Abstract
Allosteric regulation is a fundamental mechanism of biological control. Here, we investigated the allosteric mechanism by which GTP inhibits cross-linking activity of transglutaminase 2 (TG2), a multifunctional protein, with postulated roles in receptor signaling, extracellular matrix assembly, and apoptosis. Our findings indicate that at least two components are involved in functionally coupling the allosteric site and active center of TG2, namely (i) GTP binding to mask a conformationally destabilizing switch residue, Arg-579, and to facilitate interdomain interactions that promote adoption of a compact, catalytically inactive conformation and (ii) stabilization of the inactive conformation by an uncommon H bond between a cysteine (Cys-277, an active center residue) and a tyrosine (Tyr-516, a residue located on a loop of the β-barrel 1 domain that harbors the GTP-binding site). Although not essential for GTP-mediated inhibition of cross-linking, this H bond enhances the rate of formation of the inactive conformer.
Keywords: protein conformation, GTP inhibition, transamidase activity
Allosteric regulation of enzymes by conformational change is an important means of biological control, involving residues that functionally couple ligand binding at the allosteric site to modification of the catalytic site. Transglutaminase type 2 (TG2), also known as tissue TG or Gh (high molecular weight GTP-binding protein), is a multifunctional protein that is allosterically regulated by calcium and GTP (1). TG2 catalyzes calcium-dependent transamidation reactions, resulting in posttranslational amine modification of proteins or cross-linking of interchain glutamine and lysine residues to form Nε(γ-glutamyl)lysine isopeptide bonds, which confer rigidity and protease resistance on protein complexes (2). TG2 is also a GTPase (3) and mediates intracellular signaling by various G protein-coupled receptors (4–6).
GDP-bound human TG2 (7) is comprised of four domains: an N-terminal β-sandwich, a core domain in which the transamidase active site catalytic triad (Cys-277, His-335, and Asp-358) and transition-state stabilizing residue (Trp-241) (8) are buried and inaccessible to substrate, and two β-barrels. Nucleotide binds mainly to residues from the first and last strands (amino acids 476–482 and 580–583) of β-barrel 1 and to two core domain residues (Lys-173 and Phe-174) that protrude on a loop to meet β-barrel 1 (7, 9, 10). This is postulated to stabilize two β-barrel 1 loops that block access to the catalytic site (7). One of these loops protrudes into the core domain localizing Tyr-516 within hydrogen-bonding distance of Cys-277 (7). This is postulated to prevent Cys-277 interaction with the substrate (7, 11, 12). Calcium-activated TG2 has unique conformational epitopes (13) and is less compact (14–16) and less resistant to protease digestion (1, 10, 14) than GTP-bound TG2. Allosteric mechanisms governing the conformational switch between transamidase and GTPase functions have yet to be elucidated.
Mutation of Arg-579 in rat TG2 (Arg-580 in human TG2) to Ala reduces GTP affinity by 100-fold and uncouples GTP-dependent allosteric inhibition of transamidase activity, resulting in dysregulated intracellular cross-linking (10). R579A is also more resistant to μ-calpain digestion than wild-type TG2 (WT) in the absence of GTP (10), suggesting an important conformational role for this residue. Here, we show that Arg-579 is a conformationally destabilizing residue that is masked by GTP binding. Further, we provide evidence that Tyr-516 is not directly involved in transamidase catalysis but, by forming a H bond with Cys-277, stabilizes and enhances the rate of conversion to the compact, transamidase-inactive conformation. Thus, Arg-579 and Tyr-516 form an important part of the allosteric mechanism that functionally couples GTP binding to inhibition of cross-linking at the distal TG2 active site.
Results
GTP Stabilizes a Compact Conformation of TG2.
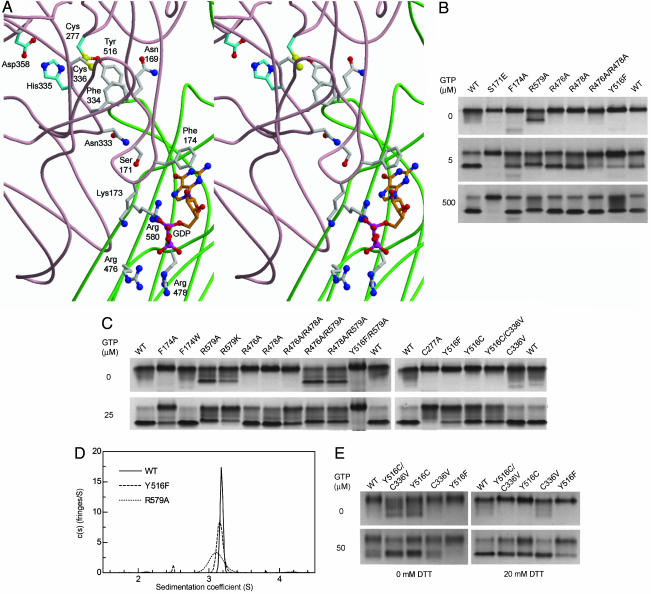
As shown by agarose gel electrophoresis (17), the addition of GTP caused a shift in WT mobility in native PAGE (nPAGE) to a faster-migrating species (Fig. 1B) that could be specifically labeled with [α-32P]GTP (data not shown). Point mutants of core (Ser-171 and Phe-174) and barrel 1 (Arg-476, Arg-478, and Arg-579) residues (Fig. 1A) showed GTP-dependent shifts in mobility (Fig. 1 B and C) in order from most to least sensitive: WT≈F174W > R476A≈R478A > R476A/R478A≈F174A > R579A≈R579K > S171E. This order is consistent with their relative reduction in GTP photolabeling and GTP-dependent inhibition of transamidase activity relative to WT (9, 10).
