Abstract
An important question in evolutionary and physiological genetics is how the control of flux-base phenotypes is distributed across the enzymes in a pathway. This control is often related to enzyme-specific levels of activity that are reported to be in excess of that required for demand. In glycolysis, metabolic control is frequently considered vested in classical regulatory enzymes, each strongly displaced from equilibrium. Yet the contribution of individual steps to control is unclear. To assess enzyme-specific control in the glycolytic pathway, we used P-element excision-derived mutagenesis in Drosophila melanogaster to generate full and partial knockouts of seven metabolic genes and to measure tethered flight performance. For most enzymes, we find that reduction to half of the normal activity has no measurable impact on wing beat frequency. The enzymes catalyzing near-equilibrium reactions, phosphoglucose isomerase, phosphoglucomutase, and triosephosphate isomerase fail to show any decline in flight performance even when activity levels are reduced to 17% or less. At reduced activities, the classic regulatory enzymes, hexokinase and glycogen phosphorylase, show significant drops in flight performance and are nearer to saturation. Our results show that flight performance is canalized or robust to the activity variation found in natural populations. Furthermore, enzymes catalyzing near-equilibrium reactions show strong genetic dominance down to low levels of activity. This implies considerable excess enzyme capacity for these enzymes.
Identifying the causes of the differing patterns of molecular evolution among genes is a compelling problem in biology. In Drosophila and yeast, genome-wide studies have emphasized gene-specific variables, such as codon bias, expression level, dispensability, and connectedness, to establish correlations with the levels of purifying selection (1–7). One complex but unexplored cause of variation is the relative difference among enzymes that functional variation imparts to phenotype. When the phenotypic variation reflects the flux rate through a pathway, this relationship between enzyme activity and phenotype is defined by the level of flux control. For enzyme steps with little flux control, activity differences will have no impact on phenotypic variation and will have consequently no effect on phenotype-associated fitness. For this reason, the relative level of control at an enzyme step is expected to be a predictor of molecular evolution for a gene. The goal of this study is to evaluate the control of flight performance associated with activity variation for a number of enzymes of the glycolytic pathway.
Flux control is expected to differ among enzymes as a result of pathway context, equilibrium status, and the presence or absence of steady-state conditions (8). The classical view of metabolic flux control proposes that unique steps, such as those under allosteric control and strongly displaced from equilibrium, control the pathway flux. These steps represent the textbook regulatory enzymes and are often said to be rate-limiting. However, control must be examined in a whole-pathway context. Certainly, the best example of this dissection of pathway control is the study of lactose metabolism in Escherichia coli (9, 10). In constitutively expressed strains, the three sequential steps of porin, permease, and β-galactosidase show markedly different levels of control over flux. Under these conditions, the diffusion-limited porin step possesses by far the greatest control, followed by the permease and β-galactosidase steps, and these differences predict different dissimilar evolutionary responses in both genetic variation and control. Unfortunately, the estimation of control in multicellular organisms is complex and difficult to measure in a biologically relevant fashion in most systems.
The glycolytic pathway is the best understood in biology. For this pathway, it is proposed that individual enzyme levels are matched to the maximum metabolic flux demands of a particular cell type (11). An extreme example of this essential matching is insect flight, where the highest mass-specific oxygen consumption rates among animals are observed. In the flight muscle, high enzyme levels are needed for the rapid turnover of ATP associated with the 100-fold or greater increase in oxygen consumption over resting states (12). Wing beat frequency (WBF) is closely correlated with oxygen consumption (13–15), and it directly reflects the rate of ATP hydrolysis and glycolytic flux. Glycogen, trehalose, and hexoses fuel dipteran flight (16), and the sole route for metabolism of these fuels is glycolysis (17). This tight connection among fuels, glycolysis, pathway flux, and ATP turnover makes the measurement of WBF an ideal system for studying, albeit indirectly, the control of flight metabolism.
For this reason, several studies in Drosophila melanogaster have examined the contribution of genetic variation in enzyme levels to flight performance (14, 18). These studies find significant line-specific variation in WBF, but they cannot separate trans-acting from cis-acting genetic effects on enzyme activity. This shortcoming prevents the measurement of enzyme-specific control of flight performance. The solution lies in the direct genetic perturbation of enzyme levels in controlled isogenic backgrounds (19).
In this study, we examine the control of several enzymes in the glycolytic pathway on flight performance. We use P-element excision-derived knockouts in D. melanogaster to experimentally lower the in vivo activity levels of seven enzymes and examine individual enzyme influence on tethered flight performance measured as WBF. The modified genes include three that encode enzymes that catalyze near-equilibrium reactions [i.e., Pgi, phosphoglucose isomerase (PGI); Pgm, phosphoglucomutase (PGM); and Tpi, triosephosphate isomerase (TPI)], and three considered to catalyze nonequilibrium reactions (or regulatory reactions) [i.e., Hex-A, muscle-specific hexokinase (HEX-A); PyK, pyruvate kinase; and GlyP, glycogen phosphorylase (GLYP)]. We refer to these two sets as nonequilibrium and near-equilibrium enzymes, respectively. We examined the effects of reduced activity for trehalase (Treh) because it degrades trehalose, the insect hemolymph storage disaccharide. These genes are homozygous lethal as full knockouts. However, we have also produced genotypes for GlyP, Pgi, Pgm, Tpi, and Hex-A possessing enzyme levels that range from trace amounts to <23% of the normal amount.
