Abstract
Chemotaxis of bacteria requires regulated methylation of chemoreceptors. However, despite considerable effort in the 1980s, transmethylation has never been established as a component of eukaryotic cell chemotaxis. S-adenosylhomocysteine (SAH), the product formed when the methyl group of the universal donor S-adenosylmethionine (SAM) is transferred to an acceptor molecule, is a potent inhibitor of all transmethylation reactions. In eukaryotic cells, this inhibition is relieved by hydrolysis of SAH to adenosine and homocysteine catalyzed by SAH hydrolase (SAHH). We now report that SAHH, which is diffuse in the cytoplasm of nonmotile Dictyostelium amoebae and human neutrophils, concentrates with F-actin in pseudopods at the front of motile, chemotaxing cells, but is not present in filopodia or at the very leading edge. Tubercidin, an inhibitor of SAHH, inhibits both chemotaxis and chemotaxis-dependent cell streaming of Dictyostelium, and chemotaxis of neutrophils at concentrations that have little effect on cell viability. Tubercidin does not inhibit starvation-induced expression of the cAMP receptor, cAR1, or G protein-mediated stimulation of adenylyl cyclase activity and actin polymerization in Dictyostelium. Tubercidin has no effect on either capping of Con A receptors or phagocytosis in Dictyostelium. These results add SAHH to the list of proteins that redistribute in response to chemotactic signals in Dictyostelium and neutrophils and strongly suggest a role for transmethylation in chemotaxis of eukaryotic cells.
Keywords: chemotaxis, Dictyostelium, neutrophils, tubercidin
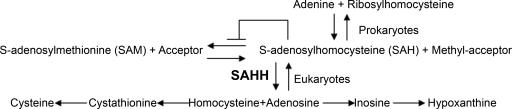
S-adenosylhomocysteine (SAH), the product of the transfer of the methyl group from S-adenosylmethionine (SAM) to DNA, RNA, phospholipids and many small molecules (Fig. 1), is a strong inhibitor of transmethylation (1). The inhibition is relieved in prokaryotes by hydrolysis of SAH to adenine and ribosylhomocysteine (2), and in eukaryotes by hydrolysis of SAH to adenosine and homocysteine, catalyzed by SAH hydrolase (SAHH) (3). Because hydrolysis of SAH is reversible, and more favored in the synthetic direction, efficient transmethylation from SAM in eukaryotes also requires removal of adenosine and homocysteine (Fig. 1).
Fig. 1.
Schematic representation of the universal methylation pathway illustrating the removal of the product inhibitor SAH by conversion to adenine and ribosylhomocysteine in bacteria, and, in eukaryotic cells, by hydrolysis to homocysteine and adenosine by SAHH and the subsequent metabolism of these products.
Stimulated by the discovery in the mid-1970s that bacterial chemotaxis is initiated by SAM-dependent transient carboxyl-O-methylation of glutamic acid residues in chemotactic ligand receptor proteins (4, 5), the possibility that transmethylation reactions have a role in the chemotactic response of Dictyostelium and leukocytes was investigated in the early 1980s. Investigators asked whether chemotactic factors increase methylation of proteins, phospholipids, and/or nucleic acids, and whether inhibition of methylation by inhibiting SAHH or adenosine deaminase inhibits chemotaxis. The results of these studies were inconclusive and often contradictory (see Discussion).
In the contemporary view of chemotaxis of Dictyostelium and leukocytes, chemoattractants mediate their effects by binding to transmembrane receptors coupled to heterotrimeric G proteins. Upon receptor activation the Gβγ-subunits are released and initiate a cascade of events that results in the redistribution of specific proteins to either the front or the rear of the polarized cell (6–9). Ultimately, the protrusive force of Arp2/3 complex-controlled assembly of F-actin at the front of the cell drives the leading edge forward, while assembly and activation of myosin II filaments at the back and sides of the cell localizes actomyosin contraction to those regions, initiating retraction and preventing pseudopod formation.
Although current models of chemotaxis have no role for transmethylation, we thought it likely that methylation would be required for the function of one or more of the multiple molecules involved in the signaling or motile events, and that SAHH would be required for efficient transmethylation. Dictyostelium SAHH, a tetramer of 47,000-Da subunits, accounts for ≈2% of the soluble protein in vegetative amoebae (10). There is a single SAHH gene with a deduced amino acid sequence 75% identical to human SAHH (11, 12) and with very similar catalytic activity. Kishi et al. (13) reported that SAHH is sequestered with actin bars in Dictyostelium spores, but that SAHH is diffuse, and not associated with F-actin, in vegetative amoebae (13, 14). Because most of the components of the chemotactic pathway are spatially and temporally localized, we thought that determining the localization of SAHH in polarized chemotaxing cells might provide evidence for the involvement of transmethylation during chemotaxis.