Fig. 1.
Mutations of residues involved in GTP-binding or the Cys-277–Tyr-516 H bond affect TG2 conformation. (A) Model of GDP-bound human TG2 showing the transamidase active center (Cys-277, His-335, and Asp-358) and other key residues [(Protein Data Bank ID code 1KV3) (7); core domain backbone, pale pink; β-barrel 1 domain, green]. Carbon atoms are gray, except those of active site residues (cyan), and GDP (orange). Other atoms include oxygen, red; nitrogen, deep blue; sulfur, yellow; phosphorous, bright pink. Arg-580 in human TG2 is equivalent to Arg-579 in rat TG2. (B and C) Samples (3 μg) of WT and mutant TG2 were incubated (3 h at 25°C) in buffer (50 mM Tris·HCl, pH 7.5/50 mM NaCl/0.5 mM EDTA/1.0 mM MgCl2/5 mM DTT/GTP as indicated). nPAGE was performed with 0.5 mM MgCl2, and the indicated GTP concentrations were included in gel solutions and running buffer. Data are representative of two to eight experiments. (D) Sedimentation coefficient distributions, cM(s), for 13 μM WT (solid line), Y516F (dashed line), and R579A (dotted line) in 10 mM Tris·HCl pH 7.5/100 mM NaCl/0.5 mM EDTA/1 mM DTT/500 μM GTP at 4°C. Molar mass was fixed at 78.0 kDa for 2.5–3.8 S species. Data are representative of three to four experiments. (E) Disulfide trapping of Cys-277 with a cysteine substituted for Tyr-516. Samples (3 μg) of WT and mutant TG2 were oxidized (2 h at 25°C) in buffer (50 mM Tris·HCl, pH 7.5/50 mM NaCl/200:800 μM Cu2+:phenanthroline/50 μM GTP), then incubated (1 h at 25°C) in 0.5 mM MgCl2, without or with 20 mM DTT, as indicated. nPAGE was performed as in B. Data are representative of two experiments.
Arg-579 Destabilizes the Compact TG2 Conformation.
Independent of GTP addition, R579A migrated as a fast-migrating species (Fig. 1 B and C) and had a dominant effect on migration when combined with R476A or R478A in the double mutants, R476A/R579A and R478A/R579A (Fig. 1C). A similar migration pattern was observed for R579K, which retains the charge and much of the bulk of the arginine side chain, but like R579A is impaired in GTP binding (10). Of note, the fast-migrating species observed for Arg-579 mutants migrated slightly more slowly than GTP-bound WT or other mutants.
The homogeneity and approximate shape of GTP-bound WT and R579A was evaluated in sedimentation velocity experiments at 4°C [see supporting information (SI) Fig. 6]. The accuracy of the molar mass estimate (Table 1) of the major GTP-bound WT species (Fig. 1D) (within 4% of the calculated molar mass of 78.0 kDa for rat TG2 with one GTP bound) indicates GTP-bound TG2 adopts a stable conformation. Similar results, i.e., s20,w of 4.94 S and a molar mass of 74.8 kDa, were obtained for WT in 500 μM GTP at 24°C, indicating the GTP-bound conformation is stable at higher temperatures. R579A consistently sedimented as a single species in the presence of GTP and at a slightly slower rate than GTP-bound WT (Fig. 1D and Table 1). This difference was not due to incomplete saturation of R579A with GTP as a 10-fold increase in GTP concentration did not affect the R579A s value (Table 1). The compact R579A conformation, therefore, is slightly more asymmetric or expanded than GTP-bound WT (dimensions of equivalent prolate ellipsoids are compared in Table 1). The fast-migrating species displayed by Arg-579 mutants (Fig. 1 B and C), thus, is not due to charge but most likely to GTP-independent isomerization to a semicompact conformation. This indicates that Arg-579 destabilizes the compact TG2 conformation in the absence of GTP.
Table 1.
Hydrodynamic size of TG2 at 4°C
| TG2 | s20,w, S | Molar mass,* kDa | Frictional ratio† | Prolate ellipsoid,‡ nm |
|---|---|---|---|---|
| WT (500 μM GTP) | 4.93 | 81.1 (80.5, 82.0) | 1.33 | 15.5 × 4.3 |
| Y516F (500 μM GTP) | 4.90 | 76.5 (76.0, 77.0) | 1.34 | 15.9 × 4.3 |
| R579A (500 μM GTP) | 4.82 | 80.1 (79.0, 82.0) | 1.36 | 16.6 × 4.2 |
| R579A (5 mM GTP) | 4.78 | 82.0 (78.0, 88.0) | 1.37 | 17.1 × 4.1 |
*Data fitted with hybrid discrete/continuous model. Confidence limits (parentheses) were calculated by using F statistics at confidence level of 1 SD.