Throughout this investigation, we address several questions: Is there canalization of flight metabolism against reduction of activity in the enzymes of the central metabolic pathway? Does canalization imply excess enzymatic capacity at some steps? Is there a relationship among enzyme status, classified as catalyzing nonequilibrium or near-equilibrium reactions, canalization, and excess capacity?
Results
Our results show the enzyme-specific effects of reduced activity variation on flight performance (Table 1). Throughout Results and Discussion, we will refer to the “control” exercised by an enzyme. Although in metabolic control analysis control is formally defined by the flux control coefficient (the ratio of fractional changes in flux and enzyme activity) (20), we interpret control informally here as a percentage change in WBF for a given pair of activity ratios. Furthermore, we are not directly measuring flux per se (e.g., by measuring oxygen consumption), but we are using WBF as a proxy for this flux. First, for five of seven enzymes, activity reduction to 50% of the “normal” activity has no statistically significant effect on WBF. Only GLYP and PGM show statistically significant reductions of WBF when activity is reduced to 50–55% of the normal activity, but a further reduction is not seen in the comparison of 60% to 10% PGM activity reduction genotypes. The use of partial knockouts in five enzymes to evaluate control in lower activity ranges shows different outcomes. Reduction of PGI and PGM to ≤10% and of TPI to ≈17% of normal activity results in no significant drop in WBF. In contrast, the nonequilibrium enzymes, HEX-A and GLYP, show significant reductions in performance. HEX-A males possessing ≈23% of the normal activity show a 15% reduction in WBF. Moreover, these HEX-A males cannot beat their wings for more than a few seconds, and free flight consists of short bursts and distances of a few centimeters. GLYP homozygotes with ≈10% normal activity show further WBF reductions of ≈6% relative to flies with 60% activity but otherwise appear able to sustain these levels for longer periods.
Table 1.
Summaries for 16 experiments on flight performance
| Genotypes (activities ratios) | nL | nH | Mean WBF nL | Mean WBF nH | ΔWBF ± SE | P |
|---|---|---|---|---|---|---|
| 6326/Pgm9, Pgm2 (50:100) | 36 | 47 | 48.3 | 49.5 | −1.14 ± 0.51 | 0.028 |
| Pgm(GlyP)/Pgm9, Pgm2 (50:100) | 47 | 59 | 48.5 | 48.7 | −0.50 ± 0.62 | 0.427 |
| PgmnGB1/Pgm9, Pgm2 (10:60) | 53 | 35 | 46.0 | 46.3 | −0.21 ± 0.30 | 0.600 |
| 6326/Pgi9, Pgi20 (50:100) | 44 | 46 | 50.5 | 49.8 | +0.66 ± 0.58 | 0.195 |
| Pgi4/Pgi20, Pgi9 (10:60) | 32 | 56 | 46.3 | 46.0 | +0.24 ± 0.69 | 0.726 |
| Treh77/Treh108, Treh77 (50:100) | 23 | 45 | 47.7 | 47.7 | -.036 ± 1.36 | 0.400 |
| 6326/Treh108, Treh77 (50:100) | 28 | 38 | 54.9 | 55.8 | −0.91 ± 0.91 | 0.420 |
| GlyP(Pgm)/GlyP3.1, GlyP9.1 (55:100) | 50 | 58 | 47.8 | 49.2 | −1.45 ± 0.60 | 0.019 |
| 6326/GlyP3.1, GlyP9.1 (55:100) | 36 | 45 | 47.5 | 47.0 | +0.50 ± 0.54 | 0.360 |
| GlyP6.1, GlyP9.1 (10:60) | 23 | 19 | 45.6 | 48.3 | −2.39 ± 0.97 | 0.015 |
| VT83/Tpi1, Tpi51 (55:100) | 36 | 39 | 48.2 | 47.0 | +1.18 ± 0.79 | 0.130 |
| EY03361/Tpi1, Tpi51 (17:70) | 36 | 39 | 47.5 | 47.0 | +0.52 ± 0.72 | 0.387 |
| 6326/PyK1, PyK24 (55:100) | 34 | 32 | 48.7 | 47.3 | +1.37 ± 1.18 | 0.065 |
| 6326/Hex-A74, Hex-A79 (63:100) | 40 | 35 | 45.6 | 46.5 | −0.95 ± 0.85 | 0.320 |
| Hex-A79/Hex-A79, Hex-A74 (67:100) | 35 | 44 | 46.1 | 44.8 | +1.29 ± 0.94 | 0.150 |
| Hex-A74, Hex-A79 (23:100) | 24 | 36 | 40.6 | 47.4 | −6.82 ± 1.19 | >0.001 |
Shown are genotypes, relative activities, sample sizes (with nL being the number of low-activity flies tested and nH the number of high-activity flies tested), WBF (per 200 msec), WBF difference with standard deviation, and probabilities associated with the analyses of covarience. Entries shown in bold are significant.
