for very accurate measurements of concentrations directly from sizes of plugs. To identify which series of plugs contained which reagent, we relied on plugs of the spacer fluid separating these series of plugs.
Implementing the Hybrid Method to Handle Solutions of Membrane Proteins.
We chose crystallization of membrane proteins to validate this hybrid approach. We chose this problem both because of its relevance and the challenges in fluid handling it presents. This validation was enabled by two technical developments: the use of perfluoroamines as carrier fluids and the use of Teflon capillaries for the formation, transport, and storage of plugs (Fig. 2).
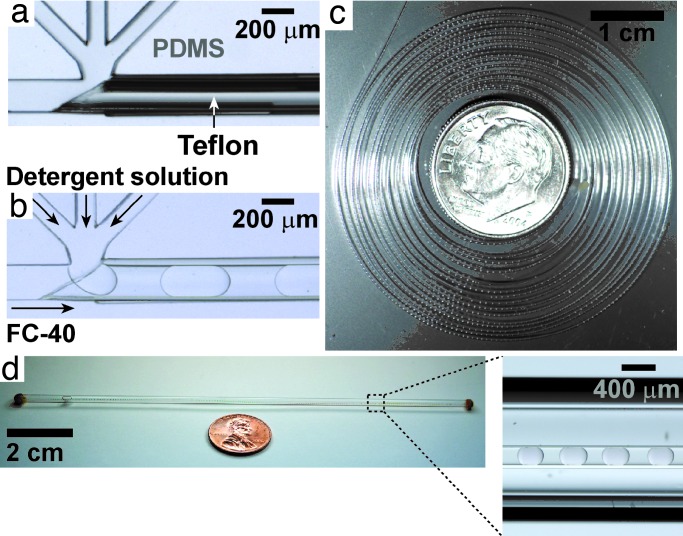
Fig. 2.
Microfluidic devices for forming and storing plugs of solutions of membrane proteins in aqueous detergents. (a) Microphotograph of an empty four-inlet PDMS device coupled to Teflon capillary. (b) Microphotograph of plugs of an aqueous solution of detergent [0.1% (wt/vol) LDAO in water] forming directly into a piece of Teflon capillary. (c) A photograph of ≈1,000 plugs stored in a 1-m-long piece of Teflon capillary wound around a dime. (d) Photograph of plugs inside Teflon capillary stored in a glass capillary filled with FC-70 to prevent evaporation of solutions and allow long-term storage.
Formation of plugs occurs at low values of the dimensionless capillary number, Ca = Uμ/γ, where U (m·s−1) is the flow velocity, μ (kg·m−1·s−1) is the dynamic viscosity, and γ (kg·s−2) is the surface tension at the interface between the aqueous phase and the carrier fluid (30). Perfluoroalkane-based carrier fluids, previously used for crystallization of soluble proteins (23), did not support formation of plugs of solutions containing membrane proteins and detergents. The high viscosity and low surface tension of these solutions increased the value of Ca above the usable range of Ca < ≈0.1. We were able to control the surface chemistry at the aqueous–fluorous interface (31) by identifying two perfluoroamines, FC-40 (a mixture of perfluoro-tri-n-butylamine and perfluoro-di-n-butylmethylamine) and FC-70 (perfluorotripentylamine), that provided a sufficiently high surface tension of 10–12 mN/m with the typical detergent solutions [e.g., 0.1% (wt/vol) lauryldimethylamine oxide (LDAO)].
To transport plugs through microchannels, the carrier fluid must wet the walls of the microchannels preferentially. Native PDMS surfaces did not support formation of plugs, because solutions of detergents wet them. Microchannels with either PDMS surfaces modified with a fluorinated silane (31) or native Teflon surfaces can be used to form plugs, because they are preferentially wet by FC-40 and FC-70. To form plugs, we used a thin-walled Teflon capillary, cut at an angle and inserted from the outlet of the PDMS device up to the plug-forming junction (Fig. 2a). Plugs formed without contacting the PDMS walls of the channel and flowed directly into the Teflon capillary (Fig. 2b). Plugs flowed reliably through long (>1 m) Teflon capillaries, because flow of plugs through channels of circular cross-section minimizes drainage of the carrier fluids and undesired coalescence of plugs (32, 33).