The results reported in this article confirm that SAHH is diffuse in the cytoplasm of both nonpolarized Dictyostelium amoebae and human neutrophils. Importantly, however, we find that SAHH is concentrated with F-actin at the front of chemotaxing amoebae and neutrophils, and that tubercidin, an inhibitor of SAHH, selectively impairs chemotaxis of Dictyostelium and inhibits streaming of chemotaxing Dictyostelium and chemotaxis of neutrophils. As the only known function of SAHH is to relieve the inhibition of SAM-mediated transmethylation, by hydrolysis of SAH, our results provide strong evidence for a role for SAM-dependent transmethylation during chemotaxis of eukaryotic cells.
Results
Localization of SAHH in Dictyostelium.
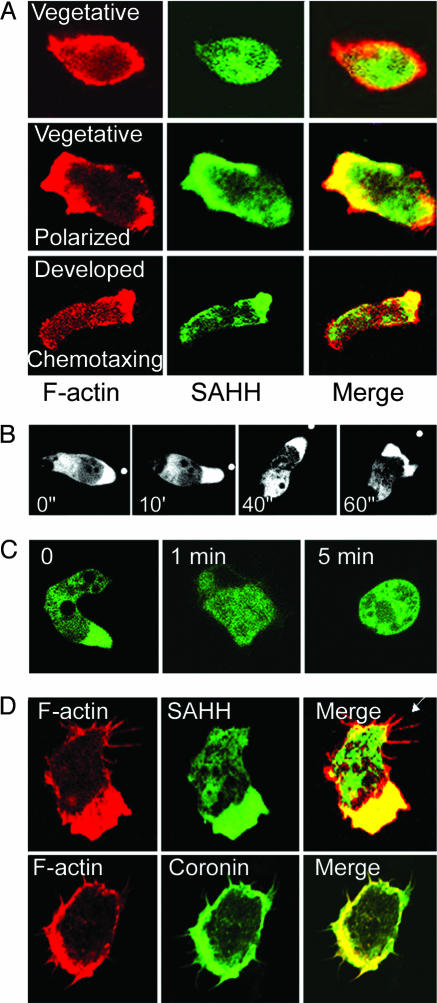
Immunolocalization of SAHH with an SAHH-specific antibody (Fig. 2) confirmed that endogenous SAHH is diffuse in the cytoplasm of vegetative amoebae, but showed localization of SAHH with F-actin in partially polarized vegetative cells, and at the front of fixed, differentiated chemotaxing cells (Fig. 3A). GFP-SAHH also localized in pseudopods at the front of living chemotaxing cells [Fig. 3B and supporting information (SI) Movie 1]. When F-actin in chemotaxing cells was depolymerized by latrunculin A, the polarized cells rounded up and SAHH rapidly diffused throughout the cytoplasm (Fig. 3C). In contrast to coronin, which also colocalizes with F-actin, SAHH is not present in filopodia or immediately under the plasma membrane at the leading edge (Fig. 3D).
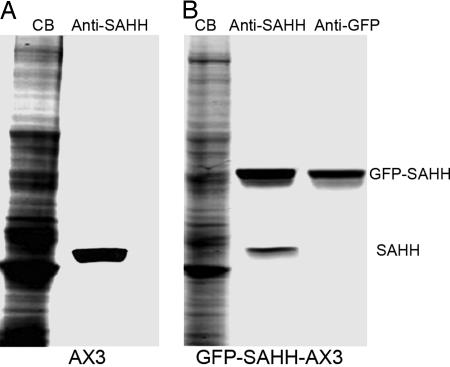
Fig. 2.
Specificity of the antibody to Dictyostelium SAHH and overexpression of GFP-SAHH in Dictyostelium. (A) Immunoblot of endogenous SAHH in an SDS/PAGE gel of total cell lysate. (B) Immunoblot of endogenous SAHH and overexpressed GFP-SAHH in a total cell lysate of cells expressing GFP-SAHH. CB, Coomassie blue.
Fig. 3.
Localization of SAHH with F-actin in pseudopods of motile Dictyostelium. (A) Localization of F-actin (rhodamine-phalloidin, red) and endogenous SAHH (anti-SAHH, green) in vegetative (Top), polarized vegetative (Middle), and developed chemotaxing (Bottom) cells. Vegetative cells are in nutrient, HL5 medium; polarized vegetative cells are motile cells in nutrient medium; and developed chemotaxing cells are cells that were pulsed for up to 8 h with cAMP in nonnutrient buffer. (B) GFP-SAHH is localized at the front of live, chemotaxing cells. White dots show the position of the tip of the micropipette filled with 1 μM cAMP. (C) GFP-SAHH becomes diffuse when F-actin in polarized cells is depolymerized by 2 μM latrunculin A. (D) Both GFP-coronin and GFP-SAHH localize with F-actin in pseudopods, but only coronin is in filopodia and immediately under the plasma membrane.
SAHH and Dictyostelium Chemotaxis.
