Abstract
κ-opioid receptor (KOR) is detected pre- and postsynaptically, but the subcellular localization, translation, and regulation of kor mRNA in presynaptic compartments of sensory neurons remain elusive. In situ hybridization detected axonal distribution of kor mRNA in primary neurons of dorsal root ganglia (DRG). The MS2-fused GFP tracked kor mRNA transport from DRG neuronal soma to axons, requiring its 5′ and 3′ UTRs. In Campenot chambers, axonal translation of kor mRNA was demonstrated for DRG neurons, which depended on its 5′ UTR and was stimulated by KCl depolarization. KCl depolarization of DRG neurons rendered redistribution of kor mRNA from the postpolysomal fraction to the translationally active polysomal fraction. This study provided evidence for mRNA transport and regulation of presynaptic protein synthesis of nonstructural proteins like KOR in primary sensory neurons and demonstrated a mechanism of KCl depolarization-stimulated axonal mRNA redistribution for localized translational regulation.
Keywords: axonal translation
Opioid receptors interact with opioid drugs and endogenous opioid ligands to affect pain sensation, consciousness, and autonomic functions. Three major opioid receptor types, μ, δ, and κ are known (1, 2), each belonging to the super family of G protein-coupled transmembrane receptors (3, 4). Gene knockout studies confirmed the functional roles for opioid receptors in mediating the pharmacological actions of various opioid compounds that are among the most commonly prescribed analgesic agents (5). Furthermore, pharmacological studies revealed that the number of opioid receptors was critical to the manifestation of opioid drugs (6). However, extensive studies of transcriptional regulation of opioid receptor genes (7, 8) failed to uncover the mechanisms underlying alteration of receptor number or its protein level in neurons that bear a physiological relevance. Recently, posttranscriptional control for the expression of this gene family has begun to be unraveled, primarily, in studying κ-opioid receptor (KOR) expression (7, 9, 10).
KOR protein is detected both pre- and postsynaptically (11–14). Like many axonal proteins, presynaptic KOR proteins were thought to be synthesized in the soma and then transported to the axons via the axonal transport mechanism (15, 16). However, using in vitro differentiated neurons (17), we have demonstrated transport of kor mRNA into nerve fibers, suggesting an intriguing possibility that kor mRNA might be transported to the axons of KOR-expressing neurons for local translation and regulation. To this end, the phenomena of mRNA transport/targeting and local protein synthesis in the dendrites of invertebrate and vertebrate neurons have been established, but the source of axonal proteins remained heavily debated (18–21). Evidence for axonal mRNA transport and local protein synthesis was provided primarily for structural proteins, although local synthesis of EphA2 receptor, a protein involved in axonal pathfinding has been demonstrated (22–29). Additionally, it was shown that intraaxonal protein synthesis could play a role in chemoattractive guidance for axons of developing Xenopus retinal ganglion and motor neurons (20, 30). Nevertheless, direct evidence for local protein synthesis and its regulation of nonstructural proteins in axons remains scarce, despite the documentation of various mRNA species in these areas by using more sensitive techniques of detection (20).
The current study was designed to address three issues with regard to presynaptic KOR protein synthesis in primary DRG neurons. We first demonstrated axonal distribution of endogenous kor mRNA in primary DRG neurons using in situ hybridization and asked whether a transport process was involved to mobilize kor mRNA into axons. Second, we detected localized translation of presynaptic KOR in the axons grown in Campenot chambers. Finally, we asked whether and how axonal translation of kor mRNA could be regulated locally and discovered a mechanism whereby KCl depolarization stimulated kor mRNA redistribution for translational activation.
Results
Axonal Localization of kor mRNA in Mouse DRG Neurons.