†Calculated from sequence molar mass and s by using SEDNTERP.
‡Two-fold major and minor axis, calculated assuming hydration of 0.402 g/g, as predicted from amino acid composition (38) by using SEDNTERP.
The Cys-277–Tyr-516 H Bond Stabilizes the Compact TG2 Conformation.
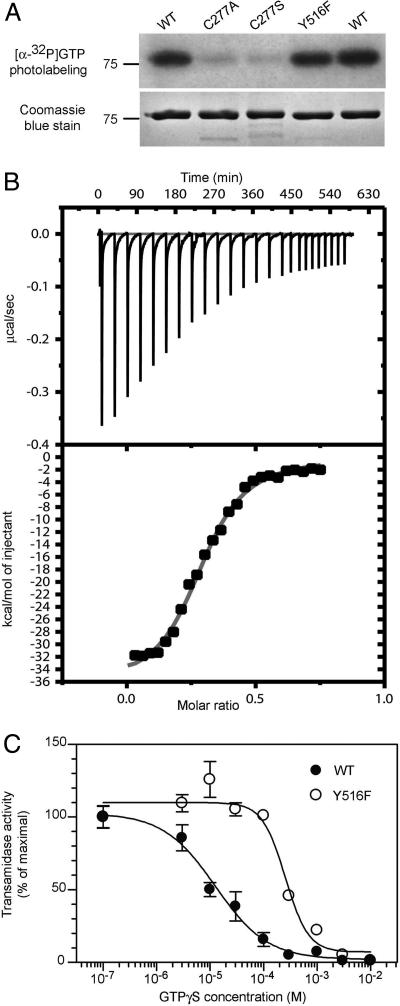
The electronegative group required for the Cys-277–Tyr-516 H bond was removed by substitution of Cys-277 with Ala or Ser or Tyr-516 with Phe. Transamidase-inactive (18) C277A (Fig. 1C) and C277S (data not shown) showed markedly weaker GTP-dependent shifts to fast-migrating species and weaker [α-32P]GTP photolabeling (Fig. 2A) compared with WT, consistent with our previous finding (19) that their affinity for 2′-(or 3′)-O-(N-methylanthraniloyl) GTP is greatly reduced. Y516F showed an impaired GTP-dependent shift to fast-migrating species (Fig. 1 B and C) and eliminated the fast-migrating species seen with R579A mutants when combined with R579A in the double mutant, Y516F/R579A (Fig. 1C). Surprisingly, Y516F photolabeled (Fig. 2A) and bound GTPγS (Fig. 2B; Kd = 0.8 ± 0.08 μM), equivalent to WT (Kd = 1.6 ± 0.09 μM; ref. 10). Thus, unlike Cys-277 mutants, decreased stability of the compact Y516F form is not due to weaker GTP binding. Isothermal titration calorimetry of WT, F174A, and R579A showed sharp peaks indicating rapid binding (10). However, GTPγS titration of Y516F (Fig. 2B) indicated a slow interaction process, with heat liberated after each injection taking >30 min to reach baseline. This indicates either slow interconversion of Y516F to a conformation competent to bind GTP, or a rapid, low-affinity interaction with GTP followed by a slow conformational change to high-affinity binding (20).
Fig. 2.
Y516F mutation does not reduce GTP affinity but weakens GTP inhibition of transamidase activity. (A) WT or mutant TG2 (1.3 μM) were photoaffinity-labeled with [α-32P]GTP and size fractionated by SDS/PAGE. Radiolabeling was detected by autoradiography (Upper), and proteins were visualized with Coomassie blue (Lower). Data are representative of two experiments. (B) Isothermal titration calorimetry of GTPγS binding to Y516F. GTPγS (150 μM) was titrated into solutions of 29 μM Y516F. (B Upper) Raw data from injections. (B Lower) Peaks were integrated to yield injection-associated heat change and buffer control was subtracted. Data are representative of three experiments. (C) GTPγS inhibition of transamidase activity was assayed at ≈20% maximal transamidase activity. Activities as a percentage of maximal were 25% for WT and 22% for Y516F at 50 μM free Ca2+. Data are means of triplicate determinations ± SE and are representative of two experiments.
GTP-bound Y516F sedimented as a major species (Fig. 1D) with a very slight decrease in s value relative to WT (Table 1), indicating that in contrast to R579A, compact GTP-bound Y516F is not measurably different from WT. This is consistent with equal migration of GTP-bound forms of WT and Y516F (Fig. 1 B and C). The minor peak at ≈2.5 S (Fig. 1D) suggests an expanded Y516F form, consistent with decreased stability of the compact form (Fig. 1 B and C).
The Cys-277–Tyr-516 Interaction Enhances Allosteric Inhibition of TG2 by GTP.