For short-term incubation, we used a 1-m-long capillary to store >1,000 crystallization trials in a compact format and at a cost of <1 cent per trial (Fig. 2c). Both the carrier fluid and water evaporated through 25-μm thin walls of amorphous Teflon, with a loss of 48% of water over 12 days. This evaporation could be used to increase the concentration of the components inside plugs to drive crystallization (20, 23) in analogy to a vapor-diffusion trial. For long-term incubation, Teflon capillaries were placed inside glass tubes (Fig. 2d) filled with the carrier fluid, reducing evaporation of the aqueous phase to below detection limit (0.5%) over 60 days. Switching between incubation with and without glass tubing allows one to stop and start evaporation at will or to perform crystallizations under microbatch (34) conditions.
We quantified the formation of plugs in the hybrid method by using reaction center (RC) from Rhodopseudomonas viridis (7.0 mg/ml) in a solution of LDAO [0.05% (wt/vol) in 10 mM Tris, pH 7.8]. Three aqueous reagents with viscosities ranging from ≈1 mPa s (water) to ≈6 mPa s [50% glycerol (wt/wt)] to ≈36 mPa s [25% (wt/vol) PEG 8000] were used to mimic the viscosities of common precipitants. To visualize the hybrid method, we added absorption dyes to these solutions (Fig. 1c). To measure concentrations quantitatively (standard dilution curves shown in SI Fig. 7), we used fluorescent dyes (Fig. 1d) but used a solution of LDAO instead of RC, because fluorescence of RC obscured the fluorescence of dyes. For all solutions, regardless of viscosity, increasing concentrations of precipitants corresponded to decreasing sizes of plugs, as expected (Fig. 1 c and d).
Before setting up crystallization trials in a particular detergent, the detergent was added to the precipitants in the screen and to the buffer. We found that detergents could be lost from solutions upon contact with hydrophobic PDMS. We have not yet established whether these losses occur by adsorption to the surface, or by absorption into PDMS. We found that losses were significant when submicroliter volumes of solutions of detergents with a low critical micelle concentration (CMC), such as n-dodecyl-β-d-Maltopyranoside [CMC ≈0.17 mM (0.0087%) in water], were flowed through ≈1 cm of a PDMS microfluidic channel. To reduce these losses, we used short inlet channels in PDMS devices. In addition, when using detergents with a low CMC, we primed the inlet channels with a solution of this detergent. Solutions of detergents with higher CMC, such as LDAO [CMC ≈1 mM (0.023%) in water] did not require priming. Once the solutions formed plugs in Teflon capillary, the layer of fluorinated carrier fluid surrounding the plugs prevented the loss of detergent to hydrophobic surfaces.
We performed hybrid crystallization screens under microbatch conditions (without evaporation) by using a custom-made screening kit of 48 precipitants with concentrations approximately twice those used in traditional vapor-diffusion screens. We measured the solubility of typical precipitants in FC-40 and FC-70 using NMR (SI Table 1) and established that detergents and some alcohols (such as ethanol and 2R,3R-butanediol) had solubilities of <10−2 M [≈0.02% (vol/vol)]. Ethanol was the most soluble at 8.5 × 10−3 M in FC-40. We concluded that the loss of these detergents and precipitants into the carrier fluids should not significantly affect the crystallization experiments.
Testing Compatibility of the Hybrid Method with Crystallization of Model Membrane Proteins.
We used two model proteins to test the compatibility of the hybrid method with membrane protein crystallization. By obtaining high-resolution diffraction of crystals of these proteins, we wished to confirm that all technical hurdles were identified and overcome successfully. To test handling of precipitants with high viscosity, we used 3.5 μl of Porin from Rhodobacter capsulatus [29.8 mg/ml in 0.6% (wt/vol) n-octyltetraoxyethylene (C8E4) and 20 mM Tris pH 7.8]. We used a viscous precipitant with viscosity of ≈250 mPa s [containing 70% (wt/vol) PEG 550MME and 3 M LiCl] chosen to approximate reported (35) crystallization conditions. We used two plugs of this precipitant in a hybrid screen together with seven other precipitants (SI Table 2). Using 3.5 μl of the protein, starting with a premade array of precipitant plugs, we set up ≈450 crystallization trials in 6 min. Only the smallest plugs [30% (wt/vol) PEG 550MME, 1.9% (wt/vol) C8E4, and 1.3 M LiCl] produced Porin crystals, whereas plugs of lower concentration (larger size) were all clear. Crystals (Fig. 3b) formed in both series of plugs containing PEG 550MME, confirming that fluid handling was performed correctly and that the system was compatible with crystallization of this protein.
