Abstract
It has been considered a “law” that a lost structure cannot reappear in evolution. The common explanation, that genes required for the development of the lost structure degrade by mutation, remains largely theoretical, however. Additionally, the extent to which this mechanism applies to systems of repeated parts, where individual modules are likely to exhibit few unique aspects of genetic control, is unclear. We investigated reversibility of evolution in one such system, the vertebrate dentition, using as a model loss of oral teeth in cypriniform fishes, which include the zebrafish. This evolutionary event, which occurred >50 million years ago, has not been reversed despite subsequent diversification of feeding modes and retention of pharyngeal teeth. We asked whether the cis-regulatory region of a gene whose expression loss parallels cypriniform tooth loss, Dlx2b, retains the capacity for expression in oral teeth. We first created a zebrafish reporter transgenic line that recapitulates endogenous dlx2b expression. We then showed that this zebrafish construct drives reporter expression in oral teeth of the related characiform Astyanax mexicanus. This result, along with our finding that Dlx genes are required for normal tooth development, suggests that changes in trans-acting regulators of these genes were responsible for loss of cypriniform oral teeth. Preservation of oral enhancer function unused for >50 million years could be the result of pleiotropic function in the pharyngeal dentition. If enhancers of other genes in the tooth developmental pathway are similarly preserved, teeth lost from specific regions may be relatively easy to reacquire in evolution.
Keywords: Astyanax, Dollo's Law, zebrafish
Dollo's Law of the Irreversibility of Evolution states that an organism can never exactly return to a previous evolutionary state (1, 2). A more restricted version, often attributed to O. Abel, is that once an anatomical feature is lost, it is difficult or impossible to regain (3, 4). The mechanism preventing reappearance of lost structures is thought to be the accumulation of mutations, in the absence of selection, in the genes responsible for their development (5). However, the genes regulating the development of morphological structures tend to exhibit extensive pleiotropy (6), a feature that may preserve them from mutational degradation even if one of the structures for which they are essential is lost (5).
The vertebrate dentition is an excellent system in which to investigate which types of lost morphological structures can or cannot reappear in evolution and why. For example, complete tooth loss, such as has occurred in birds and turtles, appears to be irreversible, possibly because of the degeneration of genes coding for tooth proteins not used elsewhere in the body (5, 7, 8). At the other extreme, individual teeth within a region can be regained in evolution, such as the reappearance of the second lower molar in the felid Lynx lynx after an absence of 20 million years (9, 10). However, between the extremes of complete dentition loss and loss of some but not all members of a tooth class, the maximum extent of reversible regional tooth loss remains unknown.
Comparisons between different classes of mammalian teeth (e.g. molars and incisors; ref. 11) and between oral and pharyngeal teeth of fishes (12–14) show extensive similarities in the genes involved in their development. Where differences in expression between different types of developing teeth have been documented, these involve genes that participate in the development of other organs. Therefore, pleiotropy is likely to maintain the coding regions of all known genes involved in tooth development after even extensive regional tooth loss. In contrast to coding regions of developmental regulatory genes, however, the cis-regulatory regions of these genes are thought to exhibit a high degree of modularity (6). Although little is known about enhancer function in tooth development (15), use of different enhancers in different regions of the dentition could render regional tooth loss irreversible because of the lack of pleiotropic effects associated with their mutational degradation.
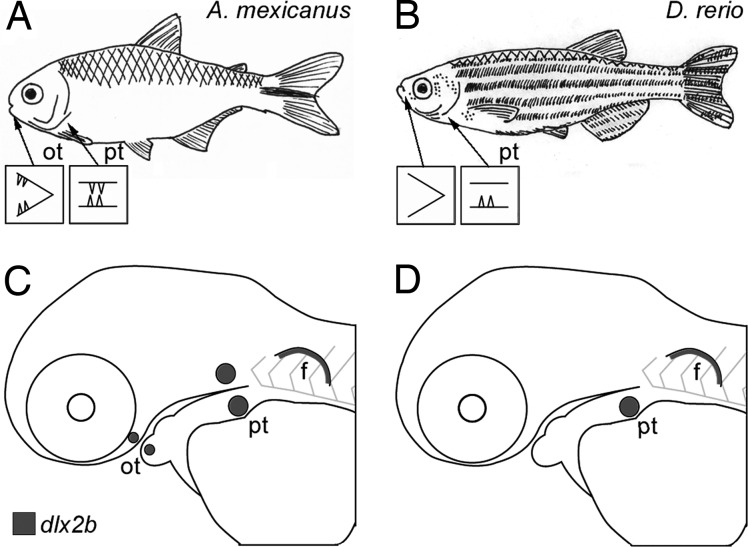
We have used the loss of oral teeth in cypriniform fishes, which include the zebrafish model species, to study the potential for reversibility of regional tooth loss. Pharyngeal teeth are retained in the zebrafish and most other cypriniforms (Fig. 1), whereas oral teeth were lost in the group at least 50 million years ago and have never been regained despite extensive diversification of species and feeding modes (16, 17). We have previously shown that expression of Dlx2a and Dlx2b, members of the Distal-less-related (Dlx) family of homeodomain transcription factors (18), was lost from tooth-forming oral epithelium of cypriniforms in association with oral tooth loss (Fig. 1 and ref. 19). Dlx genes are expressed at numerous stages of tooth development in the zebrafish (20) and the mouse (21), and loss of Dlx function leads to early arrest of tooth development in the mouse (22). We have argued previously that Dlx genes are likely downstream of the cause of cypriniform oral tooth loss (19) and are thus a good model for the fate of developmental pathways once an initiation signal has been modified in evolution.
Fig. 1.