Y516F showed similar dose-dependent calcium-stimulated transamidase activity to WT (see SI Fig. 7), and the same Km for the glutaminyl substrate, dimethyl casein (0.095% versus 0.093% wt/vol for WT), indicating that Tyr-516 is not involved in substrate binding or catalysis. GTPγS inhibition of transamidase activity, evaluated at ≈20% maximal activity (Fig. 2C), resulted in a 20-fold higher IC50 for Y516F (255 μM compared with 13 μM for WT), demonstrating that the loss of the Cys-277–Tyr-516 H bond substantially weakens, but does not prevent, GTP inhibition of transamidase activity. Together, this data indicate that transition to the compact, transamidase-inactive form is enhanced by GTP binding and that the Cys-277–Tyr-516 H bond is important for conformer stability.
A Disulfide Bond Between Cys-277 and a Cys Substituted for Tyr-516 Stabilizes the Compact TG2 Conformation.
Cys was substituted for Tyr-516 to evaluate whether oxidative trapping of Y516C in a disulfide bond with Cys-277 can mimic the native Cys–Tyr H bond to stabilize the compact form. Cys-336, which is within bonding distance of Cys-277 (Fig. 1A), was mutated to Val (the equivalent residue in TG3), either alone (control) or in combination with Y516C, to eliminate the possibility of a competing Cys-277–Cys-336 disulfide bond (12). Maximal calcium-stimulated transamidase activities of Y516C, C336V, and Y516C/C336V were 76%, 83% and 71% of WT, respectively.
To maximize disulfide bond formation between Cys-277 and Cys-substituted Tyr-516, proteins were oxidized in the presence of GTP. After GTP removal by nPAGE (Fig. 1E Upper Left), Y516C and Y516C/C336V, but not WT, C336V, or Y516F, displayed a range of fast-migrating species that were eliminated by DTT reduction before nPAGE (Fig. 1E Upper Right), demonstrating stabilization of these species by a disulfide bond, likely between Cys-277 and Cys-516. Oxidation, followed by nPAGE in the presence of GTP (Fig. 1E Lower Left), showed a marked increase in compact GTP-bound Y516C and Y516C/C336V relative to WT and C336V, indicating Cys-277–Cys-516-mediated stabilization of the fully compact GTP-bound conformation. Oxidation followed by DTT reduction before nPAGE in the presence of GTP (Fig. 1E Lower Right) completely reversed the effects of oxidation (compare to untreated mutants in Fig. 1C). The increase in the relative proportion of compact WT, C336V, and Y516F forms (Fig. 1E, Lower Right compared with Lower Left) indicates that this form is more stable under reducing conditions. In contrast, stability of the compact form of Y516C mutants already was maximized under oxidizing conditions, likely due to the Cys-277–Cys-516 disulfide bond.
Transition Between GTP-Free and GTP-Bound Forms Is Faster for R579A.
Undiluted samples of WT, R579A, and Y516F (Fig. 3) showed a major GTP-bound species of ≈3.16 S for WT and Y516F (equivalent to s20,w ≈4.9 S) and ≈3.1 S for R579A (equivalent to s20,w ≈4.8 S). The 3.9 S R579A species is likely a dimer. Upon sample dilution, the proportion of GTP-bound species decreased and species at 0.5–1.5 S and 1.5–2.5 S increased. Free GTP sediments predominantly at <0.25 S (data not shown). Species at 1.5–2.5 S have a frictional ratio of ≈1.9 and are equivalent to a prolate ellipsoid with a long axis (36 nm) approximately double that of GTP-bound TG2 (Table 1). These expanded form(s) may be similar to calcium-bound forms described in a Monte Carlo simulation of small-angle x-ray scattering data (16), where the β-barrels rotated up to 130° away from the core domain. The 0.5–1.5 S species are extremely expanded/asymmetric, equivalent to a prolate ellipsoid with an axial ratio of >30:1 or a cylinder with a length-to-diameter ratio >10.2, and may be partially unfolded. This may point to an important role for GTP in stabilizing TG2 tertiary structure.
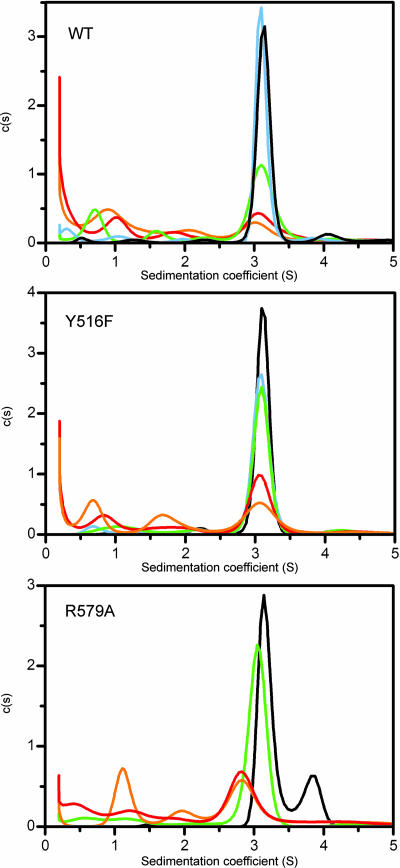
Fig. 3.