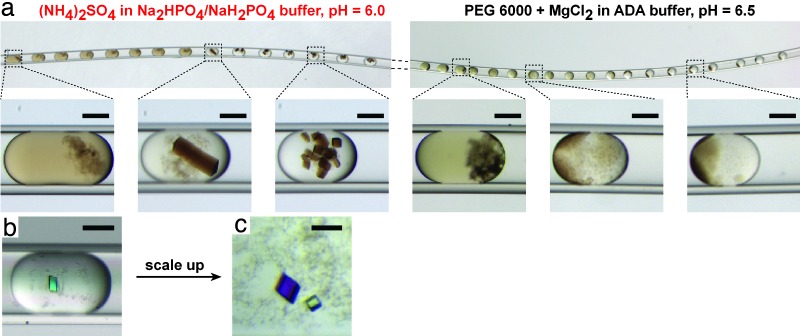
Fig. 3.
Hybrid screens of crystallization conditions for membrane proteins, illustrated with two model proteins. (a) Microphotographs of two regions in a Teflon capillary containing plugs from a hybrid screen performed for RC from R. viridis [provided by Nina Ponomarenko and James R. Norris (University of Chicago)]. As the concentration of one precipitant is increased (Left), a transition is observed from slight precipitation to large, single crystals to small microcrystals. For another precipitant (Right), a transition from precipitation to phase separation is seen. (b) An initial small crystal obtained in a hybrid screen of Porin from R. capsulatus (protein provided by the Philip D. Laible group at Argonne National Laboratory). (c) Larger crystals of Porin were obtained by scaling up the trials into 600-μm glass capillaries. (Scale bars: 100 μm.)
To test whether the conditions could be scaled up to obtain larger crystals, we repeated the screen in a larger device fitted with a silanized glass capillary 600 μm in diameter. Larger plugs gave larger crystals (Fig. 3c). In situ x-ray data collection (36–38) directly in capillaries was carried out at 4°C by using synchrotron radiation. Single shots showed that the Porin crystals diffracted up to 1.95 Å (Fig. 4a) with mosaicity <0.3°, establishing that crystals of high-quality could be formed and that these crystals could be manipulated in plugs without any damage.
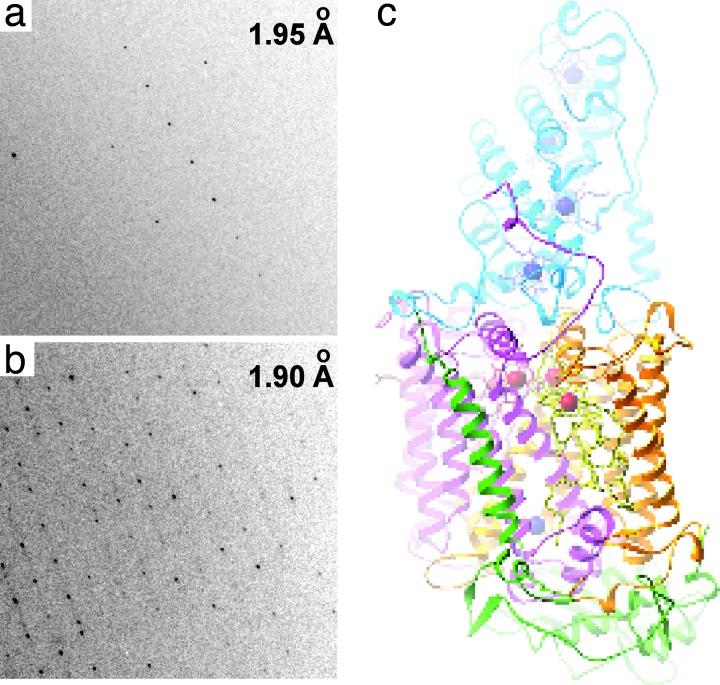
Fig. 4.