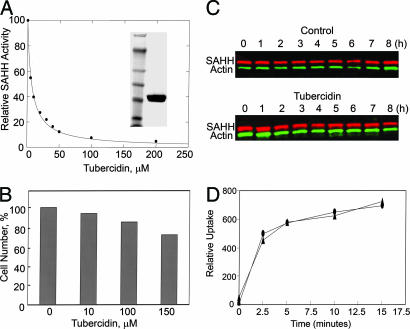
Because, thus far, all attempts to knock out or knock down SAHH have failed, we investigated the effects of tubercidin (7-deazaadenosine), an inhibitor of mammalian SAHH (15), on Dictyostelium chemotaxis. Tubercidin inhibited highly purified FLAG-tagged Dictyostelium SAHH with an IC50 of 7 μM (Fig. 4A). In vivo, 150 μM tubercidin was only mildly toxic to cell growth (Fig. 4B), did not change the level of endogenous SAHH (Fig. 4C), and did not inhibit phagocytosis of polystyrene-latex beads (Fig. 4D) or capping of Con A receptors (data not shown).
Fig. 4.
Tubercidin inhibits Dictyostelium SAHH but does not inhibit cell growth, expression of SAHH or phagocytosis. (A) Tubercidin inhibition (IC50, 7 μM) of hydrolysis of SAHH by highly purified FLAG-SAHH (uninhibited activity, 1 mol/mg per min) is shown. (Inset) SDS/PAGE of FLAG-SAHH (Right) and mass markers (Left) of 250, 148, 98, 64, 50, 36, and 22 kDa. As expected, the purified FLAG-SAHH was a tetramer by gel filtration and analytical ultracentrifugation. (B) Percent of viable cells after exposure of Dictyostelium to tubercidin for 24 h is shown. The concentration of control cells was 4 × 106 per ml. (C) Tubercidin does not inhibit SAHH expression: cells (2 × 107 per ml), in the absence and presence of 100 μM tubercidin, were pulsed with 75 μM cAMP (final concentration) at 6-min intervals and samples were taken every hour for SDS/PAGE. Gels were immunoblotted with anti-SAHH and anti-actin. (D) Phagocytosis of fluorosceine isothiocyanate-conjugated, 1-μm polystyrene-latex bead by control cells (●) and cells treated with 100 μM tubercidin (▴) is shown.
Importantly, however, although tubercidin-treated cells migrated individually toward the chemoattractant (cAMP) in the micropipette assay (Fig. 5A and SI Movies 2 and 3), the cells were less polarized, exhibited more lateral pseudopods, and moved more slowly than control cells. Moreover, in both the micropipette assay and the under-agarose assay (Fig. 5B), the cells failed to align into the head-to-tail streams characteristic of uninhibited cells. Inhibition of cell streaming was not caused by inhibition of expression of the cAMP receptor (cAR1) during the 5-h starvation period (Fig. 5C), although, interestingly, tubercidin-treated cells showed a sustained expression of cAR1 and the presence of a cAR1 doublet (the phosphorylated and unphosphorylated forms of the receptor). Tubercidin-treated cells also showed the same cAMP-mediated increase in F-actin (Fig. 5D) and the same adenylyl cyclase activity (Fig. 5E) as control cells under basal conditions and with addition of Mn2+ (a measure of nonregulated activity) or GTPγS. Taken together, these findings establish that tubercidin is not generally toxic to Dictyostelium cells and does not affect cell differentiation. Tubercidin does, however, impair chemotaxis of individual cells and cell streaming.
Fig. 5.
Tubercidin inhibits Dictyostelium cell streaming, but does not inhibit cAR1 expression, actin polymerization, or adenylyl cyclase expression. (A) Micropipette chemotaxing assay with control cells (Upper) and cells treated with 150 μM tubercidin (Lower). The black dots show the positions of the tips of micropipettes filled with 1 μM cAMP. The control cells formed long streams, whereas the tubercidin-treated cells chemotaxed individually and formed aggregates at the micropipette tip. (B) Under-agar assay of chemotaxis in which wells contained 5 μl of differentiated amoebae (2 × 107 per ml) without (Left) and with 150 μM tubercidin (Right). (Center) This well contained 1 μM cAMP. The control cells formed streams of chemotaxing amoebae, but the tubercidin-treated cells chemotaxed individually with no streams. (C) cAR1 expression: cells (2 × 107 per ml), in the absence (Upper) and presence (Lower) of 100 μM tubercidin, were pulsed with 75 μM cAMP (final concentration) at 6-min intervals. Samples were taken every hour for SDS/PAGE, and gels were stained with Coomassie blue for actin or immunoblotted with anti-cAR1 antibodies. (D) The time course of the change in F-actin content of cells after stimulation by 1 μM cAMP, in the absence or presence of 100 μM tubercidin, as determined by quantifying the staining of fixed, permeablized cells with rhodamine-phalloidin. (E) Basal and Mn2+- and GTPγS-stimulated adenylyl cyclase activity of cells in the absence (Left) or presence (Right) of 100 μM tubercidin.
SAHH and Chemotaxis in Neutrophils.
