Abstract
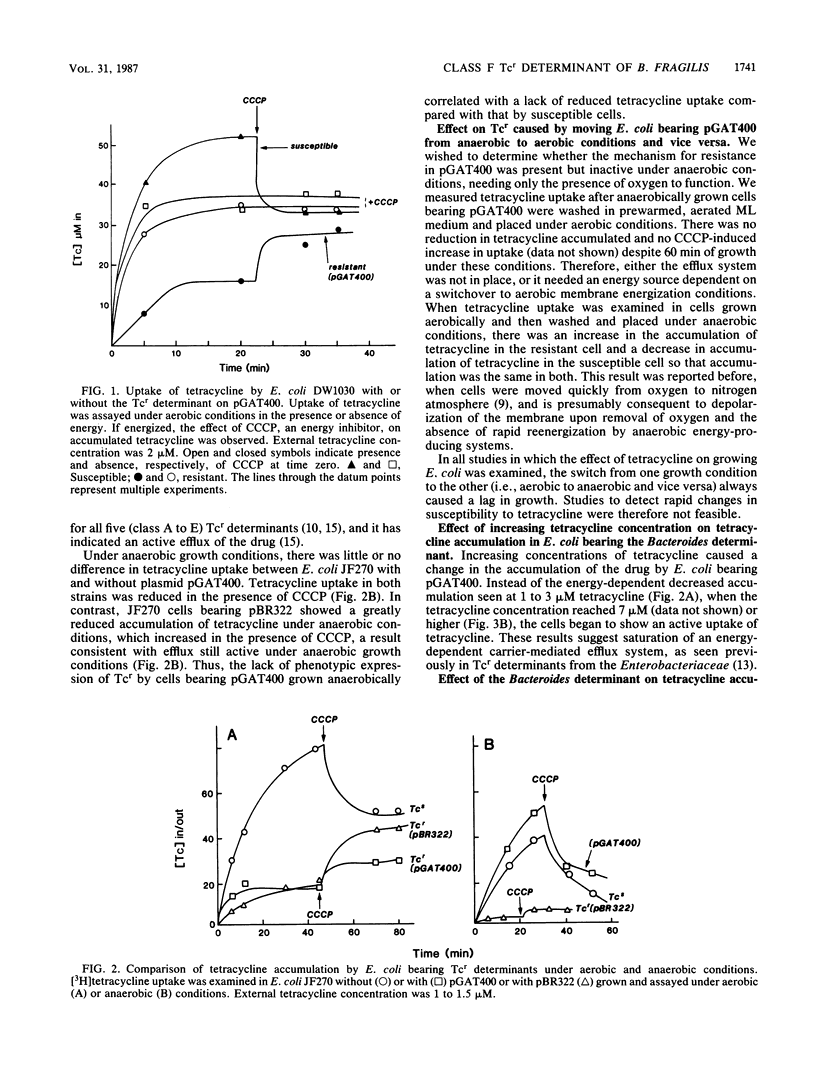
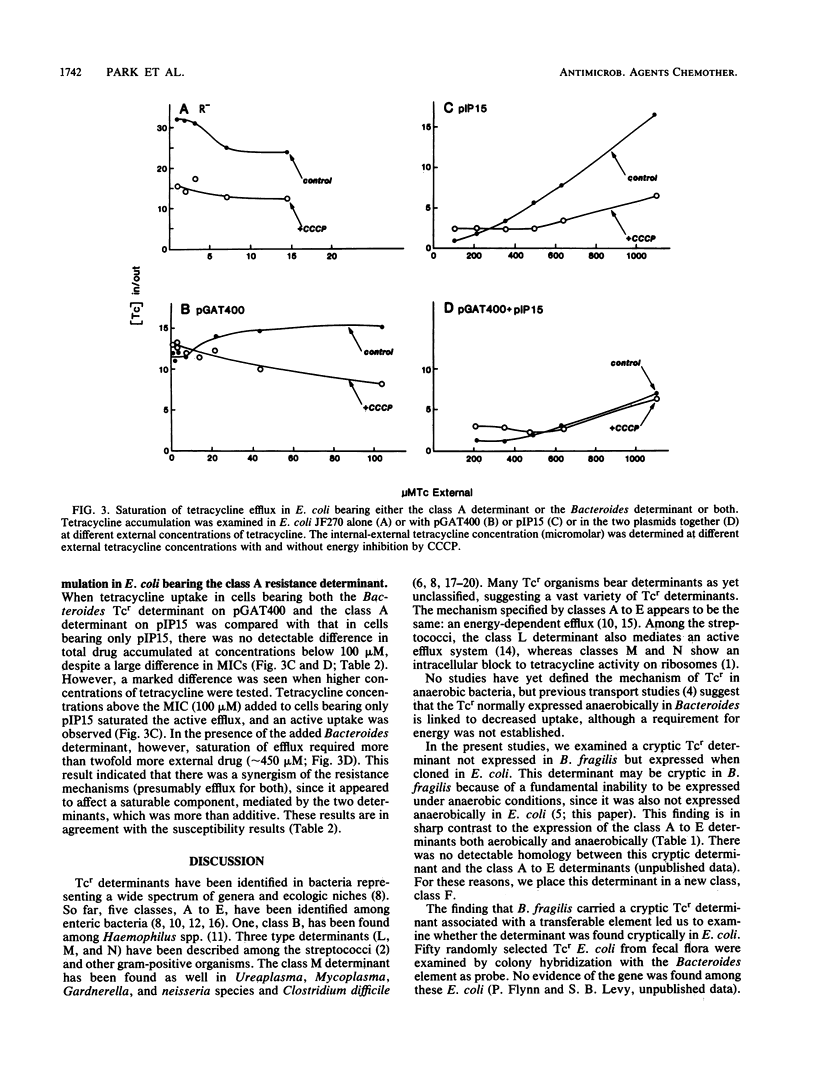
Escherichia coli bearing a cryptic tetracycline resistance determinant from Bacteroides fragilis expressed low-level constitutive resistance to tetracycline under aerobic, but not anaerobic, growth conditions and accumulated less tetracycline aerobically than did isogenic susceptible cells. This decreased uptake was energy dependent and reversible by increased concentrations of tetracycline, suggesting a saturable carrier-mediated active efflux mechanism. Decreased uptake was not seen when the cells were grown and assayed anaerobically. Other tetracycline resistance determinants (classes A to E) isolated from gram-negative enteric bacteria expressed resistance and generated active efflux of tetracycline under anaerobic as well as aerobic conditions. When the Bacteroides determinant was placed in the same cell with any of the class A to E tetracycline resistance determinants, there was an increase in resistance under aerobic conditions of as much as 48% more than was projected by adding the resistances expressed by the determinants individually. In cells bearing the class A determinant together with the Bacteroides determinant, saturation of the active efflux system required over twofold more exogenous tetracycline than did cells bearing the class A determinant alone. We have designated this new tetracycline resistance determinant class F.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burdett V., Inamine J., Rajagopalan S. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol. 1982 Mar;149(3):995–1004. doi: 10.1128/jb.149.3.995-1004.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J Bacteriol. 1986 Feb;165(2):564–569. doi: 10.1128/jb.165.2.564-569.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiale M. S., Levy S. B. Two complementation groups mediate tetracycline resistance determined by Tn10. J Bacteriol. 1982 Jul;151(1):209–215. doi: 10.1128/jb.151.1.209-215.