Using x-ray diffraction to characterize quality of crystals of model membrane proteins obtained with the hybrid method. (a and b) High-resolution parts of a diffraction pattern of Porin from R. capsulatus (a) and RC from R. viridis (b). The average signal-to-noise (S/N) ratio exceeds 10 for the row of reflections centered at 1.95 Å and 1.9 Å for Porin (a) and RC (b) crystals, respectively. (c) Structure of RC from R. viridis (refined at 1.96-Å resolution) obtained from crystals grown with the hybrid method.
We conducted a hybrid screen of RC (22 mg/ml/0.08% LDAO/7% heptane-triol/4.5% triethylammonium phosphate solution in 20 mM Na2HPO4/NaH2PO4 buffer, pH 6.0) using a total of 20 reagent plugs of 18 precipitants (SI Table 3). In addition to 16 precipitants from the custom-made kit, we used 2 precipitants, both in duplicate plugs: one was a positive control based on the original crystallization condition (39, 40) [4M (NH4)2SO4 in 50 mM Na2HPO4/NaH2PO4 buffer pH 6.0], and another was a modified crystallization condition at higher pH [3.6 M (NH4)2SO4 in 50 mM Tris, pH 7.8]. The duplicates were placed at random positions within the array of precipitants. By using 10 μl of protein for these 18 precipitants, 1,300 crystallization trials were set up within 20 min. Both the positive control and the modified condition induced crystallization. As the concentration of the ammonium sulfate increased from plug to plug (detected by the decrease in the sizes of plugs), we observed the expected trend: a mixture of the protein solution with light precipitation, progressing to single crystals, and then progressing to microcrystals (Fig. 3a). Reagents that did not lead to crystals also showed characteristic transitions. For example, PEG 6000-based precipitant caused precipitation at low concentrations, whereas it caused phase separation at higher concentrations (Fig. 3a). The transition was reproducible within pairs of duplicates, confirming the correlation between size and concentration as well as a lack of cross-contamination.
We assessed the quality of the crystals of RC obtained in a hybrid screen and tested the compatibility of this method with cryocrystallography. Crystals were slowly (to reduce shear) flowed out of the Teflon capillary and into a solution of paraffin oil as the cryoprotectant. We did not observe any crystals adhering to the walls of the Teflon capillary, presumably because of the layer of the fluorinated carrier fluid separating the plugs from the wall (24, 36). Crystals were then looped and flash-frozen in liquid nitrogen. X-ray data were collected at 100 K by using synchrotron radiation. The RC crystal grown at the original conditions (pH 6.0) diffracted up to 2.5 Å. Before data collection, this crystal was stored in Teflon capillary surrounded by FC-70 in a glass capillary for ≈2 months, confirming that long-term storage of crystals in plugs does not damage them. The modified condition (pH 7.8) gave better crystals, diffracting to 1.9 Å (Fig. 4b). Two of these crystals were used to collect a complete (96.5%) x-ray data set. Because of radiation damage, the resolution cutoff was 1.96 Å. The average mosaicity was <0.3°. The RC structure was refined to R factor of 17.2% (Rfree 18.6%) with well defined electron density for protein and solvent parts of the structure (Fig. 4c). This experiment confirmed that high-quality crystals can be grown directly in ≈10-nl plugs of the hybrid screen and extracted from the capillary without damage for traditional data collection at cryogenic temperatures.