Zebrafish lack oral teeth and oral expression of the tooth marker dlx2b. (A and B) Sketches of adult A. mexicanus (A) and zebrafish (B; D. rerio), illustrating that whereas characiforms like A. mexicanus have both oral and pharyngeal teeth, cypriniforms completely lack oral teeth and retain only lower pharyngeal teeth. (C and D) Diagrams of lateral views of developing larvae of these species, illustrating the locations of dlx2b expression in developing teeth and in the pectoral fin (data from refs. 12, 19, and 29). f, pectoral fin; ot, oral tooth germs or teeth; pt, pharyngeal tooth germs or teeth.
We here test whether the cis-regulatory region of one of these genes, dlx2b, retains in cypriniforms the capability of driving expression in oral tooth-forming epithelium. To do so, we isolated such a genomic region from zebrafish dlx2b and found it to be capable of driving reporter gene expression in pharyngeal teeth of transgenic individuals of this species. We then used the same zebrafish construct in transient expression assays in one of the closest relatives to cypriniforms retaining oral teeth, the characiform Astyanax mexicanus (23–25). We found that the zebrafish cis-regulatory region was capable of driving expression in oral teeth of A. mexicanus, consistent with our initial hypothesis that loss of cypriniform dlx2b expression occurred through the modification of an upstream regulator. Most importantly, this result indicates that the cis-regulatory region of dlx2b has retained the ability to be expressed in oral teeth at least 50 million years after they were lost. A potential mechanism for this preservation of function is pleiotropy of enhancer activity among oral and pharyngeal teeth, and possibly other organs where dlx2b is expressed, such as fins. Furthermore, if other components of the tooth developmental pathway have been similarly preserved, it is possible that oral teeth could be regained in cypriniform fishes through one or a few genetic changes in tooth initiation signals.
Results
The Genomic Region 5′ of Zebrafish dlx2b.
We cloned a 4,136-bp fragment immediately 5′ to the zebrafish dlx2b transcription start site into a GFP reporter vector because the corresponding region from mouse Dlx2 is known to drive expression in developing teeth (15). Zebrafish have duplicated semiorthologs of mouse Dlx2: dlx2a and dlx2b. We chose the region from dlx2b over dlx2a because dlx2b is not expressed in surrounding tissues and is thus a more specific marker of tooth development (12).
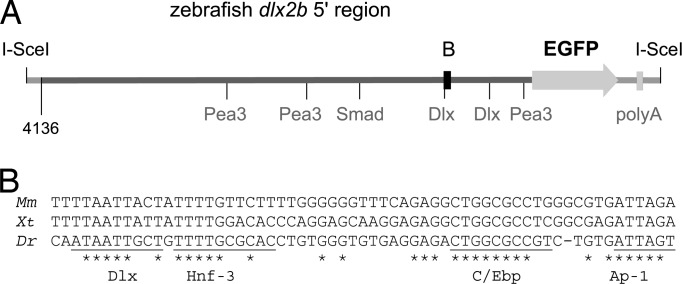
Using data obtained from the zebrafish sequencing project (The Wellcome Trust Sanger Institute, Cambridge, U.K.), we identified several potential transcription factor binding sites for proteins in developmental pathways known to interact with Dlx2, including the fibroblast growth factor (Fgf; Pea3 transcription factor) and bone morphogenetic protein (Bmp; Smad transcription factor) pathways, and Dlx itself (Fig. 2A). However, comparative phylogenetic analysis of the region revealed only a short, 61-bp domain of potential conservation with the 5′ genomic region near other vertebrate Dlx2 genes (Fig. 2B), indicating that this 4-kb region has undergone much sequence divergence since the split of ray-finned and lobe-finned fishes. This finding is consistent with a recent study that has shown that enhancer function may be retained between fish and mammalian gene orthologs with no apparent sequence conservation (26). Interestingly, the small domain of conservation we do find includes a Dlx consensus binding sequence that may represent an important binding site for Dlx regulation, such as has been found near other Dlx paralogs (27). Further comparative analysis of this 4-kb region using several sequence alignment and transcription factor binding site alignment methods revealed no additional conservation (see Materials and Methods).
Fig. 2.
The sequence of the zebrafish dlx2b 4-kb 5′ region contains a phylogenetically conserved domain and several potential transcription factor binding sites. (A) Diagram of the zebrafish dlx2b:GFP construct. Pea3 (Fgf), Smad (Bmp), and Dlx consensus binding sites are indicated. The region conserved with tetrapods is highlighted with a black box. (B) Alignment of the conserved 61-bp region between mouse (Mm), frog (Xenopus tropicalis, Xt), and zebrafish (Dr) sequences; ∗ indicates sequence identity. Potentially conserved transcription factor binding sites are underlined and labeled.
The Zebrafish dlx2b: GFP Reporter Drives Expression in Zebrafish Pharyngeal Teeth But not in Oral Epithelium.