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayolle F., Privitera G., Sebald M. Tetracycline transport in Bacteroides fragilis. Antimicrob Agents Chemother. 1980 Oct;18(4):502–505. doi: 10.1128/aac.18.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Jr, Hasegawa P., Davis C. E. Expression in Escherichia coli of cryptic tetracycline resistance genes from bacteroides R plasmids. Plasmid. 1984 May;11(3):248–252. doi: 10.1016/0147-619x(84)90031-3. [DOI] [PubMed] [Google Scholar]
- Hächler H., Kayser F. H., Berger-Bächi B. Homology of a transferable tetracycline resistance determinant of Clostridium difficile with Streptococcus (Enterococcus) faecalis transposon Tn916. Antimicrob Agents Chemother. 1987 Jul;31(7):1033–1038. doi: 10.1128/aac.31.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock G., Unger B., Gatz C., Hillen W., Altenbuchner J., Schmid K., Schmitt R. Heterologous repressor-operator recognition among four classes of tetracycline resistance determinants. J Bacteriol. 1985 Jan;161(1):326–332. doi: 10.1128/jb.161.1.326-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B., Morrissey S., Flynn P., Levy S. B. A new tetracycline-resistance determinant, class E, isolated from Enterobacteriaceae. Gene. 1986;50(1-3):111–117. doi: 10.1016/0378-1119(86)90315-x. [DOI] [PubMed] [Google Scholar]
- Marshall B., Roberts M., Smith A., Levy S. B. Homogeneity of transferable tetracycline-resistance determinants in Haemophilus species. J Infect Dis. 1984 Jun;149(6):1028–1029. doi: 10.1093/infdis/149.6.1028. [DOI] [PubMed] [Google Scholar]
- Marshall B., Tachibana C., Levy S. B. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob Agents Chemother. 1983 Dec;24(6):835–840. doi: 10.1128/aac.24.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L. M., Cullinane J. C., Levy S. B. Transport of the lipophilic analog minocycline differs from that of tetracycline in susceptible and resistant Escherichia coli strains. Antimicrob Agents Chemother. 1982 Nov;22(5):791–799. doi: 10.1128/aac.22.