Discussion
The microfluidic hybrid method described in this article provides a simple and economical procedure for simultaneous screening and optimization in nanoliter volumes. The use of plugs simplifies this approach: multiple distinct reagents are introduced serially without cross-contamination, reagents and substrate are combined by using a single passive microfabricated feature (a junction of channels), producing hundreds of plugs, with each plug representing an independent experiment. These plugs are flowed into an inexpensive piece of capillary and stored and monitored there without evaporation or damage to crystals. Overall, we found this method to be effective and now routinely use it in our laboratory. Within 1 h, using simple syringe pumps and only 17 μl of a substrate solution, a single researcher has set up a hybrid screen for 48 different reagents (≈40 plugs per reagent) to sample a total of ≈1,900 experimental conditions. We expect this method to operate on smaller scales as well, because the capillary number that governs fluid flow in these systems is not explicitly sensitive to the dimensions of the channels or plugs. Such miniaturization would require arrays of reagents in smaller plugs (41) and reliable pumping at lower flow rates.
A volume of ≈10 nl proved to be the optimal volume for plugs used for crystallization of model membrane proteins: it saved sample but contained enough protein to produce crystals of sufficient size and quality for high-resolution diffraction. Extracting and manipulating crystals grown in plugs was also straightforward, making this system compatible with both cryo (4) and in situ (36–38) x-ray diffraction. This method overcame the challenges associated with handling of solutions of membrane proteins and provided control of wetting, evaporation, and loss of detergents. Here, we have shown that this hybrid method is compatible with crystallization of membrane proteins. To establish its ultimate usefulness, one would need to screen a panel of 20–30 new, but biochemically well characterized, membrane proteins. Our laboratory does not yet have access to such a panel of proteins, but there is a significant effort to produce them under the Protein Structure Initiative and under the National Institutes of Health Roadmap (www.nigms.nih.gov/Initiatives/PSI/Centers/ and http://nihroadmap.nih.gov/structuralbiology/index.asp). We welcome both disseminating this technology and receiving such panels of samples for testing in our laboratory.
Having successfully handled solutions of membrane proteins, it was not surprising that this method had no problems handling aqueous solutions free of detergents. We have used this method to implement crystallization screens with commercial kits from Hampton Research (Aliso Viejo, CA) and Emerald Biostructures (Bainbridge Island, WA). We do not anticipate problems performing enzymatic assays with this method (22, 28), but it remains to be established whether this method is compatible with screening of samples containing live cells, an area that could also benefit from a nanoliter hybrid screen. We used optical detection in all of the experiments, by using a microscope scanning stage or by pulling the capillary with plugs through the microscope's field of view. This method would benefit from simple automated systems for detection in plugs, either optical or based on mass spectrometry (26). Beyond crystallization, we believe this method would find applications in a number of areas (5, 42–46) of chemistry, biochemistry, molecular biology, and material science that require simultaneous screening and optimization economically and in small volumes.
Materials and Methods
See SI Supporting Text for detailed procedures and additional characterizations and control experiments.
Microfluidic Hybrid Experiments.
Preformed arrays (41) and microfluidic devices (28, 31) were prepared as described previously, by using mask design and dimensions as in SI Fig. 8. A Teflon capillary (OD 250 μm, i.d. 200 μm; Zeus, Orangeburg, SC), cut at an angle (≈45°) to facilitate the transfer of plugs, was inserted into the junction of microchannels of the hybrid device through the outlet microchannel. Connections of Teflon and PDMS were sealed with capillary wax (Hampton Research). Teflon capillaries outside the PDMS device were put in standard wall glass tubing (Chemglass, Vineland, NJ) filled with FC-70 (Acros Organics, Morris Plains, NJ) to prevent evaporation. The ends of the glass tubing were sealed with capillary wax. Aqueous phases and carrier fluids were loaded into 1700 series Gastight syringes (Hamilton, Reno, NV) with removable 27-gauge needles and 30-gauge Teflon tubing (Weico Wire & Cable, Edgewood, NY). Protein was loaded into syringes with no losses (SI Fig. 9). PHD 2000 infusion syringe pumps (Harvard Apparatus, Hollistion, MA) controlled with a custom LabView subroutine were used to drive flows (subroutine provided in SI Supporting Text).
Crystallization of RC from R. viridis and Porin from R. capsulatus.
Crystallization details are in the SI Tables 2 and 3.
X-Ray Diffraction and Structure Determination.
X-ray data collection details are given in SI Table 4 and Fig. 10. A detailed flow chart outlining the procedures for the hybrid experiments is shown in SI Fig. 11.