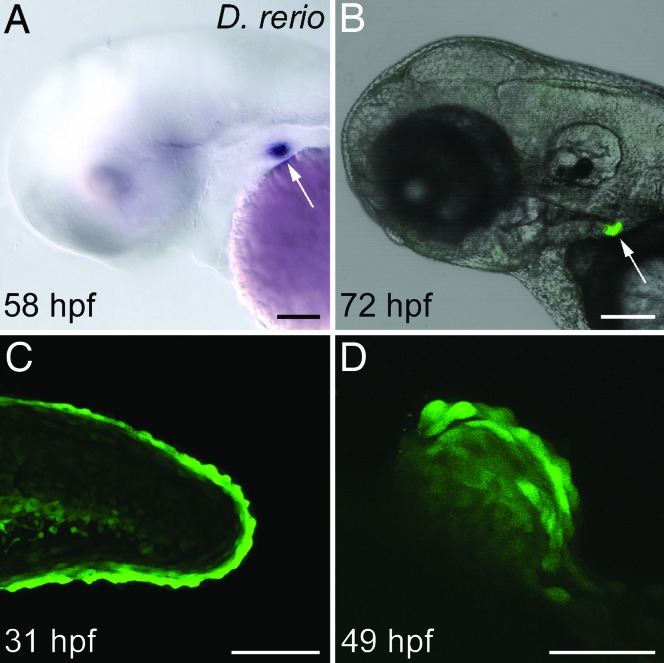
Transient assays of the dlx2b:GFP construct in zebrafish displayed pharyngeal tooth expression, and from these fish we isolated a stable transgenic line, designated Tg(dlx2b:EGFP)wj1. In this line, we first detect GFP mRNA expression at 52 hours postfertilization (hpf) in the first developing pharyngeal tooth germ (4V1; ref. 28), and by 58 hpf, mRNA expression is strong relative to background (Fig. 3A). This timing of expression is very similar to that of the endogenous dlx2b mRNA in developing pharyngeal teeth (12). GFP fluorescence is detectable by 60 hpf and is very strong by 72 hpf in what by this stage is a cluster of developing pharyngeal teeth (Fig. 3B). Expression appears to continue in each developing tooth until at least the attachment stage (data not shown).
Fig. 3.
The zebrafish dlx2b 5′ 4-kb genomic region recapitulates the endogenous dlx2b pattern with expression in pharyngeal tooth germs and pectoral fins, but not oral epithelium. Shown are whole-mount, lateral views of the Tg(dlx2b:EGFP)wj1 transgenic line, anterior to the left. (A) 58 hpf mRNA in situ hybridization against the GFP reporter message showing expression in a pharyngeal tooth germ (arrow). (B) GFP expression in a living 72 hpf larva in pharyngeal tooth germs (arrow). (C) Reporter expression in the margin of the caudal fin at 31 hpf. (D) Pectoral fin expression at 49 hpf. (Scale bars: A–C, 100 μm; D, 50 μm.)
We also observed nontooth-related GFP expression in Tg(dlx2b:EGFP)wj1. The reporter was strongly expressed in two domains where dlx2b mRNA is known to be found: in the margin of developing median fin fold and pectoral fins (Fig. 3 C and D and ref. 29), and in the hindgut (data not shown). Additional expression was seen in domains not previously identified as expressing dlx2b mRNA, possibly as a result of a chromosomal position effect, the absence of a repressor element, or weak expression that had previously been overlooked. This additional expression was seen in a subset of hindbrain and spinal neurons, and in a weak, nonhomogeneous expression pattern in much of the rest of the body during all of the stages we examined (data not shown).
In the oral region, we carefully searched for reporter expression in epithelium homologous to that which in other fishes expresses Dlx2b and gives rise to teeth. This search was to investigate whether the dlx2b:GFP reporter construct may be missing a repressor that normally turns off dlx2b expression orally or whether in situ studies failed to detect an endogenous oral domain of dlx2b. We examined larvae at ≈12-h intervals from 72 to 144 hpf, and while expression was found laterally in jaw mesenchyme in what may represent dlx2b expression in developing bone (30), there was no expression in epithelium where teeth would be expected to develop (data not shown). This finding suggests that the dlx2b:GFP reporter recapitulates the endogenous zebrafish expression pattern of dlx2b in pharyngeal tooth germs and its absence from oral epithelium.
Zebrafish dlx2b: GFP Drives Expression in Characiform Oral Teeth.
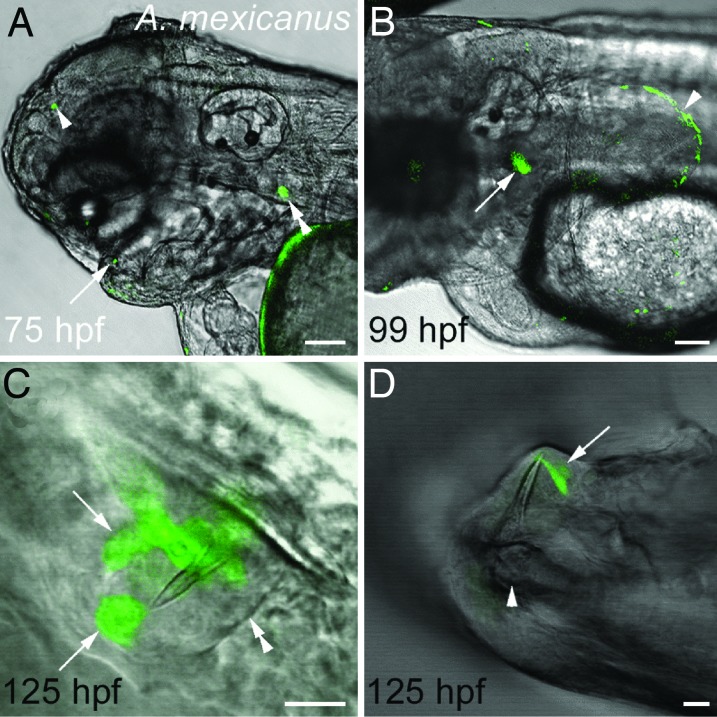
Injection of the zebrafish dlx2b:GFP construct into one-cell embryos of the characiform A. mexicanus generated transient expression in a very similar pattern to that seen with zebrafish transient assays. Expression was mosaic as is typically seen with DNA injection in zebrafish (31), but was repeatedly found in both developing pharyngeal tooth germs (n = 5/177) and the margin of pectoral fins (n = 10/89; Fig. 4A and B). In addition, strong GFP expression was seen in multiple cells in the epithelium of developing oral teeth (n = 5/152; Fig. 4 C and D). Expression was also noted in the median fin margin and the adhesive organ, both sites of endogenous dlx2b expression in A. mexicanus (data not shown). Occasional single GFP-expressing cells were seen in other, seemingly random locations (e.g., arrowhead in Fig. 4A), but we interpret these as likely artifacts of transient DNA injection.