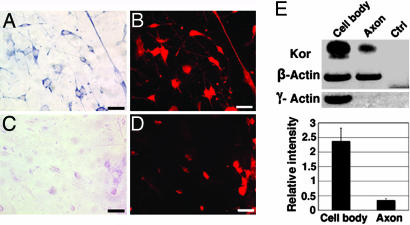
Primary cultures of mouse DRG neurons were examined by nonradioactive in situ hybridization to determine whether the endogenous kor mRNA could be localized in the axons of these sensory neurons. As shown in Fig. 1A, kor mRNA was clearly detected in the axons in addition to the cytoplasmic compartment of the soma. A sense probe corresponding to the same coding region of kor mRNA was used as a negative control, which detected no specific signals as shown in Fig. 1C. Fig. 1 B and D showed the staining patterns of a neuronal marker τ on the same sections as shown in Fig. 1 A and C, respectively.
Fig. 1.
Localization of kor mRNA in axons of primary DRG neurons. (A) In situ hybridization with an antisense kor-specific probe. (B) anti-τ immunohistochemistry of A. (C) In situ hybridization with a sense probe to kor. (D) anti-τ- protein immunostaining of C. (Scale bars, 25 μm.) (E) RT-PCR, followed by Southern blot analysis (Upper). A statistical analysis from multiple independent experiments is shown (Lower). (P = 0.0059325; n = 5). Blue stains, kor mRNA; red stains, τ protein. Ctrl, control.
To further validate the localization of kor mRNA in the axons and to gain a semiquantitative measure, DRG neurons were cultured on a two-surface culture device (31) to allow axon extension. This device consists of two different surfaces firmly bound by matrigel and sealed all around with paraffin. The plating surface is a nucleopore polycarbonate filter with a pore diameter of 3 μm that is bound to the second surface (a coverslip). On this device, axons grow into matrigel, and cell bodies stay on the surface of the filter. Arabinosylcytosine (5 μM) was added to inhibit glia in the cultures. At the end of culturing for 5 more days, axons and cell bodies were dissected from the two separated surfaces, and RNA was analyzed with one-step RT-PCR, followed by Southern blot analyses of kor mRNA levels in cell bodies versus axons. As shown in Fig. 1E, kor mRNA was detected in both the cell bodies and the axons, albeit at a lower (for approximately 6-folds) level as compared with β-actin in the axons. Clean axonal preparation was confirmed by the negative control γ-actin, which was not detected in the axons of the same preparation. These two experiments unambiguously demonstrated the presence of kor mRNA in the axons of mouse DRG neurons.
Translocation of kor mRNA to the Axons of DRG Neurons.
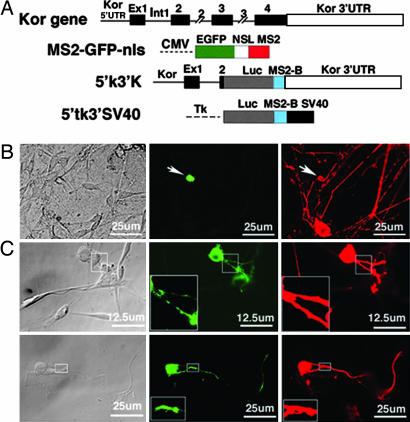
Axonal localization of kor mRNA in primary DRG neurons would suggest that kor mRNA should be translocated to axons from cell bodies. To test this possibility, an MS2-tagged RNA-tracking system was used (Fig. 2A). This system consists of an MS2-fused GFP containing a nuclear localization signal (MS2-GFP-nls) and kor mRNA containing its intact 5′ and 3′ UTRs (5′K3′K) and tagged with the MS2-binding site. An MS2-fused mRNA negative control contains the identical MS2-binding site fused to the 5′ UTR derived from the thymidine kinase reporter and the 3′ UTR/poly(A) of simian virus 40 (SV40) (5′tK3′SV).
Fig. 2.
Translocation of kor mRNA into axons of primary DRG neurons. (A) Maps of the constructs including kor gene, MS2-GFP-nls, 5′K3′K, and 5′tk3′SV40. CMV, CMV promoter; Ex, exon; Int, intron; Luc, luciferase; MS2, single-stranded RNA phage capsid protein MS2; MS2-B, MS2-binding site; NSL, nuclear localization signal; SV40, SV40 promoter; Tk, thymidine kinase promoter. (B) Cotransfection with MS2-GFP-nls and 5′tk3′SV40 (the negative control). (C) Cotransfection with MS2-GFP-nls and 5′K3′K. Two separate fields are shown. Areas of inserts are enlarged by three times and are shown at the lower left corners. Bright-field image. GFP signal is shown in green, and τ- immunoreactivity is shown in red.