Supplementary Material
Acknowledgments
We thank Nina Ponomarenko and James R. Norris [University of Chicago (UC)] for samples of RC from R. viridis, the Philip D. Laible group at Argonne National Laboratory (ANL), UC/ANL Collaborative Seed Funding for samples of Porin R. capsulatus, Cory J. Gerdts for the image in Fig. 2c and for helpful discussions, and Jessica Price for contributions in editing and writing this manuscript. Use of the ANL Structural Biology Center beamlines, BioCARS beamlines, and the National Institute for General Medical Sciences (GM) and National Cancer Institute (CA) GM/CA beamlines at the Advanced Photon Source was supported by Department of Energy Grant W-31-109-Eng-38. GM/CA-Collaborative Access Team (CAT) has been funded in whole or in part by National Cancer Institute Grant Y1-CO-1020 and National Institute of General Medical Sciences (NIGMS) Grant Y1-GM-1104. Use of the BioCARS Sector 14 was supported by National Institutes of Health (NIH) National Center for Research Resources (NCRR) Grant RR07707. We thank ATCG3D funded by the NIGMS and NCRR under the PSI-2 Specialized Center program (U54 GM074961) for partial support of V.T. and deCode Biostructures for providing the custom screening kit for membrane protein crystallization. Undergraduate research was supported by the NIH Roadmap Physical and Chemical Biology training program at UC (D.M.). This work was supported in part by NIH Roadmap for Medical Research Grant R01 GM075827-01).
Abbreviations
- FC-40
a mixture of perfluoro-tri-n-butylamine and perfluoro-di-n-butylmethylamine
- FC-70
perfluorotripentylamine
- LDAO
lauryldimethylamine oxide
- PDMS
poly(dimethlysiloxane)
- RC
reaction center from R. viridis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2I5N).
This article contains supporting information online at www.pnas.org/cgi/content/full/0607502103/DC1.
References
- 1.Kuhn P, Wilson K, Patch MG, Stevens RC. Curr Opin Chem Biol. 2002;6:704–710. doi: 10.1016/s1367-5931(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 2.Sauer S, Lange BMH, Gobom J, Nyarsik L, Seitz H, Lehrach H. Nat Rev Genet. 2005;6:465–476. doi: 10.1038/nrg1618. [DOI] [PubMed] [Google Scholar]
- 3.Hansen C, Quake SR. Curr Opin Struct Biol. 2003;13:538–544. doi: 10.1016/j.sbi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 4.McPherson A. Crystallization of Biological Macromolecules. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. [Google Scholar]
- 5.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Proc Natl Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perozo E, Rees DC. Curr Opin Struct Biol. 2003;13:432–442. doi: 10.1016/s0959-440x(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 7.Khademi S, O'Connell J, Remis J, Robles-Colmenares Y, Miericke LJW, Stroud RM. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 8.Lee SY, Lee A, Chen JY, MacKinnon R. Proc Natl Acad Sci USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang YX, Lee A, Chen JY, Ruta V, Cadene M, Chait BT, MacKinnon R. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 10.Bass RB, Strop P, Barclay M, Rees DC. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 11.Sanders CR, Myers JK. Annu Rev Biophys Biomol Struct. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]
- 12.Drews J. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 13.Rees DC, Chang G, Spencer RH. J Biol Chem. 2000;275:713–716. doi: 10.1074/jbc.275.2.713. [DOI] [PubMed] [Google Scholar]
- 14.MacKinnon R. Science. 2005;307:1425–1426. doi: 10.1126/science.1110525. [DOI] [PubMed] [Google Scholar]