Fig. 4.
The zebrafish dlx2b:GFP reporter drives expression in developing A. mexicanus oral teeth, pharyngeal teeth, and pectoral fins. (A) Lateral view of the head at 75 hpf with expression in an oral tooth germ (arrow), upper pharyngeal tooth germ (double arrowhead), and a small ectopic location (arrowhead). (B) Lateral view of pharyngeal tooth (arrow) and pectoral fin (arrowhead) expression at 99 hpf. (C) Oral tooth forming in the upper jaw adjacent to the developing premaxillary bone at 125 hpf. The boundary of the dental epithelium can be seen on one side (double arrowhead) and two groups of dlx2b:GFP reporter expressing cells on the other (arrows). (D) GFP expression in dental epithelial cells (arrow) surrounding a mineralized tooth attached to the dentary bone of the lower jaw (arrowhead) at 125 hpf. (Scale bars: A and B, 100 μm; C and D, 10 μm.)
Knockdown of Multiple Dlx Genes Results in Altered Tooth Morphology.
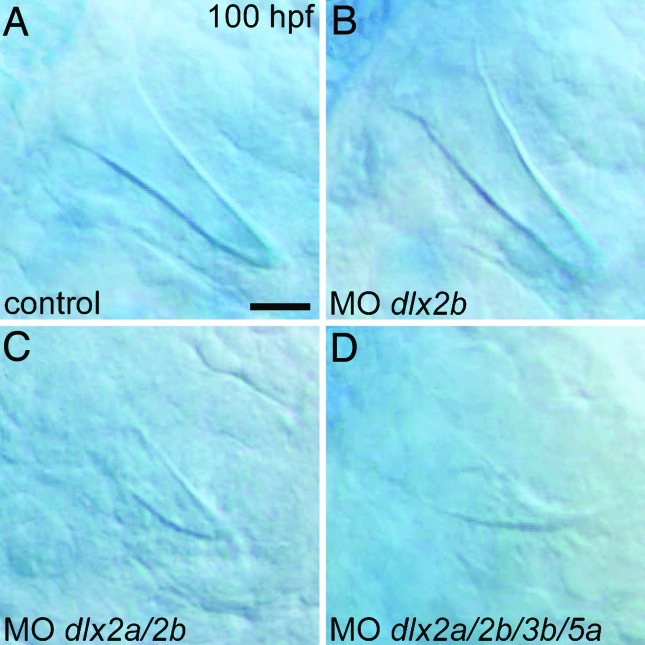
To assess whether the loss of cypriniform oral Dlx2 expression may have been a causal factor in the evolutionary loss of oral teeth, we investigated the requirement of Dlx genes during zebrafish pharyngeal tooth development by antisense inhibition of translation with morpholino (MO) oligonucleotides (Gene Tools, Philomath, OR). We found that injection of a dlx2b MO alone had no effect on tooth development, as judged by the morphology of mineralized teeth (n = 10, Fig. 5B). However, the simultaneous injection of MOs targeting dlx2a and dlx2b caused the majority of fish to develop smaller than normal teeth (n = 10/17; Fig. 5C) and inhibited pharyngeal cartilage development (data not shown). This latter result is consistent with the expression of dlx2a in cranial neural crest (32). The combined injection of dlx2a, dlx2b, dlx3b, and dlx5a MOs resulted in fish with very small and morphologically altered pharyngeal teeth (n = 7; Fig. 5D), reduced cartilages, and the fused otoliths previously observed after dlx3b inhibition (data not shown; ref. 33). Because the development of teeth is relatively late, MO injection amounts were by necessity calibrated to prevent severe abnormalities in early development. Because such treatments are unlikely to eliminate Dlx expression completely, these genes may be required for additional aspects of tooth development. At a minimum, however, they are required for proper tooth morphology.
Fig. 5.
Dlx gene knockdown perturbs zebrafish pharyngeal tooth morphogenesis. (A) High magnification view of a pharyngeal tooth at 100 hpf in a control injected with the injection solution and no MO. (B) A normal-looking tooth from an identically treated specimen injected with a dlx2b MO. (C) After injection of combined dlx2a and dlx2b MOs, pharyngeal tooth size is reduced (representative example shown). (D) Pharyngeal teeth are even smaller, and are now misshapen, after simultaneous injection with MOs against dlx2a, dlx2b, dlx3b, and dlx5a. (Scale bar: 10 μm.)
Fgf Signaling Acts Positively on the Zebrafish dlx2b Tooth Enhancer Region.
We have previously shown that inhibition of Fgf signaling with the compound SU5402 arrests zebrafish pharyngeal tooth development and prevents the expression of dlx2b (12). Similar SU5402 treatment of Tg(dlx2b:EGFP)wj1 from 30 hpf through 72 hpf completely eliminated GFP reporter expression from the region where pharyngeal teeth develop (Fig. 6; n = 8). Pectoral fins are inhibited from forming by this SU5402 treatment, so that the effect of Fgf inhibition on reporter expression in this domain could not be assayed. Hindgut and median fin fold expression was maintained after this treatment, indicating either that dlx2b expression in these locations is Fgf-independent or that expression was initiated before the inhibitor was applied (data not shown). Pharyngeal tooth GFP expression in DMSO carrier controls was normal (n = 6). This result suggests that Fgf pathway factors act positively on this dlx2b enhancer region and not in a more indirect way such as by alleviating repression from a cis-silencer (e.g., ref. 34).
Fig. 6.