GTP-bound TG2 is in a slow reversible equilibrium with multiple GTP-free conformers. Sedimentation coefficient distributions c(s) of WT (Top), Y516F (Middle), and R579A (Bottom) TG2 with equimolar GTPγS in 20 mM Tris·HCl, pH 7.5/100 mM NaCl/0.5 mM EDTA/1 mM MgCl2/2 mM DTT at 4°C: black, undiluted (≈25 μM) TG2/GTP; blue, 3-fold dilution; green, 10-fold dilution; red, 33-fold dilution; orange, 100-fold dilution. Distributions, calculated by using 300 s values between 0.2 and 15 S, are shown normalized for unit area. Data are representative of two experiments.
The absence of a concentration-dependent shift of the ≈3.2 S peak upon WT and Y516F dilution (Fig. 3) indicates a very slow exchange between this and slower sedimenting species at 4°C (koff ≈10−4/s or slower) (21). This is consistent with a significant GTP-dependent conformational change requiring a relatively large activation energy. In contrast, R579A dilution showed progressive decrease of the main boundary from 3.1 S to an endpoint (observed with 33- and 100-fold dilution) of 2.8 S. This indicates a reaction on the time scale of the experiment and likely reflects faster GTP dissociation from R579A, consistent with its lower GTP affinity (10). Moreover, GTP dissociation is accompanied by conformational change, because reduction from 3.1 to 2.8 S is much greater than expected from loss of one GTP molecule, using the 2/3 rule (s ≈ M2/3). The semicompact form of R579A mutants (Fig. 1 B and C) may correspond to a rapidly exchanging mixture of GTP-free compact (≈2.8 S) and GTP-bound compact (≈3.1 S) R579A species (Fig. 4).
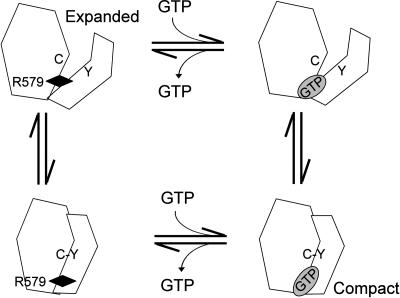
Fig. 4.
Model of allosteric inhibition of TG2 by GTP. Transition from expanded, GTP-free TG2 (upper left) to compact, GTP-bound TG2 (lower right) may occur via an induction mechanism (clockwise) in which GTP binding to the destabilizing Arg-579 (black diamond) triggers a conformational change to the compact form, which is stabilized by Cys-277–Tyr-516 interaction, or a selection mechanism (counterclockwise) in which an unstable conformational intermediate facilitated by Cys-277–Tyr-516 interaction (GTP-free compact, lower left) is stabilized by GTP binding. Removal of the destabilizing Arg-579 by substitution stabilizes the GTP-free compact form in Arg-579 mutants.
The overall weight average for all R579A species (excluding dimer) decreased to 2.5 S upon 100-fold dilution, compared with 2.3 S for WT and 2.1 S for Y516F (see SI Fig. 8). This suggests, in agreement with nPAGE migration (Fig. 1B), that compact R579A forms are more stable at low GTP than those of WT, whereas compact Y516F forms are less stable.
Discussion
We provide evidence that Arg-579 is critical for TG2 transition to the compact, catalytically inactive conformation and, thus, functions as a conformational switch residue that is essential for allosteric regulation of transamidase activity by guanine nucleotide. Substitution with Ala or Lys removes the destabilizing effect of Arg-579 and allows TG2 to adopt a GTP-independent semicompact conformation (Fig. 1 B–D) that is resistant to allosteric inhibition by GTP and results in dysregulated intracellular transamidase activity (10). Binding of GTP by Arg-579 is required for TG2 to adopt the most compact, transamidase-inactive, and protease-resistant (10) conformation. Association of the allosteric ligand, GTP, with TG2 likely promotes the compact conformation by (i) neutralizing the destabilizing effect of Arg-579 and (ii) facilitating core and β-barrel 1 domain interaction.
We further demonstrate that allosteric regulation of TG2 by GTP involves a stabilizing H bond between Cys-277 and Tyr-516. Substitution of Cys-277 with Ala or Ser weakened both GTP binding (Fig. 2A) and compact TG2 stability (Fig. 1C), reinforcing the importance of GTP binding for compact conformer stabilization. Tyr-516 substitution, in contrast, did not affect GTP affinity (Fig. 2 A and B). Despite this, GTP inhibition of Y516F transamidase activity was impaired (Fig. 2C), and, like Cys-277 mutants, the compact GTP-bound conformation of Tyr-516 mutants was less stable than WT (Fig. 1 B and C). Disulfide trapping of Cys-277 with Cys-substituted Tyr-516 stabilized compact TG2 conformations, including fully compact GTP-bound TG2 (Fig. 1E). Together, this demonstrates that inhibition of transamidase activity requires GTP binding, but not the Cys-277–Tyr-516 H bond per se. The Cys-277–Tyr-516 interaction, however, increases the stability of the compact TG2 form and enhances GTP inhibition of transamidase activity.