Primary cultures of rat DRG neurons were cotransfected with GFP-MS2-nls and 5′K3′K or GFP-MS2-nls and the negative control 5′tk3′SV. Thirty hours later, transfected DRG neurons were detected with anti-τ. GFP- and τ-specific signals were captured simultaneously. As shown in Fig. 2C (Center), GFP was clearly detected in the axons of 5′K3′K-transfected neurons, particularly in some hot spots, indicating translocation, or transport, of kor mRNA to the axonal compartment. The same field was verified with τ-staining (Right). As predicted, the GFP in DRG neurons cotransfected with the negative control mRNA (5′tk3′SV40) was retained in the nuclei (Fig. 2B Center), supporting a specific functional role for the 5′ and 3′ UTRs of kor mRNA. These results demonstrated that kor mRNA indeed could be translocated from the soma to the axons of DRG neurons. Furthermore, the active axonal transport of kor mRNA was mediated by its 5′ and 3′ UTRs.
Local Translation of kor mRNA in the Axons of DRG Neurons.
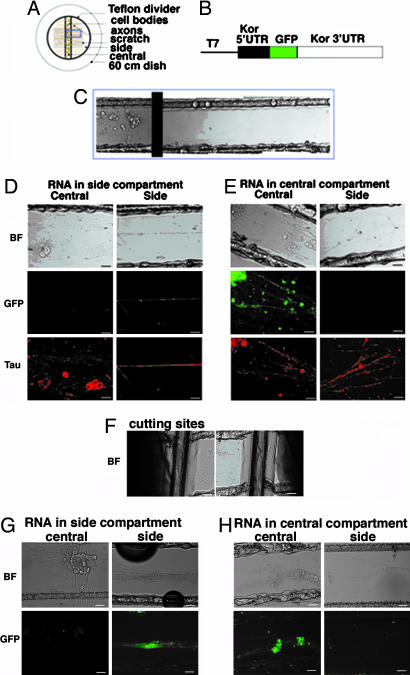
The presence of kor mRNA in the axons of DRG neurons would suggest that kor mRNA might be translated in these compartments. To directly demonstrate localized translation in the axonal compartments, Campenot chamber cultures were applied in the experiments by using a direct RNA-transfection procedure. This methodology allows direct detection of local translation of specific mRNA in various parts of living neurons, because in vitro-synthesized mRNA can be introduced directly into either the central (containing, primarily, the soma) or the side (containing only axons) chamber, followed by monitoring their translational products in situ. To assist visualization of de novo synthesized proteins, the coding sequence of the intact kor mRNA was replaced, in frame, with the GFP coding sequence, producing a translation cassette for GFP protein that entirely depends on translational regulation of the natural UTRs of kor mRNA. Thus, GFP is made according to the regulatory signals of kor mRNA translation and distributed without the potentially interfering sequences of KOR protein that may impose trafficking effects. Fig. 3A shows a Campenot chamber device used in these studies, Fig. 3B shows the construct of KOR-GFP fusion, and Fig. 3C shows the bright field of a representative slide spanning both the central and side chambers.
Fig. 3.
Localized translation of axonal kor mRNA in primary DRG neurons. (A) A Campenot chamber. (B) The construct of the intact KOR–GFP translational cassette. (C) A Campenot slide with DRG neurons seeded in the central compartment and axons extended into the side compartment. (D) RNA transfection conducted only in the side compartment. (E) RNA transfection conducted only in the central compartment. (F) A cut made near the divider in the central chamber. (G) RNA transfection conducted in side compartment after disconnecting side chamber from central chamber. (H) RNA transfection conducted in central compartment after disconnecting side chamber from central chamber. BF, bright-field image. (Scale bars, 25 μm.) GFP signal is green, and τ-immunoreactivity is in red.