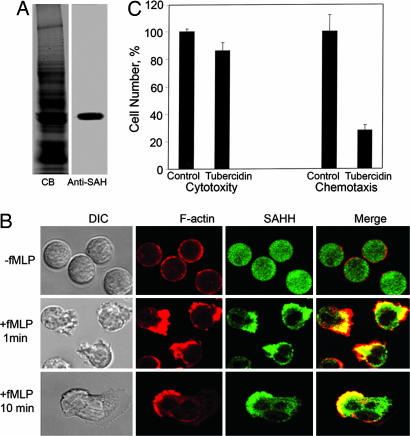
Immunolocalization with an antibody specific for human SAHH (Fig. 6A) showed that, as in Dictyostelium, SAHH is diffuse in the cytoplasm of nonpolarized neutrophils (Fig. 6B), concentrates with F-actin at the front of polarized neutrophils that are exposed to a uniform increase of the chemoattractant formyl-methionyl-leucyl-phenylalanine (fMLP) (Fig. 6B), but does not localize with F-actin immediately under the plasma membrane (Fig. 6B). Exposure of neutrophils to 100 μM tubercidin for 2 h at 37°C had only a small effect on cell viability (Fig. 6C), but strongly inhibited chemotaxis (Fig. 6C).
Fig. 6.
Localization of SAHH with F-actin at the front of chemotaxing human neutrophils and tubercidin inhibition of neutrophil chemotaxis. (A) SDS/PAGE gel of total lysate of neutrophils stained with Coomassie blue (CB; Left) and immunoblotted with anti-SAHH antibody (Right). (B) Immunolocalization of actin and SAHH in neutrophils before and after stimulation of chemotaxis with fMLP. SAHH is concentrated with F-actin in pseudopods but not immediately under the plasma membrane at the leading edge (merge). (C) Chemotaxis: 100 μM tubercidin had little effect on cell viability, but substantially inhibited chemotaxis, as measured in a modified Boyden chamber. (Left) In the cytotoxicity assay, the concentration of control cells was 2 × 107 per ml. (Right) In the chemotaxis assay, 4 × 105 cells (20% of the total) migrated from the upper to the lower chamber in the absence of tubercidin.
Discussion
We have shown that SAHH localizes at the front of chemotaxing neutrophils and Dictyostelium cells, and that tubercidin, an inhibitor of SAHH, impairs chemotaxis in both cell types. Moreover, we found that tubercidin severely inhibits streaming of Dictyostelium cells. Streams of cells are formed during chemotaxis in response to secreted cAMP signals, which have been proposed to occur at the rear of cells where adenylyl cyclase is enriched (16). Cells align in a head-to-tail fashion as the signal is propagated through neighboring cells. Interference with this signaling cascade, for example, in adenylyl cyclase null cells (17), inhibits streaming with minor effects on chemotaxis of individual cells. As we have shown that tubercidin does not affect adenylyl cyclase activity, we reason that it may block streaming by interfering with either the proper localization of adenylyl cyclase or the secretion of cAMP at the rear of polarized Dictyostelium cells. Tubercidin may have additional effects because chemotaxis of individual cells appears to be impaired more by tubercidin than in adenylyl cyclase null cells. Speculatively, chemotaxis of neutrophils, as measured in this study, could also be enhanced by similar signaling pathways that are blocked by tubercidin.
As the only known function of SAHH is to facilitate transmethylation, the unequivocal localization of SAHH with F-actin at the front of chemotaxing cells and the inhibition of chemotaxis by the SAHH-inhibitor tubercidin are strong circumstantial evidence for a role for methylation in chemotaxis in both Dictyostelium and neutrophils. However, what is methylated and by which transmethylase(s) have yet to be determined.
More than 20 years ago, it was suggested that cAMP and other chemotactic agents stimulate transient methylation of phospholipids (18) in Dictyostelium, but it was also reported (18), to the contrary, that cAMP stimulates transient demethylation of phospholipids. Similarly, carboxylmethylation of proteins was reported both to be (19–22) and not to be (23) associated with chemotaxis. Since then, the possible role of methylation in Dictyostelium chemotaxis has remained dormant.
In leukocytes, some, but not all, inhibitors of phospholipid methylation were found to inhibit fMLP-induced chemotaxis (24–28), but, paradoxically, fMLP itself was found to inhibit (not enhance) phospholipid methylation (29–31). Although fMLP has been reported to have no effect on the level of protein carboxylmethylation in leukocytes and macrophages (27, 28), except perhaps transiently (25, 26, 32, 33), some inhibitors of protein carboxylmethylation have been reported to inhibit chemotaxis (25, 26, 33), whereas others did not (26, 27). In at least one study, neither chemotactic agents nor transmethylation inhibitors affected methylation of nucleic acids (25). From these varied results, it would seem that if, as our results indicate, methylation is essential for normal chemotaxis of Dictyostelium and neutrophils, it is likely to be methylation of a specific phospholipid, protein or nucleic acid that is obscured by methylation of a larger pool that is unaffected by chemotaxis.