Slow interconversion between WT and Y516F GTP-free and GTP-bound conformations indicates a significant GTP-dependent conformational change (Fig. 3). The required activation energy is greater for Y516F (Fig. 2B), indicating an important role for the Cys-277–Tyr-516 interaction in decreasing the activation barrier by stabilizing one or more conformational intermediates. Consistent with this, combining Y516F with R579A in the double mutant Y516F/R579A abolished the semicompact conformation of Arg-579 mutants (Fig. 1C). Together, this indicates that although GTP binding is necessary and sufficient to stabilize the compact, transamidase-inactive TG2 conformation, the Cys-277–Tyr-516 interaction greatly increases the rate of conformational transition to the transamidase-inactive form.
The conformational effect of Arg-579, Cys-277, and Tyr-516 mutations may be articulated through an allosteric communication network (Fig. 1A), whereby GTP binding to TG2 neutralizes the destabilizing effect of Arg-579 and anchors β-barrel 1 to the core domain GTP-binding loop. This loop forms part of a highly conserved interface (19) with a β-strand of the core domain that not only places the active site His-335 near Cys-277, but also interacts with Tyr-516. Aromatic stacking between side chains of Phe-334 and Tyr-516, together with backbone H bonding of Tyr-516 with Phe-334 and Asn-333, likely promotes Cys-277–Tyr-516 H bond formation and, consequently, stabilization of the compact TG2 form. It remains unclear whether transition of the expanded, GTP-free TG2 form to the compact, GTP-bound conformation involves binding of GTP to Arg-579, triggering isomerization and Cys-277–Tyr-516-facilitated stabilization of the compact form or, alternatively, Cys-277–Tyr-516-facilitated isomerization followed by GTP binding to the destabilizing Arg-579 (Fig. 4).
GDP-bound TG2 (7) and nucleotide-bound [GTPγS and GDP (22) or GMP (23)] TG3 are all very similar to nucleotide-free, calcium-activated TG3 (24), with tertiary structure superposition showing that Cys-277 in TG2 (Cys-272 in TG3) is within H bonding distance of Tyr-516 in TG2 (Tyr-525 in TG3). Arg-587 in the TG3 nucleotide-binding site (equivalent to Arg-579 in rat TG2) is occupied by β-octyl glucoside in calcium-activated TG3. It is tempting to speculate that Arg-587 also may be destabilizing in TG3 and that nucleotide or β-octyl glucoside binding to mask Arg-587 promotes crystallization in the compact conformation.
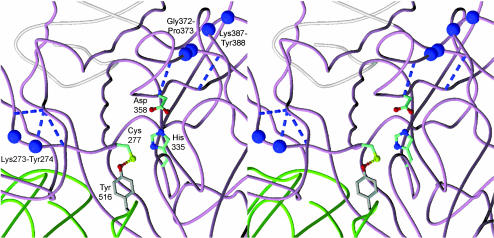
Robust transamidase activity of Y516F and Y516C (SI Fig. 7 and Results) is contrary to recent molecular modeling data that suggest Tyr-516 functions in transition-state stabilization (25) but is entirely consistent with our earlier work demonstrating that Trp-241 is a transition-state stabilizing residue (8). In addition, calcium dependence of Y516F transamidase activity shows that loss of the Cys-277–Tyr-516 bond, and destabilization of the compact form alone, is not sufficient to activate transamidase activity; rather, calcium binding is required. Molecular details of this activation are as yet unknown, however, active site access to substrate would disrupt ≈3,000 Å2 of buried surface area between the core and β-barrel 1 (26). Cis/trans isomerization of three energetically unfavorable cis-peptide bonds (two of which are nonproline), found in all transglutaminases crystallized to date, may reduce the estimated 11 kcal/mol of binding energy (26). A schematic HERA diagram (27) of TG2 secondary structure (data not shown) indicates that two of the cis-peptide bonds (Lys-273-Tyr-274 and Gly-372-Pro-373) are components of β-bulges (28), important determinants of protein stability and function (29) that have been associated with conformational isomerization (30). Gly-372-Pro-373 is in the β-sheet containing the active site Asp-358 and is close to the nonproline cis-peptide bond Lys-387-Tyr-388 (Fig. 5). Lys-273-Tyr-274 is in a small parallel β-sheet immediately N-terminal to the α-helix containing Cys-277. Abrogation of the Cys-277–Tyr-516 H bond may promote the initial step(s) in a sequence of cis/trans-peptide bond isomerizations of Lys-273-Tyr-274, Gly-372-Pro-373, and Lys-387-Tyr-388, leading to destabilization of the compact TG2 form.
Fig. 5.
Structural relationship between β-bulges and conserved cis-peptide bonds in TG2. GDP-bound human TG2, colored as in Fig. 1A, with residues linked by cis-peptide bonds shown as purple spheres. The proline cis-peptide bond, Gly-372-Pro-373, is located within a W β-bulge (37), and together with the Lys-387-Tyr-388 cis-peptide bond, clusters with the active site Asp-358. The Lys-273-Tyr-274 cis-peptide bond is part of a PC β-bulge (37) and is N-terminal to the active site Cys-277. Dashed lines indicate β-bulge-associated H bonds.