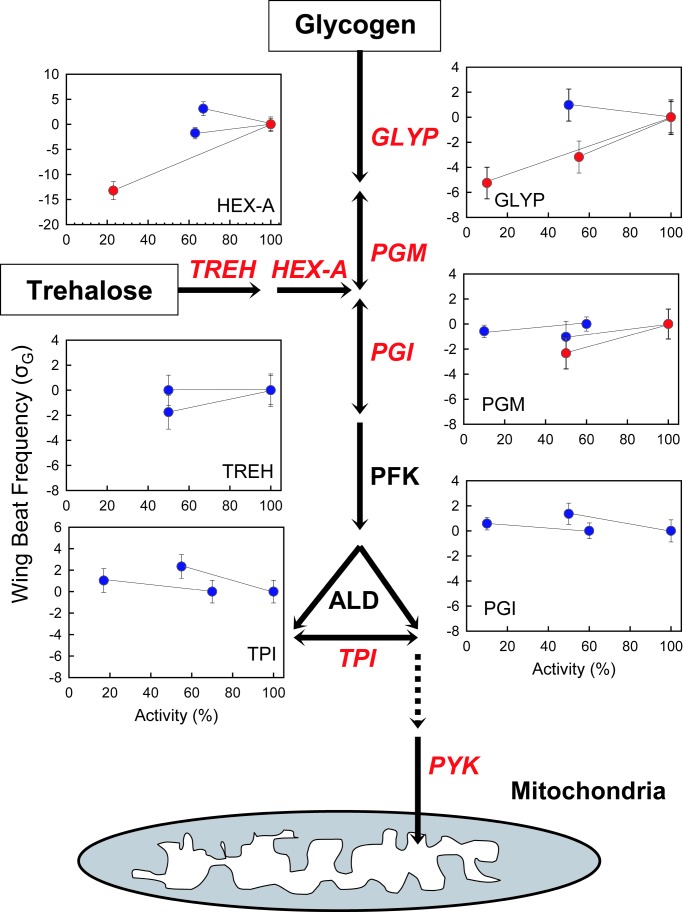
There are two reasons why these modest enzyme-specific effects on WBF are biologically important. First, consideration of the importance of effects using the full WBF scale is irrelevant; a reduction of WBF by only one-third results in a loss of sufficient lift to sustain flight (13). Second, in D. melanogaster, the genetic coefficient of variation for WBF is small. Therefore, when scaled against reported natural variation, these differences between genotypes are relatively large (14). Fig. 1 places our results in the context of pathway position and rescales the relative percent changes in genetic (or line) standard deviations of WBF. Even the small significant difference observed for Pgm (−1.14 ± 0.51) is two and a half times this standard genetic deviation.
Fig. 1.
The upper glycolytic pathway and the summary results of 15 flight experiments using knockouts. The seven enzymes addressed in this study are shown in red italics: GLYP, HEX (muscle-specific form HEX-A), PGM, PGI, trehalase (TREH), TPI, and pyruvate kinase (PYK). Within each graph, a line connects the results for a single experiment. Significant genotype differences are shown in red, and nonsignificant cases are shown in blue. The y axis is WBF transformed into units of the standard deviation of the line variance component (σG) for second chromosomes reported in Curtsinger and Laurie-Ahlberg (14). Confidence bars span one SE, and the normal activity genotype mean is set as zero.
Discussion
This study uses genetic means to systematically perturb in vivo enzyme levels across a number of steps in a well known pathway and to measure the effect on a flux-dependent physiological phenotype. The results have implications both with respect to questions of the enzyme-specific control of metabolic flux and to whether individual enzymes appear in excess of flux demand. First, from a genetic perspective, we show most study genes display strong dominance with respect to flight metabolism under these test conditions. For these enzymes, heterozygotes of knockout and full activity alleles have activity reductions of 50% and show little reduction, if any, in WBF. This observation demonstrates that flight metabolism is canalized against levels of a naturally occurring activity variation (21). Second, from the perspective of flight physiology, only GLYP is strictly near “capacity.” The rest, as shown by no reduction in WBF with genotypes with only 10% activity, have apparent “excess capacity.” It must be pointed out that the concepts of genetic dominance and excess capacity, as used in the fields of genetics and physiology, describe the same phenomenon.
Both of the nonequilibrium enzymes, GLYP and HEX-A, show some measure of control over flight performance in this activity range. Hex-A flies with low activity (23%) are able to beat their wings only at a rate sufficient enough to provide lift for a few seconds, tethered or untethered. At the initiation of flight, the immediate depletion of ATP is believed to be buffered by the phosphogen arginine-phosphate and the adenylate kinase reactions (22, 23). This response may operate for a few seconds, after which ATP is resupplied through glycolysis. Therefore, the short pulse in WBF measured for HEX-A low-activity genotypes may simply reflect this depletion of the arginine-phosphate pool and the subsequent inability to resupply ATP through glycolysis.