Recently, two distinct actin zones, with different properties and functions, have been recognized at the front of migrating epithelial cells (34): the lamellipodium, a narrow band of rapidly polymerizing and depolymerizing actin at the leading edge and in filopodia, which may serve to explore and respond to the environment, and the lamella, an overlapping larger area whose expansion may provide the driving force for movement (35). It is noteworthy that, in both Dictyostelium and neutrophils, SAHH is absent from the region that may correspond to the lamellipodium of epithelial cells, but localizes with actin in the region that may correspond to the lamella.
Because both SAHH and Tyr-53-phosphorylated actin are associated with actin bars in Dictyostelium spores, but neither is associated with F-actin in vegetative cells, it was reasonably speculated (13, 14) that SAHH may bind to Tyr-53-phosphorylated actin. However, we find that purified GFP–SAHH does not bind to either phosphorylated or unphosphorylated actin (unpublished observations), and we find no phosphorylated actin in chemotaxing Dictyostelium (36).
In summary, our findings establish that SAHH is asymmetrically distributed in polarized, chemotaxing cells. Furthermore, an inhibitor of SAHH, strongly, and specifically, impairs both chemotaxis of neutrophils and chemotaxis and chemotaxis-dependent cell streaming in Dictyostelium. Therefore, transmethylation of substrate(s) at the front of chemotaxing cells, a process that appears to be conserved throughout eukaryotic evolution, appears to be required for proper chemotaxis.
Materials and Methods
Plasmids.
Standard methods were used for synthesis of SAHH cDNA by using SAHH-specific primers based on the sequence published by Kasir et al. (12), with additional SacI and XhoI restriction sites on the ends: forward primer, GCGGAGCTCATGACTAAATTACACTACAAAGTT; reverse primer, GCGCTCGAGTTAATATCTGTAGTGATCAACTTTGTATGG. The cDNA was subcloned into pTX-FLAG and pTX-GFP vectors, which have N-terminal FLAG and GFP, respectively (37). All sequences were confirmed. The plasmids were introduced into WT Dictyostelium AX3 cells using a gene pulser (Bio-Rad, Hercules, CA) (38).
Cells.
Dictyostelium AX3 cells were grown in HL5 medium containing 60 μg/ml penicillin and streptomycin at 21°C (39), and transformed cells were selected in the presence of 12 μg/ml G418 in the same medium. Peripheral human neutrophils were isolated from heparinized whole blood as described (40), using dextran sedimentation and differential ultracentrifugation over Histopaque 1077 (Sigma/Aldrich, St. Louis, MO). Residual red blood cells were removed with three rounds of hypotonic lysis with 0.2% and 1.3% saline. Purified neutrophils were treated for 20 min with 2 mM diisoflurophosphate and kept on ice in modified Hanks' balanced salt solution (10 mg/ml glucose, 150 mM NaCl, 4 nm KCl, 1.2 mM MgCl2, 20 mM Hepes, pH 7.5, containing protease inhibitors) until use.
Electrophoresis and Immunoblots.
SDS/PAGE was conducted by standard procedures (41); neutrophil samples were ultrasonicated for 1 min to reduce their viscosity before SDS/PAGE. Gels were stained with Coomassie blue or proteins were transferred electrophoretically to nitrocellulose membranes that were immunoblotted with one or two antibodies: 10-fold diluted monoclonal SAHH antibody supernatant (Developmental Studies Hybridoma Bank, Iowa City, IA) for Dictyostelium SAHH; 1,000-fold diluted rabbit anti-GFP (AB 3080; Chemicon, Temecula, CA); 1,000-fold diluted rabbit anti-actin (A-2066; Sigma); 10-fold diluted rabbit anti-cAR1 antibody (42); 10,000-fold diluted mouse anti-human SAHH (gift from M. S. Hershfield, Duke University, Durham, NC). Secondary antibodies goat IRDye800 anti-rabbit IgG (Rockland Immunochemicals, Gilbertsville, PA) and Alexa Fluor 680 goat anti-mouse IgG (Molecular Probes, Carlsbad, CA) were used at 10,000-fold dilution and visualized with the Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE) (36).
To analyze the time course of expression of cAMP receptor, cAR1, during development, cells were washed and resuspended in developmental buffer (5 mM Na2HPO4, 5 mM KH2PO4, 2 mM MgSO4, 200 μM CaCl2, pH 6.2) at 2 × 107 cells per ml. The cells were pulsed with 75 nM cAMP (final concentration) at 6-min intervals (43), and samples were taken at 1-h intervals for SDS/PAGE.
Fluorescence Microscopy.