In conclusion, GTP-mediated conformational transition of TG2 to its compact, transamidase-inactive form involves masking of a destabilizing Arg-579 residue by GTP binding and formation of a stabilizing Cys-277–Tyr-516 H bond.
Materials and Methods
Protein Preparation, Enzyme, and Binding Assays.
Site-directed mutants were constructed in rat TG2 cDNA (19) by using GST-TG2/pGEX2T (31) as a template. Escherichia coli-expressed, thrombin-cleaved TG2-GST fusion proteins were purified by using affinity and ion exchange chromatography (10). Transamidase activity assays (9), GTP photolabeling, and isothermal titration calorimetry (10) were performed as described. Approximately 30% of Y516F (0.3 Y516F:1 GTPγS binding stoichiometry) was available to bind GTP, likely due to residual nucleotide binding as observed for WT and other mutants (10).
nPAGE.
This was performed on 8–10% resolving gels (100 V at 4°C) by using Laemmli buffers (32) without SDS, with pH adjusted at 4°C. Loading buffer (0.5 M Tris/20% glycerol/bromophenol blue/20 mM DTT, pH 6.8 at 4°C) was added to ice-chilled samples in a 3:1 sample:buffer ratio. Gels were stained with 0.5% Coomassie brilliant blue R-250 in 40% methanol/10% acetic acid and destained in 10% methanol/10% acetic acid.
Disulfide Bond Formation.
The redox catalyst Cu(II)(1,10-phenanthroline)3 (made by diluting 20 mM CuSO4/20 mM 1,10-phenanthroline in buffer) was used to trigger oxidation by ambient dissolved oxygen (33).
Sedimentation Velocity Experiments.
TG2 was dialyzed against buffer containing GTP or GTPγS at indicated concentrations. Interference optical fringe displacement data were acquired (30- to 45-sec intervals) at 201,600 × g by using a Beckman-Coulter (Fullerton, CA) Optima XL1 ultracentrifuge fitted with an AnTi60 rotor. Temperature-equilibrated cells were loaded with sample (350 μl) and dialysate (350 μl). Data were analyzed by the c(s) method (and variations thereof) with SEDFIT (34) and SEDPHAT (35, 36) programs. For further details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sara Holman for protein purification. This work was supported by Australian National Health and Medical Research Council Grant 142000, Australian Research Council Grant DP0665422, and a National Heart Foundation Postdoctoral Fellowship (to G.E.B.).
Abbreviation
- nPAGE
native PAGE
Footnotes
The authors declare no conflict of interest.
References
- 1.Achyuthan KE, Greenberg CS. J Biol Chem. 1987;262:1901–1906. [PubMed] [Google Scholar]
- 2.Lorand L, Graham RM. Nat Rev Mol Cell Biol. 2003;4:140–157. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 3.Lee KN, Birckbichler PJ, Patterson MK., Jr Biochem Biophys Res Commun. 1989;162:1370–1375. doi: 10.1016/0006-291x(89)90825-5. [DOI] [PubMed] [Google Scholar]
- 4.Nakaoka H, Perez DM, Baek KJ, Das T, Husain A, Misono K, Im MJ, Graham RM. Science. 1994;264:1593–1596. doi: 10.1126/science.7911253. [DOI] [PubMed] [Google Scholar]
- 5.Park ES, Won JH, Han KJ, Suh PG, Ryu SH, Lee HS, Yun HY, Kwon NS, Baek KJ. Biochem J. 1998;331:283–289. doi: 10.1042/bj3310283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vezza R, Habib A, FitzGerald GA. J Biol Chem. 1999;274:12774–12779. doi: 10.1074/jbc.274.18.12774. [DOI] [PubMed] [Google Scholar]
- 7.Liu S, Cerione RA, Clardy J. Proc Natl Acad Sci USA. 2002;99:2743–2747. doi: 10.1073/pnas.042454899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iismaa SE, Holman S, Wouters MA, Lorand L, Graham RM, Husain A. Proc Natl Acad Sci USA. 2003;100:12636–12641. doi: 10.1073/pnas.1635052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iismaa SE, Wu MJ, Nanda N, Church WB, Graham RM. J Biol Chem. 2000;275:18259–18265. doi: 10.1074/jbc.M000583200. [DOI] [PubMed] [Google Scholar]
- 10.Begg GE, Holman SR, Stokes PH, Matthews JM, Graham RM, Iismaa SE. J Biol Chem. 2006;281:12603–12609. doi: 10.1074/jbc.M600146200. [DOI] [PubMed] [Google Scholar]