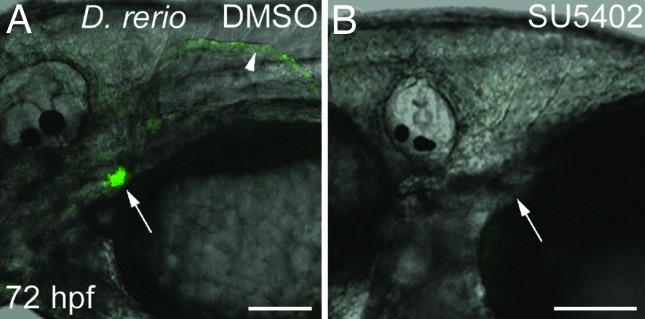
Fgf inhibition eliminates dlx2b:GFP reporter expression in zebrafish pharyngeal tooth germs. Tg(dlx2b:EGFP)wj1 treated from 30–72 hpf with either a DMSO control solution (A) or the Fgf inhibitor SU5402 (B). Pharyngeal tooth expression of the dlx2b:GFP reporter is eliminated with this treatment (arrows). Pectoral fin expression (arrowhead) is also absent, but no pectoral fin bud develops and this therefore may be the result of earlier Fgf requirements in fin development. (Scale bars: 100 μm.)
Discussion
Regulatory Change Developmentally Upstream of dlx2b Is Implicated in Cypriniform Oral Tooth Loss.
The ability of the zebrafish dlx2b:GFP reporter construct to drive expression in A. mexicanus oral tooth germs indicates that loss of oral Dlx2b expression in cypriniforms resulted from changes in one or more trans-acting regulators rather than in the cis-regulatory regions of this gene. This interpretation is consistent with the coordinate loss of the closely related paralog Dlx2a from cypriniform oral epithelium, a gene likely to be regulated by many of the same trans-acting factors (19). Some of these changes might involve other members of the Dlx gene family. Eight Dlx genes are present in the zebrafish (35) and six of these are expressed in zebrafish pharyngeal teeth, with considerable spatial and temporal overlap (20). Redundancy of Dlx function has been documented in mammals (36), and our finding that increasingly severe phenotypes are produced by knocking down combinations of Dlx genes (Fig. 5) suggests redundancy in the teleost dentition as well. Interestingly, despite the expression of Dlx genes during zebrafish pharyngeal tooth initiation (20) and the fact that dlx2b prefigures the epithelium from which oral teeth arise in A. mexicanus (19), none of the phenotypes we obtained from gene knockdown involved the early arrest of tooth initiation, as seen in the mouse (22). This result might be explained by the more extensive redundancy of Dlx gene expression during tooth initiation in the zebrafish relative to the mouse (20). Alternatively, because the doses of MOs we used had to be adjusted to allow the larvae to develop to the ages at which teeth appear, we are likely to have produced only partial losses of function. Nevertheless, our demonstration that Dlx genes are necessary for normal tooth development supports the hypothesis that the trans-acting factors responsible for Dlx2b expression loss are candidates for causing cypriniform tooth loss.
Although the upstream regulators responsible for oral Dlx2b expression loss remain unknown, two candidates are the Bmp and Fgf signaling pathways. Wise and Stock (14) found that loss of oral bmp2b expression is associated with cypriniform oral tooth loss. Dlx2 expression is known to be regulated by Bmp4 in mouse tooth development (15), but whether dental Dlx2b expression in teleosts is regulated by Bmp2b remains to be determined. Fgf signaling has been shown to be upstream of Dlx2b expression in both the oral and pharyngeal dentition of teleosts (12, 19). In addition, inhibition of Fgf signaling in A. mexicanus prevents oral tooth development and produces a partial phenocopy of the zebrafish oral gene expression profile (19). Although comparisons of expression of these signaling pathway components between the zebrafish and teleosts retaining oral teeth remain to be carried out, it is also possible that trans-acting regulators of Dlx genes have undergone protein rather than expression modification during cypriniform evolution.
The Cypriniform Oral dlx2b Enhancer Has Been Preserved for >50 Million Years.
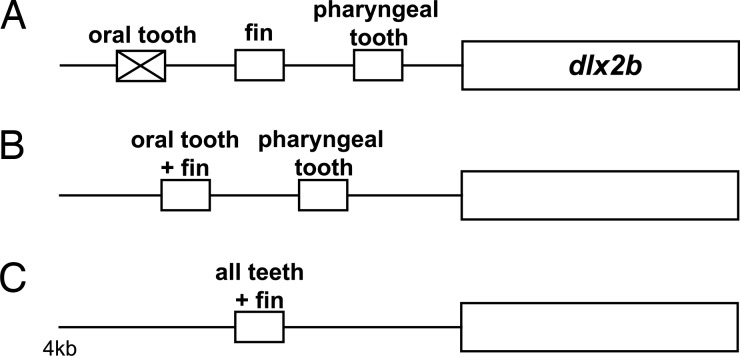
Oral teeth are likely to have been lost in cypriniforms after their divergence from characiforms and other otophysans, but before the divergence of extant families (ref. 16 and D.W.S., unpublished work). The fossil record provides minimum estimates of these events as 100 million and 50 million years ago (mya), respectively, whereas molecular clocks suggest considerably older dates of 250 mya and 180 mya (37). Regardless of which of these dates is correct, the ability of the zebrafish dlx2b:GFP reporter to drive oral expression in A. mexicanus is remarkable. It is possible that mutational degradation of enhancer elements driving oral tooth expression of Dlx2b in cypriniforms was prevented by the pleiotropic function of these elements. Pleiotropic function of the oral tooth enhancer is suggested by the ability of the 4-kb cis-regulatory region to additionally drive expression in pharyngeal teeth and pectoral fins (Fig. 7). A prediction of this hypothesis is that it will not be possible to separate oral tooth expression from expression in one or both of these other domains by transgenic dissection of the 4-kb region examined in this study. However, because the specific transcription factor binding sites involved are still unknown, we cannot rule out that they are small enough to have escaped mutational degradation by chance. Characterization of the size and composition of these key regulatory sequences is required to determine the most likely mechanism of enhancer preservation.