Indirect immunofluorescence microscopy was performed as described (44). For localization of SAHH in Dictyostelium, cells were fixed in −20°C methanol containing 1% formalin for 5 min and incubated with undiluted SAHH hybridoma antibody, followed by 750-fold diluted FITC-conjugated goat anti-mouse IgG (Molecular Probes). To compare the localization of F-actin and coronin, and F-actin and SAHH, cells expressing GFP-SAHH or GFP-coronin (gift from P. N. Devreotes, The Johns Hopkins University, Baltimore, MD) were fixed and incubated with 500-fold diluted rhodamine-phalloidin (R-415; Molecular Probes).
For localization of SAHH in neutrophils, cells in modified Hanks' balanced salt solution were plated in a chambered coverglass that had been precoated with 0.05% gelatin, incubated at room temperature for 10 min, and 10−6 M fMLP (BioChemika 47729; Sigma/Aldrich) was added to induce differentiation. Cells were fixed with 1% formaldehyde, 0.1% glutaraldehyde, and 0.01% Triton X-100 in 5 mM Na2HPO4, 5 mM NaH2PO4, pH 6.2, at room temperature for 15 min, washed three times with PBS containing 10% FBS and 0.02% sodium azide for 5 min, incubated for 60 min at 37°C with anti-human SAHH antibody diluted in the same buffer supplemented with 0.2% saponin, and washed, and 750-fold diluted FITC-conjugated goat anti-mouse IgG (Molecular Probes) was added. For colocalization of SAHH and F-actin, after staining with antibodies, the cells were incubated with 500-fold diluted rhodamine-phalloidin. Confocal microscopy was performed on a LSM-510 laser scanning fluorescence microscope (Zeiss, Thornwood, NY) equipped with a Plan apoX63 oil objective.
Tubercidin Treatment.
For Dictyostelium viability, 10–150 μM tubercidin (T-0642; Sigma) was added to cells at a concentration of 2 × 106 cells per ml, and cells were counted in a hemocytometer after 24 h. For neutrophil viability, 2 × 107 cells per ml were incubated in 100 μM tubercidin on ice for 2 h, and at 37°C for 1.5 h, and diluted 10-fold before counting. For all biological assays, Dictyostelium and neutrophils were pretreated with 150 μM tubercidin for 24 and 2 h, respectively, and kept at the same concentration of tubercidin throughout the assay. All assays were repeated at least three times.
Chemotaxis Assays.
The micropipette assay of cAMP-induced Dictyostelium chemotaxis was performed as described (45). Cells in developmental buffer were repeatedly pulsed with 75 nM cAMP (final concentration for each pulse), while shaking at 100 rpm, harvested and resuspended in phosphate buffer, and placed in a small spot on a chambered coverslip, and a chemoattractant gradient was generated with a microinjector (Eppendorf, Hamburg, Germany) connected to a micropipette filled with 1 μM cAMP. Migration was recorded at 10-s intervals with the time-lapse acquisition software of a LSM-510 and a laser scanning fluorescence microscope (Zeiss) or an inverted Axiovert S100 microscope (Zeiss) and processed with IP-Lab software. The under-agarose assay (0.5% agarose in developmental buffer containing 1 mm caffeine) of Dictyostelium chemotaxis was as described (46, 47) with cells and streams visualized with a DM IL stereoscope (Leica, Deerfield, IL).
Neutrophils were incubated on ice for 2 h at 37°C with and without 100 μM tubercidin and chemotaxis assayed as described (48). Neutrophils (100 μl, 2 × 107 cells per ml) were placed in the upper chambers of a modified Boyden chamber (Corning catalog no. 3421; Transwell, Acton, MA), and 600 μl of buffer, with or without 100 nM fMLP and/or 100 μM tubercidin, was placed in the lower chamber. After 1.5 h at 37°C, the number of neutrophils that had migrated through the filter from the upper to the lower chamber was counted in a Coulter (Fullerton, CA) counter.
Phagocytosis.
The rate of uptake of fluorosceine isothiocyanate-conjugated, 1-μm polystyrene-latex beads (Polysciences, Warrington, PA) was quantified by measuring the fluorescence intensity at excitation and emission wavelengths of 485 and 520 nm, respectively, using a LS55 luminescence spectrometer (PerkinElmer, Wellesley, MA) (49, 50).
Actin Polymerization.
The time course of actin polymerization after addition of 1 μM cAMP to differentiated cells (3 × 107 per ml) in 10 mM phosphate buffer (pH 6.2), 2 mM MgSO4, and 3 mM caffeine was determined as described (51). Cells were fixed and stained in buffer containing 3.7% formaldehyde, 0.15 Triton X-100, 250 nM TRITC-phalloidin, 20 mM potassium phosphate, 10 mM Pipes, 5 mM EGTA, and 2 mM MgCl2, pH 6.8. F-actin was quantified by measuring TRITC-phalloidin fluorescence with a LS55 luminescence spectrometer.
Purification of SAHH.
Dictyostelium FLAG-SAHH was purified by FLAG-affinity chromatography on an anti-FLAG antibody-conjugated resin (Sigma) eluted with 0.1 mg/ml FLAG-peptide (EZ Biolab, Inc., Westfield, IN) (52).