- 15.Loll PJ. J Struct Biol. 2003;142:144–153. doi: 10.1016/s1047-8477(03)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- 17.Nollert P. Prog Biophys Mol Biol. 2005;88:339–357. doi: 10.1016/j.pbiomolbio.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. Acta Crystallogr D. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 19.Weselak M, Patch MG, Selby TL, Knebel G, Stevens RC. In: Macromolecular Crystallography. Carter CW, Sweet RM, editors. Vol. 368. San Diego: Academic; 2003. pp. 45–76. [Google Scholar]
- 20.Hansen CL, Classen S, Berger JM, Quake SR. J Am Chem Soc. 2006;128:3142–3143. doi: 10.1021/ja0576637. [DOI] [PubMed] [Google Scholar]
- 21.Hansen CL, Skordalakes E, Berger JM, Quake SR. Proc Natl Acad Sci USA. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng B, Ismagilov RF. Angew Chem Int Ed. 2005;44:2520–2523. doi: 10.1002/anie.200462857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng B, Roach LS, Ismagilov RF. J Am Chem Soc. 2003;125:11170–11171. doi: 10.1021/ja037166v. [DOI] [PubMed] [Google Scholar]
- 24.Song H, Tice JD, Ismagilov RF. Angew Chem Int Ed. 2003;42:768–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 25.Dittrich PS, Jahnz M, Schwille P. ChemBioChem. 2005;6:811–814. doi: 10.1002/cbic.200400321. [DOI] [PubMed] [Google Scholar]
- 26.Hatakeyama T, Chen DLL, Ismagilov RF. J Am Chem Soc. 2006;128:2518–2519. doi: 10.1021/ja057720w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H, Chen DL, Ismagilov RF. Angew Chem Int Ed. 2006;45:7336–7356. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song H, Ismagilov RF. J Am Chem Soc. 2003;125:14613–14619. doi: 10.1021/ja0354566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tice JD, Song H, Lyon AD, Ismagilov RF. Langmuir. 2003;19:9127–9133. [Google Scholar]
- 30.Stone HA. Annu Rev Fluid Mech. 1994;26:65–102. [Google Scholar]
- 31.Roach LS, Song H, Ismagilov RF. Anal Chem. 2005;77:785–796. doi: 10.1021/ac049061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olbricht WL, Kung DM. Phys Fluids A. 1992;4:1347–1354. [Google Scholar]
- 33.Stebe KJ, Lin SY, Maldarelli C. Phys Fluids A. 1991;3:3–20. [Google Scholar]
- 34.Chayen NE. J Cryst Growth. 1999;196:434–441. [Google Scholar]
- 35.Kreusch A, Weiss MS, Welte W, Weckesser J, Schulz GE. J Mol Biol. 1991;217:9–10. doi: 10.1016/0022-2836(91)90604-5. [DOI] [PubMed] [Google Scholar]
- 36.Zheng B, Tice JD, Roach LS, Ismagilov RF. Angew Chem Int Ed. 2004;43:2508–2511. doi: 10.1002/anie.200453974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McPherson A. J Appl Crystallogr. 2000;33:397–400. [Google Scholar]
- 38.Yadav MK, Gerdts CJ, Sanishvili R, Smith WW, Roach LS, Ismagilov RF, Kuhn P, Stevens RC. J Appl Crystallogr. 2005;38:900–905. doi: 10.1107/S002188980502649X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baxter RHG, Seagle BL, Ponomarenko N, Norris JR. Acta Crystallogr D. 2005;61:605–612. doi: 10.1107/S0907444905005809. [DOI] [PubMed] [Google Scholar]
- 40.Baxter RHG, Ponomarenko N, Srajer V, Pahl R, Moffat K, Norris JR. Proc Natl Acad Sci USA. 2004;101:5982–5987. doi: 10.1073/pnas.0306840101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adamson DN, Mustafi D, Zhang JXJ, Zheng B, Ismagilov RF. Lab Chip. 2006;6:1178–1186. doi: 10.1039/b604993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bach S, Talarek N, Andrieu T, Vierfond JM, Mettey Y, Galons H, Dormont D, Meijer L, Cullin C, Blondel M. Nat Biotechnol. 2003;21:1075–1081. doi: 10.1038/nbt855. [DOI] [PubMed] [Google Scholar]
- 43.Kung LA, Snyder M. Nat Rev Mol Cell Biol. 2006;7:617–622. doi: 10.1038/nrm1941. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 45.Krogan NJ, Cagney G, Yu HY, Zhong GQ, Guo XH, Ignatchenko A, Li J, Pu SY, Datta N, Tikuisis AP, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 46.Danielson E, Golden JH, McFarland EW, Reaves CM, Weinberg WH, Wu XD. Nature. 1997;389:944–948. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.