- 11.Yee VC, Pedersen LC, Le Trong I, Bishop PD, Stenkamp RE, Teller DC. Proc Natl Acad Sci USA. 1994;91:7296–7300. doi: 10.1073/pnas.91.15.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguchi K, Ishikawa K, Yokoyama K, Ohtsuka T, Nio N, Suzuki E. J Biol Chem. 2001;276:12055–12059. doi: 10.1074/jbc.M009862200. [DOI] [PubMed] [Google Scholar]
- 13.Fesüs L, Laki K. Biochemistry. 1977;16:4061–4066. doi: 10.1021/bi00637a019. [DOI] [PubMed] [Google Scholar]
- 14.Casadio R, Polverini E, Mariani P, Spinozzi F, Carsughi F, Fontana A, Polverino de Laureto P, Matteucci G, Bergamini CM. Eur J Biochem. 1999;262:672–679. doi: 10.1046/j.1432-1327.1999.00437.x. [DOI] [PubMed] [Google Scholar]
- 15.Di Venere A, Rossi A, De Matteis F, Rosato N, Agro AF, Mei G. J Biol Chem. 2000;275:3915–3921. doi: 10.1074/jbc.275.6.3915. [DOI] [PubMed] [Google Scholar]
- 16.Mariani P, Carsughi F, Spinozzi F, Romanzetti S, Meier G, Casadio R, Bergamini CM. Biophys J. 2000;78:3240–3251. doi: 10.1016/S0006-3495(00)76860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasanna Murthy SN, Lomasney JW, Mak EC, Lorand L. Proc Natl Acad Sci USA. 1999;96:11815–11819. doi: 10.1073/pnas.96.21.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee KN, Arnold SA, Birckbichler PJ, Patterson MK, Jr, Fraij BM, Takeuchi Y, Carter HA. Biochim Biophys Acta. 1993;1202:1–6. doi: 10.1016/0167-4838(93)90055-v. [DOI] [PubMed] [Google Scholar]
- 19.Prasanna Murthy SN, Iismaa SE, Begg G, Freymann DM, Graham RM, Lorand L. Proc Natl Acad Sci USA. 2002;99:2738–2742. doi: 10.1073/pnas.052715799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson G, Owen D, Chalk PA, Lowe PN. Biochemistry. 1998;37:7885–7891. doi: 10.1021/bi980140+. [DOI] [PubMed] [Google Scholar]
- 21.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Biophys J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahvazi B, Boeshans KM, Idler W, Baxa U, Steinert PM, Rastinejad F. J Biol Chem. 2004;279:7180–7192. doi: 10.1074/jbc.M312310200. [DOI] [PubMed] [Google Scholar]
- 23.Ahvazi B, Boeshans KM, Steinert PM. J Biol Chem. 2004;279:26716–26725. doi: 10.1074/jbc.M403481200. [DOI] [PubMed] [Google Scholar]
- 24.Ahvazi B, Kim HC, Kee S-H, Nemes Z, Steinert PM. EMBO J. 2002;21:2055–2067. doi: 10.1093/emboj/21.9.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chica RA, Gagnon P, Keillor JW, Pelletier JN. Protein Sci. 2004;13:979–991. doi: 10.1110/ps.03433304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss MS, Metzner HJ, Hilgenfeld R. FEBS Lett. 1998;423:291–296. doi: 10.1016/s0014-5793(98)00098-2. [DOI] [PubMed] [Google Scholar]
- 27.Hutchinson EG, Thornton JM. Proteins. 1990;8:203–212. doi: 10.1002/prot.340080303. [DOI] [PubMed] [Google Scholar]
- 28.Richardson JS, Getzoff ED, Richardson DC. Proc Natl Acad Sci USA. 1978;75:2574–2578. doi: 10.1073/pnas.75.6.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Axe DD, Foster NW, Fersht AR. J Mol Biol. 1999;286:1471–1485. doi: 10.1006/jmbi.1999.2569. [DOI] [PubMed] [Google Scholar]
- 30.Steadman DJ, Zhao PS, Spencer HT, Dunlap RB, Berger SH. Biochemistry. 1998;37:7089–7095. doi: 10.1021/bi9725428. [DOI] [PubMed] [Google Scholar]
- 31.Iismaa SE, Chung L, Wu MJ, Teller DC, Yee VC, Graham RM. Biochemistry. 1997;36:11655–11664. doi: 10.1021/bi970545e. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Kobashi K. Biochim Biophys Acta. 1968;158:239–245. doi: 10.1016/0304-4165(68)90136-0. [DOI] [PubMed] [Google Scholar]
- 34.Schuck P. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuck P. In: Analytical Ultracentrifugation: Techniques and Methods. Scott DJ, Harding SE, Rowe AJ, editors. Cambridge, UK: R Soc Chem; 2005. pp. 26–49. [Google Scholar]
- 36.Boukari H, Nossal R, Sackett DL, Schuck P. Phys Rev Lett. 2004;93:098106. doi: 10.1103/PhysRevLett.93.098106. [DOI] [PubMed] [Google Scholar]
- 37.Chan AW, Hutchinson EG, Harris D, Thornton JM. Protein Sci. 1993;2:1574–1590. doi: 10.1002/pro.5560021004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunz ID. J Amer Chem Soc. 1971;93:516–518. doi: 10.1021/ja00731a037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.