Enzyme Assays.
SAHH activity was assayed by the rate of homocysteine production determined by the conversion of dinitrothionitrobenzoic acid to thionitrobenzoic acid (53). The data were fit to zero-order kinetics by using an extinction coefficient of 13,600 M−1·cm−1 for thionitrobenzoic acid. Adenylyl cyclase activity was stimulated in Dictyostelium cells with pulses of 75 nM cAMP (final concentration after each pulse), and adenylyl cyclase activity was assayed (54) by the conversion of [α-32P]ATP to radiolabeled cAMP purified by sequential chromatography over Dowex 50W X-4 (Dow Chemical, Midland, MI) and alumina columns (55).
Supplementary Material
Acknowledgments
We thank Dr. M. S. Hershfield for anti-human SAHH antibodies; Dr. T. T. Egelhoff (Case Western Reserve University, Cleveland, OH) for GFP and FLAG plasmids; Dr. P. N. Devreotes for Dictyostelium expressing GFP-coronin; Drs. G. Piszczek and A. Ginsburg (National Heart, Lung, and Blood Institute) for analytical ultracentrifugal characterization of FLAG-SAHH; Dr. A. Peterkofsky (National Heart, Lung, and Blood Institute) for gel filtration chromatography; Drs. X. Wu (National Heart, Lung, and Blood Institute), A. Bagorda, F. I. Comer, and P. Kriebel for assistance in microscopy; and the National Institutes of Health Department of Transfusion Medicine, and the many blood donors, for the neutrophils. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Abbreviations
- fMLP
formyl-methionyl-leucyl-phenylalanine
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- SAHH
SAH hydrolase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609385103/DC1.
References
- 1.Gibson KD, Wilson JD, Udenfriend S. J Biol Chem. 1961;236:673–679. [PubMed] [Google Scholar]
- 2.Duerre JA. J Biol Chem. 1962;237:3737–3741. [Google Scholar]
- 3.de La Haba G, Cantoni GL. J Biol Chem. 1959;234:603–608. [PubMed] [Google Scholar]
- 4.Kort EN, Goy MF, Larsen SH, Adler J. Proc Natl Acad Sci USA. 1975;72:3939–3943. doi: 10.1073/pnas.72.10.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Springer WR, Koshland DE., Jr Proc Natl Acad Sci USA. 1977;74:533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parent CA, Devreotes PN. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 7.Chung CY, Funamoto S, Firtel RA. Trends Biochem Sci. 2001;26:557–566. doi: 10.1016/s0968-0004(01)01934-x. [DOI] [PubMed] [Google Scholar]
- 8.Kimmel AR, Parent CA. Science. 2003;300:1525–1527. doi: 10.1126/science.1085439. [DOI] [PubMed] [Google Scholar]
- 9.Parent CA. Curr Opin Cell Biol. 2004;16:4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Hohman RJ, Guitton MC, Véron M. Arch Biochem Biophys Acta. 1984;233:785–795. doi: 10.1016/0003-9861(84)90507-1. [DOI] [PubMed] [Google Scholar]
- 11.Guitton MC, Part D, Véron M. Biochimie. 1988;70:835–840. doi: 10.1016/0300-9084(88)90115-0. [DOI] [PubMed] [Google Scholar]
- 12.Kasir J, Askanit RR, Backland PS, Jr, Cantoni GL. Biochem Biophys Res Commun. 1988;153:359–364. doi: 10.1016/s0006-291x(88)81231-2. [DOI] [PubMed] [Google Scholar]
- 13.Kishi Y, Sugo T, Mahadeo D, Cotter D, Sameshima M. FEBS Lett. 2001;508:433–437. doi: 10.1016/s0014-5793(01)03105-2. [DOI] [PubMed] [Google Scholar]
- 14.Yamada Y, Sameshima M. FEMS Microbiol Lett. 2004;238:417–422. doi: 10.1016/j.femsle.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Fabianowska-Majewska K, Duley JA, Simmonds HA. Biochem Pharmacol. 1994;48:897–903. doi: 10.1016/0006-2952(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 16.Mahadeo DC, Parent CA. Curr Top Dev Biol. 2006;73:115–140. doi: 10.1016/S0070-2153(05)73004-0. [DOI] [PubMed] [Google Scholar]
- 17.Kriebel PW, Barr VA, Parent CA. Cell. 2003;112:549–560. doi: 10.1016/s0092-8674(03)00081-3. [DOI] [PubMed] [Google Scholar]
- 18.Alemany S, Gil MG, Mato JM. Proc Natl Acad Sci USA. 1980;77:6996–6999. doi: 10.1073/pnas.77.12.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mato JM, Marín-Cao D. Proc Natl Acad Sci USA. 1979;76:6106–6109. doi: 10.1073/pnas.76.12.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Waarde A. FEBS Lett. 1982;149:266–270. doi: 10.1016/0014-5793(82)81114-9. [DOI] [PubMed] [Google Scholar]