Fig. 7.
Cypriniform oral dlx2b enhancer function may have been preserved in evolution because of pleiotropic function. (A) If the oral tooth enhancer did not drive expression in a separate, required region it may have been lost because of mutation, although such loss could also be prevented by the requirement of only a small number of nucleotides for function. (B and C) Two possible scenarios (of many) for preserving the oral enhancer include that it is also responsible for pectoral fin expression (B) or that there is a single enhancer element that drives expression in both oral and pharyngeal teeth, and fins (C). Small boxes indicate enhancer elements; larger boxes indicate coding region.
Is Cypriniform Tooth Loss Reversible?
It is likely that complete loss of the dentition is irreversible if the condition persists for several million years (5). The mechanism preventing reversal is thought to be the mutational degradation of the coding regions of tooth-specific genes. Supporting this theoretical argument is the apparent absence of the enamel-specific variant of the SCPP protein in the chicken genome (8). In addition, mutations or experimental manipulations that produce tooth rudiments in the chicken fail to generate enamel (38–40). In contrast, there is evidence that loss of individual teeth can be reversed even after a long absence. The second lower molar appears as a frequent atavism in the felid Lynx lynx, after having been absent for ≈20 million years (9, 10). It is possible that reappearance of this tooth resulted from a simple increase in the level of signaling in tooth initiation pathways (5, 9). Supporting this idea, experimentally adjusting the level of Bmp and ectodysplasin signaling can increase or decrease the number of molars in mice (41–43).
Between the extremes provided by the above examples of tooth loss, the threshold beyond which regional tooth loss becomes irreversible is poorly understood. Loss of oral teeth in cypriniform fishes is proposed to have arisen as an adaptation to suction feeding (44). However, extensive diversification of feeding modes has occurred since the initial loss event (17). These include feeding on other fishes or large aquatic plants, for which oral teeth might be expected to be adaptive (19, 45). This pattern suggests that failure to regain oral teeth is not the result of an absence of selection for their function. The specific genetic changes that caused loss of cypriniform oral teeth remain unknown (14, 19). If our finding that Dlx2b retains the capacity for expression in oral teeth is representative of the genetic pathways controlling tooth development, it is possible that the redeployment or modification of one or a few upstream signals could restore oral teeth. Resolving the apparent conflict between the failure of cypriniforms to reacquire oral teeth and the ease of re-expression of Dlx2b will require characterizing the evolution of the regulatory control of additional genes involved in tooth development. The approach used in our study, in which we assayed enhancer function in the trans-background of a species retaining the structure(s) should provide a powerful means of addressing the reversibility of character loss in this and other systems.
Materials and Methods
Fish Strains.
Zebrafish (Danio rerio) WT strains and husbandry were as reported (12). Embryos of A. mexicanus surface fish were obtained from an in-house line descended from individuals collected by W. Jeffery (University of Maryland, College Park, MD) in Balmorrhea State Park, TX (46). Embryonic stages are described in hpf, with zebrafish raised at 28.5°C and A. mexicanus raised at 25°C. The rate of zebrafish development in our laboratory is ≈10% delayed relative to the standard staging series (47).
Genomic Analysis.
In the genomic sequence 4,136 bp 5′ to the zebrafish dlx2b transcription start site, a 61-bp domain conserved with tetrapods was initially identified by using the University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu and ref. 48), and the zebrafish May 2005 (danRer3; The Wellcome Trust Sanger Institute), and human February 2006 (hg18 and ref. 49) genome assemblies. This browser uses the phastCons phylogenetic hidden Markov model to identify regions of sequence similarity (50). Subsequent alignment of this region with that from other vertebrate Dlx2 5′ genomic regions was done with Clustal X (51). Further alignments of the entire zebrafish dlx2b 4,136-bp 5′ region and corresponding regions 5′ to other fish and mammal Dlx2 genes by using Clustal X (51), blast2sequences (52), and Vista (53) revealed no additional conserved regions. Similar negative results were obtained with transcription factor binding site alignments using rVista (54) and EEL (55). For this analysis, previously reported consensus binding sequences for zebrafish Pea3 (56), Smad (57), and Dlx (58) were used.
Reporter Construction.
PCR primers to amplify a 4,136-bp fragment immediately 5′ to the zebrafish dlx2b transcription start site were designed from genomic sequence [University of California, Santa Cruz Zebrafish Genome Browser and The Wellcome Trust Sanger Institute; primers with restriction sites underlined are: DrDlx2bF4(EcoRI) CGGAATTCCGAGTCATTTTGATCTGGAGAAAGCTGATG and DrDlx2bR6(BamHI) ATGCGGATCCTTCGCAGGAAGAAGAGACTACTCAACG]. Expand Long Template System proofreading PCR (Roche, Indianapolis, IN) was used for the amplification, and a zebrafish PAC clone containing dlx2b (#209) was used as template (59). The resulting product was digested with EcoRI and BamHI and cloned into the pEGFP-1 vector (Clontech, Mountain View, CA). The 4,136-bp sequence plus the EGFP gene and polyadenylation signal sequence were then PCR-amplified (primers pGFPF2(XhoI) CCGCTCGAGTAGTTATTACTAGCGCTACC and pGFPR7a(PstI) AAACTGCAGTTTGGACAAACCACAACTAGA) and ligated into the pI-SceI vector (60). This final dlx2b:EGFP construct was checked by restriction analysis and the insert ends were checked by automated sequencing.