The different results among nonequilibrium and near-equilibrium enzymes qualitatively support observations from comparative physiological studies that use in vitro measures to predict in vivo conditions. One claim of physiological studies is that there is a large excess of activity over the amount required to support performance (24). Maximum available activities (Vmax) are typically one to two orders of magnitude higher than required. However, this excess appears to be greatly reduced under the high-flux conditions associated with flight. For insect and hummingbird flight, the general observation is that nonequilibrium enzymes operate close to the maximum Vmax (25). In flying honey bees, the required flux is estimated to be ≈76% of Vmax for HEX, and in vertebrate muscle activity levels are reported to operate close to Vmax for GLYP (26, 27). In contrast, the same study predicted the required flux was 4–5% of Vmax for PGI (26). In a further study on honey bee flight, it was predicted that PGI Vmax is 1.7- to 2.2-fold in excess of that required to support assumed in vivo flux rates (28). However, this estimate depends on in vitro measurement conditions. If measured at physiological pH and ionic strength, the estimated Vmax is only 5% greater than required to support in vivo metabolic flux under flight.
These generalities about regulation and capacity also have been explored in comparative evolution studies using independent contrasts in orchid bees. Across 28 orchid bee species, the 20-fold change in body mass results in 2-fold variation in flight metabolism. For HEX and GLYP, there are close matches between mass-specific enzyme activity levels and corresponding metabolic flux rates (29); enzyme activities increase with increasing species-specific flight metabolism. This correlation suggests that both enzymes are near maximum capacity (saturation), and enzyme activity levels must increase to match escalating flux demand. It has also been speculated that this association is required to maintain tight flux regulation. Numerous studies have implicated HEX and GLYP as regulatory enzymes in glycolysis (30–32). Our results in D. melanogaster show that as HEX-A and GLYP activities continue to be reduced, capacities are exceeded, regulation is disrupted, and flux is reduced. In contrast, the same orchid bee studies report that PGI mass-specific activities are constant across species; PGI does not evolve higher activities to match capacities with increased flux rates. Our manipulation of PGI enzyme levels finds excess in vivo activity (>10-fold) with respect to flight-based flux demand. This finding supports the physiological argument that PGI will be less responsive to a variation in overall flight metabolism, as in orchid bees. Likewise, TPI is the penultimate near-equilibrium enzyme and is often recognized as having evolved the highest specific activity of any enzyme known in nature (33, 34). However, TPI activity reduction to <18% of normal activity also shows no significant effect on WBF and predicts at least 5-fold excess capacity.
Why are there apparent excesses of protein for near-equilibrium enzymes, when reduced amounts are clearly sufficient to meet the high flux demands of flight? In D. melanogaster, the genes for Pgi, Pgm, and Tpi show high codon bias, which means that they are under natural selection to maintain high levels of expression. Protein synthesis and turnover are energetically expensive (35), and true excess activity will be costly and reduce fitness. It is possible that high enzyme levels are required for low-flux phenotypes, where steady-state conditions predominate and are near equilibrium. In the case of near-equilibrium reactions, higher activities (Vmax) are required to drive low net fluxes, at which product/substrate mass-action ratios are nearer to equilibrium (36). Although reduction of PGI activity had no effect on WBF, we observed significant reductions in larval and adult Drosophila body size for small activity reductions in Pgi genotypes (unpublished data). Body size variation is an expression of the integration of glycolytic flux over the course of larval development. The conditions present during development are likely to be different from the large transient displacements from equilibrium expected under flight (8, 17, 22). One might speculate that the high activity levels of PGI and TPI may be required to meet the near-equilibrium conditions and low flux demands of other metabolic states, such as the large resting flight muscle.
Yet, there are several reasons why the interpretation of excess enzyme capacity may be misleading. Enzyme levels that appear in excess may be available to support performance demands in nature that are outside of the test conditions used in physiological studies (24, 37). Our experiments are carried out at 24–26°C, yet D. melanogaster has a lower thermal flight limit of ≈15°C (14) and certainly must experience wide temperature variation in nature (38). The small size of Drosophila precludes the effective operation of thermogenesis like that seen in larger insects (15), and at reduced temperatures flies must maintain physiological performance by catalytic means. Unacclimated individuals may meet performance demands at lower temperatures by drawing on excess activity. In physiological studies, this excess has been explained as a “safety factor” (39, 40), but it also entered into Sewall Wright's (41) explanations for the evolution of genetic dominance. The testing of genotypic effects at a lowered temperature should be a goal of future studies. Finally, in comparison to the wing-loading demands of free flight, tethered flies may be relatively unchallenged. Although the tethered design is extensively used in flight studies, there is debate as to its relevance to free flight (15). Studies in virtual flight arenas suggest that wing-loading forces under tethered flight are conservatively ≈80% of those experienced under free flight (13). Nevertheless, our genotypes of near-equilibrium enzymes with greatly reduced activity (PGI, TPI, and PGM) disperse from release points using free flight with no apparent difficulty, albeit we have not quantified this using free flight measures (42, 43).