- 21.van Waarde A. FEBS Lett. 1983;161:45–50. [Google Scholar]
- 22.van Waade A, van Haastert PJ. J Bacteriol. 1984;157:368–374. doi: 10.1128/jb.157.2.368-374.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Waarde A, van Hoof PJM. Biochim Biophys Acta. 1985;840:344–354. [Google Scholar]
- 24.Aksamit RR, Falk W, Cantoni GL. J Biol Chem. 1982;257:621–625. [PubMed] [Google Scholar]
- 25.Bareis DL, Hirata F, Schiffman E, Axelrod J. J Cell Biol. 1982;93:690–697. doi: 10.1083/jcb.93.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carcia-Castro I, Mato JM, Vasanthakumar G, Weismann WP. J Biol Chem. 1983;258:4345–4349. [PubMed] [Google Scholar]
- 27.Askamit RR, Backlund PS, Jr, Cantoni GL. J Biol Chem. 1983;258:20–23. [PubMed] [Google Scholar]
- 28.Askamit RR, Backlund PS., Jr Surv Immunol Res. 1983;2:150–154. doi: 10.1007/BF02918574. [DOI] [PubMed] [Google Scholar]
- 29.Hirata F, Corcoran BA, Venkatsubramanian K, Schiffman E, Axelrod J. Proc Natl Acad Sci USA. 1979;76:2640–2643. doi: 10.1073/pnas.76.6.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pike MC, Kredich NM, Snyderman R. Proc Natl Acad Sci USA. 1979;76:2922–2926. doi: 10.1073/pnas.76.6.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyderman R, Pike MC, Kredich NM. Mol Immunol. 1980;17:209–218. doi: 10.1016/0161-5890(80)90073-5. [DOI] [PubMed] [Google Scholar]
- 32.O'Dea RF, Viveros OH, Axelrod J, Aswanikumar S, Schiffman E, Corcoran BA. Nature. 1978;272:462–464. doi: 10.1038/272462a0. [DOI] [PubMed] [Google Scholar]
- 33.Pike MC, Kredich NM, Snyderman R. Proc Natl Acad Sci USA. 1978;75:3928–3932. doi: 10.1073/pnas.75.8.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 35.Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, Waterman-Storer CM. J Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Shu S, Hong MS, Levine RL, Korn ED. Proc Natl Acad Sci USA. 2006;103:13694–13699. doi: 10.1073/pnas.0606321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levi S, Polakov M, Egelhoff TT. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- 38.Egelhoff TT, Lee RJ, Spudich JA. J Cell Biol. 1991;112:677–688. doi: 10.1083/jcb.112.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sussman S. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- 40.Boyum A. Tissue Antigens. 1974;4:269–274. [PubMed] [Google Scholar]
- 41.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 42.Kim JY, Borleis JA, Devreotes PN. Dev Biol. 1998;197:117–128. doi: 10.1006/dbio.1998.8882. [DOI] [PubMed] [Google Scholar]
- 43.Devreotes PN, Fontana D, Klein P, Sherring J, Theibert A. Methods Cell Biol. 1987;28:299–331. doi: 10.1016/s0091-679x(08)61653-2. [DOI] [PubMed] [Google Scholar]
- 44.Shu S, Liu X, Korn ED. Proc Natl Acad Sci USA. 2005;95:1472–1477. doi: 10.1073/pnas.0409528102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 46.Nelson E, Quie P, Simmons R. J Immunol. 1975;115:1650–1656. [PubMed] [Google Scholar]
- 47.Comer FI, Lippincott CK, Masbad JJ, Parent CA. Curr Biol. 2005;15:134–139. doi: 10.1016/j.cub.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 48.Neptune ER, Bourne HR. Proc Natl Acad Sci USA. 1997;94:14489–14494. doi: 10.1073/pnas.94.26.14489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shu S, Liu X, Parent CA, Uyeda TQP, Korn ED. J Cell Sci. 2002;115:4237–4249. doi: 10.1242/jcs.00112. [DOI] [PubMed] [Google Scholar]
- 50.Maniak M, Rauchenberger R, Albrecht R, Murphy J, Gerisch G. Cell. 1995;83:915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- 51.Chen I, Janetopoulos C, Huang YE, Iijima M, Devreotes PN. Mol Biol Cell. 2003;14:5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shu S, Liu X, Korn ED. Proc Natl Acad Sci USA. 2003;100:6499–6504. doi: 10.1073/pnas.0732155100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan CS, Ault-Riche DB, Borchardt RT. J Biol Chem. 1996;271:28009–28016. doi: 10.1074/jbc.271.45.28009. [DOI] [PubMed] [Google Scholar]
- 54.Parent CA, Devreotes PN. J Biol Chem. 1995;270:22693–22696. doi: 10.1074/jbc.270.39.22693. [DOI] [PubMed] [Google Scholar]
- 55.Salomon Y. Adv Cyclic Nucleotide Res. 1979;10:35–55. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.