Transgenic Analysis.
Injection solutions were made containing 30–130 ng/μl of the dlx2b:GFP construct, 0.1 M KCl, 0.1% phenol red, 0.5 units I-SceI (New England Biolabs, Ipswich, MA), and 0.5× I-SceI buffer, similar to I-SceI construct injection as described (60). Approximately 0.5 nl of these solutions was injected into blastomeres of one- or two-cell-stage zebrafish and A. mexicanus embryos. In both species, mosaic GFP expression was seen in these embryos as they developed, including expression in dental epithelium and fins. In zebrafish, four instances of germ-line transmission of the transgene were found in 53 injected fish raised to adulthood. One transgenic line, designated Tg(dlx2b:EGFP)wj1, displayed strong GFP expression in pharyngeal tooth germs and pectoral and caudal fin margins. The other three examples had very weak GFP expression in the fins with no tooth expression and were not investigated further.
In Situ Hybridization.
mRNA in situ hybridization was performed as described (12). The template for synthesizing the GFP riboprobe was created by excising the EGFP coding sequence from the pEGFP-1 vector with NotI and SacII and ligating the resulting fragment into pBS SK+ (Stratagene, La Jolla, CA) before probe synthesis.
Gene Knockdown.
MO antisense oligonucleotides (Gene Tools) were used to inhibit translation from target mRNAs in vivo by injection of ≈2 nl of a 0.2M KCl, 0.2% phenol red solution containing 3 mg/ml each into the yolk of 1-cell embryos. MO information: dlx2b (GCTGTACTTCCTAACAGTTAATAGT), dlx2a (TGAGGCTGTCAAAAACTCCAGTCAT), dlx3b (61), dlx5a (62). MO-injected larvae were fixed and stained with alcian green to aid in visualization of teeth (12). FGF receptor activity was inhibited by application of 25 μM SU5402 (CalBiochem, San Diego, CA; ref. 63) in 0.5% DMSO embryo medium as described (12).
Photography.
Fluorescent images were taken with a TCS SP2 confocal microscope (Leica, Exton, PA) with an acousto-optical beam splitter. Differential interference contrast bright-field images were taken with a AxioCam on an Axiovert 135 inverted compound microscope (Zeiss, Thornwood, NY). Photographs were processed with Photoshop (Adobe Systems, San Jose, CA), and diagrams were produced with Adobe Illustrator.
Acknowledgments
We thank Angel Amores (University of Oregon, Eugene, OR), Kristin Artinger (University of Colorado Health Sciences Center, Denver, CO), Chuck Kimmel (University of Oregon, Eugene, OR), and Joachim Wittbrodt (European Molecular Biology Laboratory, Heidelberg, Germany) for sharing reagents and Sarah Wise for helpful comments on the manuscript. This work was supported by National Science Foundation Grants IBN-0092487 and IBN-0446720 (to D.W.S.) and National Institutes of Health Grant 5F32DE015029 (to W.R.J.).
Abbreviations
- hpf
hours postfertilization
- Fgf
fibroblast growth factor
- Bmp
bone morphogenetic protein
- MO
morpholino.
Footnotes
The authors declare no conflict of interest.
References
- 1.Dollo L. Bull Soc Belge Géol Pal Hydr. 1893;7:164–166. [Google Scholar]
- 2.Gould SJ. J Hist Biol. 1970;3:189–212. doi: 10.1007/BF00137351. [DOI] [PubMed] [Google Scholar]
- 3.Simpson GG, editor. The Major Features of Evolution. New York: Columbia Univ Press; 1953. [Google Scholar]
- 4.Bull JJ, Charnov EL. Evolution (Lawrence, Kans.) 1985;39:1149–1155. doi: 10.1111/j.1558-5646.1985.tb00455.x. [DOI] [PubMed] [Google Scholar]
- 5.Marshall CR, Raff EC, Raff RA. Proc Natl Acad Sci USA. 1994;91:12283–12287. doi: 10.1073/pnas.91.25.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll SB. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll RL. Vertebrate Paleontology and Evolution. New York: Freeman; 1988. [Google Scholar]
- 8.Kawasaki K, Weiss KM. J Exp Zool B Mol Dev Evol. 2006;306:295–316. doi: 10.1002/jez.b.21088. [DOI] [PubMed] [Google Scholar]
- 9.Kurtén B. Comment Biol. 1963;26:1–10. [Google Scholar]
- 10.Werdelin L. J Zool Lond. 1987;211:259–266. [Google Scholar]
- 11.Tucker A, Sharpe P. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- 12.Jackman WR, Draper BW, Stock DW. Dev Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Fraser GJ, Graham A, Smith MM. Proc Biol Sci. 2004;271:2311–2317. doi: 10.1098/rspb.2004.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wise SB, Stock DW. Evol Dev. 2006;8:511–523. doi: 10.1111/j.1525-142X.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- 15.Thomas BL, Liu JK, Rubenstein JL, Sharpe PT. Development (Cambridge, UK) 2000;127:217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- 16.Nelson JS. Fishes of the World. 4th Ed. Hoboken, NJ: Wiley; 2006. [Google Scholar]
- 17.Sibbing FA. In: Cyprinid Fishes: Systematics, Biology, and Exploitation. Winfield IJ, Nelson JS, editors. New York: Chapman & Hall; 1991. pp. 377–412. [Google Scholar]
- 18.Panganiban G, Rubenstein JL. Development (Cambridge, UK) 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 19.Stock DW, Jackman WR, Trapani J. Development (Cambridge, UK) 2006;133:3127–3137. doi: 10.1242/dev.02459. [DOI] [PubMed] [Google Scholar]