The various levels of control on flight performance might result in different patterns of molecular evolution for these genes. We propose that enzymes possessing greater control will be the most conserved by purifying selection. This proposition requires a larger study to establish overall pattern, yet we can see some differences. Hex-A shows no replacement polymorphism in D. melanogaster or Drosophila simulans and no amino acid fixations in the 10 million years separating these species from Drosophila yakuba (44), indicating a high evolutionary constraint at the molecular level. We see that across the 12 Drosophila species for which there are genome sequences, the consensus amino acid substitution rates for the Hex-A and GlyP genes are among the lowest in the pathway, and the near-equilibrium enzymes, Pgi, Pgm, and Tpi, possess rates that are two to four times higher (unpublished data). In D. melanogaster, Pgm has a large excess of amino acid polymorphism and latitudinal clines in protein haplotype frequencies (45, 46). Both observations are strong evidence that Pgm is responding positively to natural selection. PGI also shows a significant excess of amino acid polymorphism in D. melanogaster (47) and is highly polymorphic for amino acid replacements in other species (48, 49). Furthermore, PGI has been implicated in selection for flight duration in Colias butterflies (50). Our results show little control of PGI for flight metabolism in Diptera, but the difference for Colias may reflect the lower flux demand of synchronous flight muscles in Lepidoptera. We speculate that excess capacity, if selectively maintained to match extreme demand states, will lead to fitness tradeoffs in near-equilibrium enzymes. This extra activity will be balanced against the fitness costs of producing and maintaining high levels of enzymes in environments where the demand is reduced or absent. In heterogeneous environments, this tradeoff might lead to the maintenance of polymorphism (51).
Our recent studies with Gpdh and the glycerol-3-phosphate shuttle show glycerol-3-phosphate dehydrogenase reduction to ≈10% of normal activity depresses WBF by ≈7% (19). Stoichiometrically, we expect that the complete blockage of the glycerol-3-phosphate shuttle will result in a loss of one-sixth of the reducing equivalents available for flight metabolism (52). However, this calculation ignores the more serious problem of maintaining the redox balance of the cell. Failure to remove NADH and restore NAD will block glycolysis at GAPDH and impact flight performance. This disruption of redox balance, along with the disruption of physical enzyme–enzyme interactions (53, 54), explains the complete loss of flight ability in homozygous-null Gpdh genotypes, and it highlights the complexity of flux regulation.
Because in vivo enzyme concentrations are varied in our genotypes, flux may change not only because of lower activity, but also because of the disruption of physical interactions with partner enzymes. In these conditions, flux rates may depart significantly from expectations of simple solution biochemistry (55), and many glycolytic enzymes may be diffusion-limited and therefore require close proximity to operate under high flux (56). Studies in Drosophila of glycerol-3-phosphate dehydrogenase, GAPDH, TPI, phosphoglycerate kinase, phosphoglycerol mutase, and aldolase point to a structured muscle cell environment in which enzymes “colocalize” (53, 54) and removal of a member causes others to disassociate. Changes in flux may emerge from stoichiometric changes in the scaffolding of enzymes and structural organization rather than activity levels of individual enzymes. This complication remains a promising, but still unexplored, question whose impact on the evolution of enzymes is unknown.
In summary, using transposon-associated knockout to directly modify in vivo enzyme levels, we show a general canalization of a metabolic performance-based phenotype. Our results find that the classic regulatory enzyme GLYP is nearest to capacity or saturation, whereas other enzymes, especially those termed near-equilibrium, possess excess capacity. These latter enzymes show extreme genetic dominance and low flux control that extends down to markedly reduced enzyme levels. The genetic results are consistent with physiological studies using in vitro estimates that predict that near-equilibrium enzymes carry excess capacity. The possibility is that this excess capacity is exploited under demand states different from the experimental test conditions.
Materials and Methods
Stocks.
The following P-element insertions were used: Pgi(EY09730), GlyP[l(2)ko7198], Pgm [l(3)s1939], Treh(EY06982), Tpi(KG03964), Tpi(EY03361), PyK(EY10213), and Hex-A[EP(X)352]. The insertion of GlyP[l(2)ko7198] possessed a linked lethal, and it was repeatedly outcrossed to line w; 6326;6326.1 for six generations. The insertion was then reconstituted as a single chromosome isolate for the excision series. Transposase sources were obtained from the Bloomington Stock Center (Bloomington, IN). Lines VT46 and VT83 are derived from inbred lines collected in 1997 in Whiting, VT. Line w;6326;6326.1 is a derivative of Bloomington line 6326 that has its X chromosome replaced with the chromosome from Bloomington stock 2475.
Screening.
P-elements were excised by using male flies in standard dysgenic crosses (19). Excision chromosomes (flies with white eyes) were isolated with the balancers TM3 and CyO. Excision lines (≈80–100) were sampled for each dysgenic cross. Lines were first screened for lethality. When possible, excision allele activity was first assessed by using allozyme screens with alternative mobility alleles (Pgi, Tpi, and Pgm). Relative function was first screened by direct spectrophotometric assay of crude mass-adjusted enzyme activity. Interline crosses were used to create heterozygotes for different test alleles. This approach permitted the assessment of additivity in allele combinations in the event that transvection effects were present (57). PCR and sequencing with flanking primers were used to determine molecular changes in the alleles. All full activity alleles were confirmed to have sequences consistent with the “precise” excision or gene conversion to a normal sequence. Reduced activity alleles possessed a spectrum of molecular changes from the deletion of entire exons to the retention of large pieces of the original P-element (19, 57). None of the alleles involved point changes in amino acid sequence, and thus, catalytic function. Using marker-assisted introgression (19), excision chromosomes with test alleles were placed in 6326.1, VT46, and VT83 isogenic backgrounds. Lines were again assayed for thoracic activity and expressed in units of activity per milligram of wet weight. We expect paired test genotypes to differ only in the gene of interest.