We first tested KOR-GFP translation in the soma (Fig. 3E) where mRNA was introduced only into the central chamber (Fig. 3E Left). Twenty hours later, axons grown in the side chamber (Fig. 3E Right) of the same slide were examined. Interestingly, GFP signals were clearly detected in many soma and proximal axons in the central chamber (Fig. 3E Middle Left), but no GFP (green color) could be detected in the axons (Fig. 3E Middle Right) that were marked by τ-staining (red color) (Fig. 3E Bottom Right) on the same slide. This result confirmed the predicted translation in the soma. We then examined translation in the axons by transfecting KOR-GFP mRNA in the side chamber (Fig. 3D). Twenty hours after transfection, GFP was evidently detected in axons (Fig. 3D Middle Right) that were also marked by τ-staining (Fig. 3D Bottom Right), but GFP was not detected in the central chamber of the same slide, although DRG neurons and proximal axons were present abundantly (Fig. 3D Left). This result showed that translation could occur not only in the soma but also in axons. To further examine whether axonal translation could occur when axons were disconnected from soma, a cut was made at the divider area after axons were fully extended to physically disconnect axons from their soma (Fig. 3F). RNA was then introduced into the side or central chamber. GFP images in central and side chambers were captured 6–8 h later. It appeared that translation indeed occurred independently even after axons were disconnected from soma (Fig. 3 G and H). These results directly demonstrated local translation of kor mRNA not only in the DRG neuronal soma but also in the axons even after they were physically disconnected from the soma.
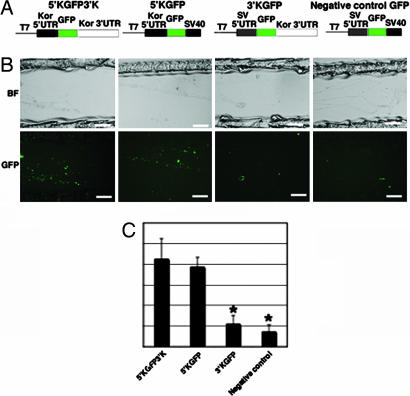
To locate important UTRs of kor mRNA in axonal translation, a panel of translation cassettes were generated with deletion in either the 5′ UTR, the 3′ UTR, or both 5′ and 3′ UTRs (Fig. 4A) and tested for their translation in the axons. As shown in Fig. 4B, the construct containing both 5′ and 3′ UTRs (5′KGFP3′K) and the construct containing only the 5′ UTR of kor (5′KGFP) were similarly effective in producing GFP in axons, whereas the construct deleted in the 5′ UTR of kor and the construct deleted in both 5′ and 3′ UTRs of kor could not be translated efficiently. These results revealed that the 5′ UTR of kor mRNA was critical for its efficient translation in the axons of DRG neurons.
Fig. 4.
The role of kor mRNA 5′ UTR in axonal translation. (A) A panel of KOR-GFP translational cassettes, including the wild type (5′KGFP3′K), the 3′-deleted (5′KGFP), the 5′-deleted (3′KGFP), and the 5′- and 3′-deleted (negative control). (B) Transfection conducted in the side compartments for each construct. (Scale bars, 25 μm.) (C) A statistic analysis of quantification of GFP signals for each construct was obtained. (For comparing 3′KGFP with 5′KGFP3′K, ∗, P = 0.00034388; n = 7. For comparing negative control with 5′KGFP3′K, ∗, P = 0.000012207; n = 7.) GFP signal is in green.
Regulation of kor mRNA Translation in the Axons of DRG Neurons.
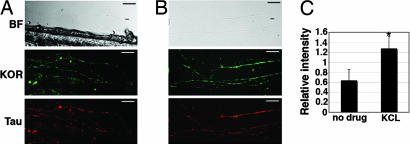
To explore whether KOR protein synthesis in axons could be regulated, we first examined the effect of neuronal activity on the expression level of endogenous KOR protein in DRG axons triggered by KCl depolarization. As shown in Fig. 5A, in control cultures, both endogenous KOR (Fig. 5A Middle) and τ (Fig. 5A Bottom) proteins were clearly detected in DRG axons grown in the Campenot chamber. Interestingly, KCl depolarization significantly stimulated the level of these presynaptic KOR proteins when KCl (45 mM) was applied into the side chamber where axons grew (Fig. 5B). The level of τ-immunoreactivity remained comparable (Fig. 5B Bottom) before and after KCl depolarization. A semiquantitative measure (Fig. 5C) of the amount of the endogenous presynaptic KOR indicated that the presynaptic KOR immunoreactivity was specifically and significantly elevated (for ≈2-folds) by localized KCl depolarization, suggesting a potential effect of KCl depolarization on the synthesis of KOR protein in these areas.