- 20.Borday-Birraux V, Van der Heyden C, Debiais-Thibaud M, Verreijdt L, Stock DW, Huysseune A, Sire JY. Evol Dev. 2006;8:130–141. doi: 10.1111/j.1525-142X.2006.00084.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, Stock D, Buchanan A, Weiss K. Dev Genes Evol. 2000;210:270–275. doi: 10.1007/s004270050314. [DOI] [PubMed] [Google Scholar]
- 22.Thomas BL, Tucker AS, Qui M, Ferguson CA, Hardcastle Z, Rubenstein JL, Sharpe PT. Development (Cambridge, UK) 1997;124:4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- 23.Fink SV, Fink WL. In: Interrelationships of Fishes. Stiassny MLJ, Parenti LR, Johnson GD, editors. New York: Academic; 1996. pp. 209–249. [Google Scholar]
- 24.Jeffery WR. Dev Biol. 2001;231:1–12. doi: 10.1006/dbio.2000.0121. [DOI] [PubMed] [Google Scholar]
- 25.Saitoh K, Miya M, Inoue JG, Ishiguro NB, Nishida M. J Mol Evol. 2003;56:464–472. doi: 10.1007/s00239-002-2417-y. [DOI] [PubMed] [Google Scholar]
- 26.Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 27.Ghanem N, Jarinova O, Amores A, Long Q, Hatch G, Park BK, Rubenstein JL, Ekker M. Genome Res. 2003;13:533–543. doi: 10.1101/gr.716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der heyden C, Huysseune A. Dev Dyn. 2000;219:486–496. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1069>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 29.Ellies DL, Stock DW, Hatch G, Giroux G, Weiss KM, Ekker M. Genomics. 1997;45:580–590. doi: 10.1006/geno.1997.4978. [DOI] [PubMed] [Google Scholar]
- 30.Verreijdt L, Debiais-Thibaud M, Borday-Birraux V, Van der Heyden C, Sire JY, Huysseune A. Dev Dyn. 2006;235:1371–1389. doi: 10.1002/dvdy.20734. [DOI] [PubMed] [Google Scholar]
- 31.Stuart GW, McMurray JV, Westerfield M. Development (Cambridge, UK) 1988;103:403–412. doi: 10.1242/dev.103.2.403. [DOI] [PubMed] [Google Scholar]
- 32.Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maroon H, Walshe J, Mahmood R, Kiefer P, Dickson C, Mason I. Development (Cambridge, UK) 2002;129:2099–2108. doi: 10.1242/dev.129.9.2099. [DOI] [PubMed] [Google Scholar]
- 34.Lin CY, Chen YH, Lee HC, Tsai HJ. Gene. 2004;334:63–72. doi: 10.1016/j.gene.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Stock DW, Ellies DL, Zhao Z, Ekker M, Ruddle FH, Weiss KM. Proc Natl Acad Sci USA. 1996;93:10858–10863. doi: 10.1073/pnas.93.20.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Depew MJ, Simpson CA, Morasso M, Rubenstein JL. J Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng Z, He S, Wang J, Wang W, Diogo R. Gene. 2006;370:113–124. doi: 10.1016/j.gene.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Zhang Y, Jiang TX, Barlow AJ, St Amand TR, Hu Y, Heaney S, Francis-West P, Chuong CM, Maas R. Proc Natl Acad Sci USA. 2000;97:10044–10049. doi: 10.1073/pnas.160245097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsiadis TA, Cheraud Y, Sharpe P, Fontaine-Perus J. Proc Natl Acad Sci USA. 2003;100:6541–6545. doi: 10.1073/pnas.1137104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris MP, Hasso SM, Ferguson MW, Fallon JF. Curr Biol. 2006;16:371–377. doi: 10.1016/j.cub.2005.12.047. [DOI] [PubMed] [Google Scholar]
- 41.Kangas AT, Evans AR, Thesleff I, Jernvall J. Nature. 2004;432:211–214. doi: 10.1038/nature02927. [DOI] [PubMed] [Google Scholar]
- 42.Plikus MV, Zeichner-David M, Mayer JA, Reyna J, Bringas P, Thewissen JG, Snead ML, Chai Y, Chuong CM. Evol Dev. 2005;7:440–457. doi: 10.1111/j.1525-142X.2005.05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kassai Y, Munne P, Hotta Y, Penttila E, Kavanagh K, Ohbayashi N, Takada S, Thesleff I, Jernvall J, Itoh N. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- 44.Gosline WA. Copeia. 1973;1973:761–776. [Google Scholar]
- 45.Portz D, Tyus H. Environ Biol Fishes. 2004;71:233–245. [Google Scholar]
- 46.Dowling TE, Martasian DP, Jeffery WR. Mol Biol Evol. 2002;19:446–455. doi: 10.1093/oxfordjournals.molbev.a004100. [DOI] [PubMed] [Google Scholar]
- 47.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 48.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 50.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tatusova TA, Madden TL. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 53.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. Nucleic Acids Res. 2004;32:273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 56.Brown LA, Amores A, Schilling TF, Jowett T, Baert JL, de Launoit Y, Sharrocks AD. Oncogene. 1998;17:93–104. doi: 10.1038/sj.onc.1201911. [DOI] [PubMed] [Google Scholar]
- 57.Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 58.Feledy JA, Morasso MI, Jang SI, Sargent TD. Nucleic Acids Res. 1999;27:764–770. doi: 10.1093/nar/27.3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amemiya CT, Zon LI. Genomics. 1999;58:211–213. doi: 10.1006/geno.1999.5827. [DOI] [PubMed] [Google Scholar]
- 60.Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. Mech Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 61.Kaji T, Artinger KB. Dev Biol. 2004;276:523–540. doi: 10.1016/j.ydbio.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker MB, Miller CT, Talbot JC, Stock DW, Kimmel CB. Dev Biol. 2006;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 63.Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]