Enzyme Assays.
For final genotype-specific measures of activity, multiple flies for each line were homogenized in a grinding buffer (0.01 M KH2PO4/1.0 mM EDTA, pH 7.4) at a “concentration” of one fly per 200 μl of buffer and were centrifuged at 9,460 × g for 5 min at 4°C to pellet all solids. The supernatant was recovered and transferred to a 96-well plate and used in all enzymatic assays. Blanks were run to establish background activity levels and remove them from the assay. Representative “wild” lines were included in assay sets to assure that precise excisions had high or full activity.
The assay buffers for the enzymes assayed in this study were as follows: PGI: 20 mM Tris·HCL/1 mM NADP/8 mM MgCl2/2.5 mM fructose-6-phosphate/0.5 units/ml G6PD, pH 7.4; PGM: 20 mM Tris·HCL/0.5 mM NADP/1 mM MgCl2/0.83 mM glucose-1-phosphate/0.62 units/ml G6PD, pH 7.4; TPI: 100 mM Tris·HCL/0.15 mM NADH/0.83 mM glyceraldehydes-3-phosphate/2 units/ml G6PD, pH 8.6; HEX-A: 20 mM Tris·HCL/1 mM NADP/2 mM MgCl2/8 mM ATP/100 mM glucose/5.0 units/ml G6PD, pH 8.0; GLYP: 50 mM KH2PO4/0.6 mM NADP/15 mM MgCl2/5 μm glucose 1,6-diphosphate/7.0 mg/ml glycogen/2 mM 5′AMP/2 units/ml PGM/2 units/ml G6PD, pH 6.8; trehalase: 50 mM Tris·HCL, 0.3 mM NADP, 50 mM trehalose, 4 mM MgCl2, 2 mM ATP, 0.7 units/ml HEX 0.6 units/ml G6PD, pH 7.1; pyruvate kinase: 50 mM imidazole/60 mM KCl/8 mM MgSO4/2 mM ADP/4 mM phosphoenolpyruvate/0.15 mM NADH/10 units/ml lactate dehydrogenase, pH 7.3.
Enzyme activity assays were carried out on a SpectraMax 384 Plus 96-well plate spectrophotometer (Molecular Designs, Memphis, TN) with 10 μl of fly extract and 100 μl of assay buffer. Optical density was measured every 9 sec for 3 min. TPI possesses extremely high activity, and a 1/10th dilution of the supernatant was used to obtain accurate measures. All activity assays were conducted at 25°C. In all experiments, replicated samples were assayed twice, and the average was used in analysis. Enzyme activity was estimated from the change in optical density and converted into nanomolar NAD+ or NADPH reduced per minute per fly, depending on the coupled assay. Initial values for appropriate pH, substrate, and cofactor concentrations for the reactions were taken from the literature and modified to give maximum enzyme activity.
Test Crosses and Genotypes.
In each flight experiment, two genotypes emerging from a common environment were created by crossing ≈100 females, heterozygous for paired full and knockout alleles, with males homozygous for high or low activity alleles and allowed to lay eggs for 24 h on standard cornmeal. Females were subsequently transferred each day to fresh bottles. For a single flight experiment (or block), emerging adult progeny were collected on a single day from a single bottle, aged for 4–7 days, assayed for WBF, and frozen for genotyping. In the case of GlyP and Hex-A, low-activity homozygous and hemizygous genotypes possess sufficiently low, relative viability to distort emerging genotype numbers and prevent balanced sample numbers. In these cases, homozygous genotypes were reared separately under equal densities and flown on the same day in random order.
Flight Assay and Statistical Analysis.
Five-day-old adult male flies were gently anesthetized by using CO2 and tethered by attaching a syringe-cleaning wire hook to the dorsal surface of their mesothorax with Permount adhesive. After recovery from the CO2, tethered flies were video recorded for 2-sec intervals with a MotionScope high-speed video camera (Red Lake, Tucson, AZ) with a shutter speed of 1,000 frames per second. Temperature was monitored but not controlled; it was constant within days (blocks) and ranged from 26°C to 28°C among days. Only flies beginning to beat their wings within 15 min of tethering were included in the experiment. No fly was recorded more than 30 min after being tethered. Video recording and playback was conducted with Red Lake MotionScope software. After the recording of WBF, each individual was removed from the wire and frozen. To determine genotype, each fly was weighed and assayed for whole-body enzyme activity by using the standard assays in the microplate reader (58). Hex-A genotypes were determined by assaying whole thoraxes. Inspecting the biplot of activity and weight allows for the unambiguous assignment of individual genotypes (19). WBF was determined by counting the number of beats in a 200-msec interval. WBF for each fly was recorded twice, and two replicates were used in analysis. WBF is approximately normally distributed. Statistical significance was determined with a linear ANOVA model applied to untransformed WBF estimates (over 200 msec) as the dependent variable with fixed genotype and random blocks (days 1 and 2, when appropriate) as independent variables. Weight was used as a covariate, but it never showed any significant effect on WBF. Statistical testes were carried out by using the JMP application (SAS Institute, Cary, NC).