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L. M., Park B. H., Burdett V., Levy S. B. Energy-dependent efflux mediated by class L (tetL) tetracycline resistance determinant from streptococci. Antimicrob Agents Chemother. 1987 Oct;31(10):1648–1650. doi: 10.1128/aac.31.10.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurry L., Petrucci R. E., Jr, Levy S. B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez B., Tachibana C., Levy S. B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980 Mar;3(2):99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- Morse S. A., Johnson S. R., Biddle J. W., Roberts M. C. High-level tetracycline resistance in Neisseria gonorrhoeae is result of acquisition of streptococcal tetM determinant. Antimicrob Agents Chemother. 1986 Nov;30(5):664–670. doi: 10.1128/aac.30.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C., Hillier S. L., Hale J., Holmes K. K., Kenny G. E. Tetracycline resistance and tetM in pathogenic urogenital bacteria. Antimicrob Agents Chemother. 1986 Nov;30(5):810–812. doi: 10.1128/aac.30.5.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C., Kenny G. E. Dissemination of the tetM tetracycline resistance determinant to Ureaplasma urealyticum. Antimicrob Agents Chemother. 1986 Feb;29(2):350–352. doi: 10.1128/aac.29.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. C., Koutsky L. A., Holmes K. K., LeBlanc D. J., Kenny G. E. Tetracycline-resistant Mycoplasma hominis strains contain streptococcal tetM sequences. Antimicrob Agents Chemother. 1985 Jul;28(1):141–143. doi: 10.1128/aac.28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robillard N. J., Tally F. P., Malamy M. H. Tn4400, a compound transposon isolated from Bacteroides fragilis, functions in Escherichia coli. J Bacteriol. 1985 Dec;164(3):1248–1255. doi: 10.1128/jb.164.3.1248-1255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters S. H., Rogowsky P., Grinsted J., Altenbuchner J., Schmitt R. The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res. 1983 Sep 10;11(17):6089–6105. doi: 10.1093/nar/11.17.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]