Fig. 5.
Specific stimulation of endogenous KOR protein level in axons of primary DRG neurons seeded on Campenot chambers. (A) Control axons. (B) Axons treated with 45 mM KCl. (C) A statistical analysis of the relative level of KOR expression, as compared with τ expression, with or without 45 mM KCl treatment (∗, P = 0.00085783; n = 6). (Scale bars, 25 μm.) KOR immunoreactivity is in green, and τ-immunoreactivity is in red.
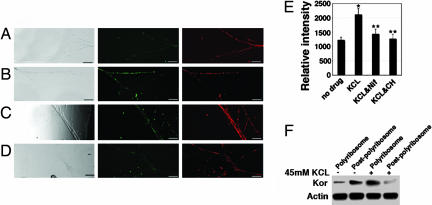
To directly demonstrate localized regulation of kor mRNA translation by KCl depolarization, we conducted direct mRNA transfection in Campenot chambers and monitored de novo synthesized GFP and endogenous τ-proteins before and after KCl depolarization. Fig. 6A shows the patterns of unstimulated KOR-GFP (Fig. 6A Center) and τ (Fig. 6A Right) of the same field. In agreement with the elevation of endogenous KOR after KCl depolarization, the same treatment also resulted in significantly intensified GFP signals, with τ-staining remained relatively constant (Fig. 6B). This result confirmed that local KCl depolarization stimulated localized translation of KOR-GFP mRNA in the axons of DRG neurons. KCl depolarization is known to increase intracellular calcium concentration by activating the L-type voltage-gated calcium channel. An L-type voltage-gated calcium channel blocker, nifedipine, should inhibit the effect of KCl. This indeed was the case as shown in Fig. 6C, where the KCl-stimulated GFP signal, but not τ, was significantly weakened after treatment with nifedipine. To verify whether the increase in GFP intensity after KCl depolarization was attributed to new translation of the introduced KOR-GFP mRNA, cycloheximide was applied. This translation blocker significantly reduced the intensity of KCl depolarization-stimulated GFP signals but not τ (Fig. 6D). To obtain a semiquantitative measure of the translation-stimulating effect of KCl depolarization, we quantified axonally translated GFP by normalizing to the axonal τ-specific immunoreactivity. The normalized GFP signals were obtained from multiple areas, and means were obtained after statistic analyses as shown in Fig. 6E. It appeared that KCl depolarization stimulated newly synthesized proteins, regulated by the UTRs of kor mRNA, in the axons to ≈2-folds, which was effectively blocked by the translational blocker cycloheximide.
Fig. 6.
KCl depolarization-stimulated local synthesis of KOR in axons (side chamber) of DRG neurons seeded in the central chamber on Campenot slides. (A) Side chamber treated with a vehicle control. (B) Side chamber treated with 45 mM KCl. (C) Side chamber treated with 45 mM KCl and 10 nM nifedipine. (D) Side chamber treated with 45 mM KCl and 2 μM cycloheximide. (Left, bright-field images; Center, GFP; Right, τ-immunoreactivity. (E) A statistical analysis of GFP signals of various experimental groups. (For comparing KCl with control, ∗, P = 0.00005702; n = 7. For comparing KCl plus cycloheximide with KCl alone, ∗∗, P = 0.00000602552; n = 7. For comparing KCl plus nifedipine with KCl alone, ∗∗, P = 0.00024525; n = 7.) (F) Redistribution of endogenous kor mRNA in polysomal and postpolysomal fractions of DRG neurons before and after KCl depolarization. (Scale bars, 25 μm.) GFP signal is in green, and τ-immunoreactivity is in red.