Acknowledgments
We thank Joe Lachance, Dan Dykhuizen, and John True for commenting on earlier versions of the manuscript; Ward Watt and an anonymous reader for contributing constructive points that significantly improved the paper; and Mike Doall and the Functional Ecology and Research Training Laboratory (under National Science Foundation Grant DEB 431-1515A) for access to the high-speed video equipment. This study was supported by U.S. Public Health Service Grant GM-45247 (to W.F.E.) and is contribution number 1146 from the Graduate Program in Ecology and Evolution, State University of New York, Stony Brook, NY.
Abbreviations
- PGI
phosphoglucose isomerase
- HEX-A
hexokinase
- GLYP
glycogen phosphorylase
- TPI
triosephosphate isomerase
- PGM
phosphoglucomutase
- GPDH
glycerol-3-phosphate dehydrogenase
- WBF
wing beat frequency.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 2.Pal C, Papp B, Hurst LD. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocha EP, Danchin A. Mol Biol Evol. 2004;21:108–116. doi: 10.1093/molbev/msh004. [DOI] [PubMed] [Google Scholar]
- 4.Subramanian S, Kumar S. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurst LD, Smith NG. Curr Biol. 1999;9:747–750. doi: 10.1016/s0960-9822(99)80334-0. [DOI] [PubMed] [Google Scholar]
- 6.Vitkup D, Kharchenko P, Wagner Genome Biol. 2006;7:R39. doi: 10.1186/gb-2006-7-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Proc Natl Acad Sci USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watt WB, Dean AM. Annu Rev Genet. 2000;622:593–622. doi: 10.1146/annurev.genet.34.1.593. [DOI] [PubMed] [Google Scholar]
- 9.Dean AM. Genetics. 1989;123:441–454. doi: 10.1093/genetics/123.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dykhuizen DE, Dean AM. Trends Ecol Evol. 1990;5:257–262. doi: 10.1016/0169-5347(90)90067-N. [DOI] [PubMed] [Google Scholar]
- 11.Hochochka PW, Somero GN. Biochemical Adaptation. New York: Oxford Univ Press; 2002. [Google Scholar]
- 12.Sacktor B. Biochem Soc Symp. 1976:111–131. [PubMed] [Google Scholar]
- 13.Lehmann FO, Dickinson MH. J Exp Biol. 1997;200:1133–1143. doi: 10.1242/jeb.200.7.1133. [DOI] [PubMed] [Google Scholar]
- 14.Curtsinger JW, Laurie-Ahlberg CC. Genetics. 1981;98:549–564. doi: 10.1093/genetics/98.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudley R. The Biomechanics of Insect Flight. Princeton: Princeton Univ Press; 2000. [Google Scholar]
- 16.Wiggleworth VB. J Exp Biol. 1949;26:150–163. doi: 10.1242/jeb.26.2.150. [DOI] [PubMed] [Google Scholar]
- 17.Sacktor B, Wormser-Shavit E. J Biol Chem. 1966;241:624–631. [PubMed] [Google Scholar]
- 18.Laurie-Ahlberg CC, Barnes PT, Curtsinger JW, Emigh TH, Karlin B, Morris R, Norman RA, Wilton AN. Genetics. 1985;111:845–868. doi: 10.1093/genetics/111.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merritt TJ, Sezgin E, Zhu CT, Eanes WF. Genetics. 2006;172:293–304. doi: 10.1534/genetics.105.047035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fell DA. Understanding the Control of Metabolism. London: Portland; 1997. [Google Scholar]
- 21.Laurie-Ahlberg CC, Wilton AN, Curtsinger JW, Emigh TH. Genetics. 1982;102:191–206. doi: 10.1093/genetics/102.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacktor B, Hurlbut EC. J Biol Chem. 1966;241:632–634. [PubMed] [Google Scholar]
- 23.Kohane MJ, Watt WB. J Exp Biol. 1999;202:3145–3154. doi: 10.1242/jeb.202.22.3145. [DOI] [PubMed] [Google Scholar]
- 24.Salvador A, Savageau MA. Proc Natl Acad Sci USA. 2003;100:14463–14468. doi: 10.1073/pnas.2335687100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suarez RK. Physiol Biochem Zool. 2000;73:765–771. doi: 10.1086/318112. [DOI] [PubMed] [Google Scholar]
- 26.Suarez RK, Lighton JRB, Joos B, Roberts SP, Harrison JF. Proc Natl Acad Sci USA. 1996;93:12616–12620. doi: 10.1073/pnas.93.22.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suarez RK, Staples JF, Lighton JRB, West TG. Proc Natl Acad Sci USA. 1997;94:7065–7069. doi: 10.1073/pnas.94.13.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staples J, Suarez R. J Exp Biol. 1997;200:1247–1254. doi: 10.1242/jeb.200.8.1247. [DOI] [PubMed] [Google Scholar]
- 29.Darveau CA, Hochachka PW, Roubik DW, Suarez RK. J Exp Biol. 2005;208:3593–3602. doi: 10.1242/jeb.01777. [DOI] [PubMed] [Google Scholar]
- 30.Kashiwaya Y, Sato K, Tsuchiya N, Thomas S, Fell DA, Veech RL, Passonneau JV. J Biol Chem. 1994;269:25502–25514. [PubMed] [Google Scholar]
- 31.Fueger PT, Bracy DP, Malabanan CM, Pencek RR, Wasserman DH. Am J Physiol. 2004;286:E77–E84. doi: 10.1152/ajpendo.00309.2003. [DOI] [PubMed] [Google Scholar]
- 32.Puigjaner J, Rais B, Burgos M, Comin B, Ovadi J, Cascante M. FEBS Lett. 1997;418:47–52. doi: 10.1016/s0014-5793(97)01347-1. [DOI] [PubMed] [Google Scholar]
- 33.Fersht A. Enzyme Structure and Mechanism. San Francisco: Freeman; 1977. [Google Scholar]
- 34.Albery JW, Knowles JR. Biochemistry. 1976;15:5631–5640. doi: 10.1021/bi00670a032. [DOI] [PubMed] [Google Scholar]
- 35.Hawkins AJS. Funct Ecol. 1991;5:222–233. [Google Scholar]
- 36.Suarez RK, Darveau CA, Hochachka PW. J Exp Biol. 2005;208:3603–3607. doi: 10.1242/jeb.01778. [DOI] [PubMed] [Google Scholar]
- 37.Salvador A, Savageau MA. Proc Natl Acad Sci USA. 2006;103:2226–2231. doi: 10.1073/pnas.0510776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehmann FO. J Comp Physiol. 1999;169:165–171. doi: 10.1007/s003600050207. [DOI] [PubMed] [Google Scholar]
- 39.Steyermark AC, Lam MM, Diamond J. Proc Natl Acad Sci USA. 2002;99:8754–8759. doi: 10.1073/pnas.132277799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam MM, O'Connor TP, Diamond J. J Physiol. 2002;542:493–500. doi: 10.1113/jphysiol.2002.023275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright S. Am Nat. 1934;68:24–53. [Google Scholar]
- 42.Marden JH, Wolf MR, Weber KE. J Exp Biol. 1997;200:2747–2755. doi: 10.1242/jeb.200.21.2747. [DOI] [PubMed] [Google Scholar]
- 43.Montooth KL, Marden JH, Clark AG. Genetics. 2003;165:623–635. doi: 10.1093/genetics/165.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duvernell DD, Eanes WF. Genetics. 2000;156:1191–1201. doi: 10.1093/genetics/156.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verrelli BC, Eanes WF. Genetics. 2000;156:1737–1752. doi: 10.1093/genetics/156.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verrelli BC, Eanes WF. Genetics. 2001;157:1649–1663. doi: 10.1093/genetics/157.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moriyama EN, Powell JR. Mol Biol Evol. 1996;13:261–277. doi: 10.1093/oxfordjournals.molbev.a025563. [DOI] [PubMed] [Google Scholar]
- 48.Wheat CW, Watt WB, Pollock DD, Schulte PM. Mol Biol Evol. 2006;23:499–512. doi: 10.1093/molbev/msj062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katz LA, Harrison RG. Genetics. 1997;147:609–621. doi: 10.1093/genetics/147.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watt WB, Carter PA, Blower SM. Genetics. 1985;109:157–175. doi: 10.1093/genetics/109.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gillespie JH. The Causes of Molecular Evolution. New York: Oxford Univ Press; 1991. [Google Scholar]
- 52.Chance B, Sacktor B. Arch Biochem Biophys. 1958;76:509–531. doi: 10.1016/0003-9861(58)90176-0. [DOI] [PubMed] [Google Scholar]
- 53.Wojtas K, Slepecky N, von Kalm L, Sullivan D. Mol Biol Cell. 1997;8:1665–1675. doi: 10.1091/mbc.8.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan DT, MacIntyre R, Fuda N, Fiori J, Barrilla J, Ramizel L. J Exp Biol. 2003;206:2031–2038. doi: 10.1242/jeb.00367. [DOI] [PubMed] [Google Scholar]
- 55.Suarez RK. J Exp Biol. 2003;206:2021–2029. doi: 10.1242/jeb.00366. [DOI] [PubMed] [Google Scholar]
- 56.Dzeja PP, Terzic A. J Exp Biol. 2003;206:2039–2047. doi: 10.1242/jeb.00426. [DOI] [PubMed] [Google Scholar]
- 57.Merritt TJ, Duvernell D, Eanes WF. Genetics. 2005;171:1707–1718. doi: 10.1534/genetics.105.048249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clark AG. Evolution (Lawrence, Kans) 1990;33:637–650. [Google Scholar]