KCl Depolarization-Stimulated Redistribution of kor mRNAs.
To examine the mechanism underlying translation-stimulating effect of KCl depolarization in DRG neurons, we examined one early step in translational control, polysome-association of the target mRNA before and after KCl depolarization in DRG neurons. The translationally active polysomal fraction and the inactive postpolysomal fraction from primary cultured DRG neurons were collected and analyzed for the association of endogenous kor mRNA as shown in Fig. 6F. Without depolarization, most endogenous kor mRNA was distributed in the postpolysomal fraction, but KCl depolarization triggered redistribution of kor mRNA to the polysomal fraction. For a comparison, the distribution of actin mRNA was monitored in parallel and appeared relatively constant before and after depolarization. This experiment revealed that KCl depolarization stimulated the redistribution of kor mRNA from the translationally inactive postpolysomal fraction to the translationally active polysomal fraction, thereby rapidly enhancing local KOR synthesis.
Discussion
Opioid receptors are detected both pre- and postsynaptically. Alteration in the binding patterns of these receptors in animal tissues, either during developmental stages or after pharmacological manipulation, has been demonstrated (32, 33). Attempts to address the expression and regulation of these receptors have been initiated from studying transcriptional regulation of their genes. However, transcriptional regulation affects the expression of receptors mostly when cells undergo differentiation, express different transcription factors, or engage in chromatin remodeling processes (34–37). It has remained a contested issue whether transcriptional control could adequately account for alteration of receptor binding or receptor protein expression in neurons. This study specifically addresses posttranscriptional regulation of KOR protein expression in the specific neuronal compartment of primary DRG neurons that bear a physiological and pharmacological relevance to opioid analgesics and provides an example of posttranscriptional regulation of presynaptic receptor production when the neurons are stimulated, such as by KCl depolarization. However, the data (Fig. 1) show that only ≈16% of total kor mRNA is localized to axons. Therefore, the functional significance of this relatively small fraction of kor mRNA in the overall pharmacological response of DRG neurons awaits further verification. To this end, it is important to determine whether KOR protein synthesized locally in the axons of DRG neurons is functional. Furthermore, it remains elusive how the targeting of kor mRNA to axons is regulated.
Three intensively debated issues in the field of neurobiology are addressed in this study specifically related to several hypothetical events that could occur in the axons of sensory neurons. These include axonal distribution/transport of mRNAs for nonstructural components such as a transmembrane drug receptor, the localized synthesis of nonstructural protein in the axons of DRG neurons, and a mechanism of depolarization-triggered, enhanced local translation in neurons by altering mRNA distribution between the polysomal and the postpolysomal fractions. The treatment of 45 mM KCl mimics the effects of neuronal activity. This treatment has also been shown to induce mRNA targeting and local protein synthesis in the dendrites (38–41). A shift of dendritic mRNAs from RNA granules to the polysomal fraction by depolarization has been proposed to provide a mechanism connecting RNA localization to translation in dendrites (39). Our finding of redistribution of kor mRNA after KCl depolarization in axons and the concurrent enhancement of kor mRNA translation in these areas strongly suggest similar mechanisms of regulation adopted for both axons and dendrites with regard to mRNA targeting and translation. These types of regulatory events in the dendrites are widely examined, and several regulatory pathways and components have been demonstrated (40–42). However, given the apparent structural divergence of axons from dendrites, it is possible that certain regulatory mechanisms can be adopted for specific axonal regulation. To this end, one important task in the future is to identify the possible regulatory components that are specific to the axonal compartment.
Because of extensive compartmentalization of neurons, the subcellular localization of specific mRNAs in neurons could play an important role in maintaining normal neuronal activity, as has been widely demonstrated and established for mRNA targeting in dendrites and the synthesis of dendritic proteins; but it remains largely debatable for axonal proteins. The classical view of axonal transport focuses on the cargo-binding mechanism of protein complexes (43). Active transport of microtubules is also established (44). Although it may be more efficient in terms of the amount of proteins that cargos can deliver, this type of transport may not be effective when a certain degree of plasticity is needed, such as when neuronal activity is transiently elicited or specific receptors and signaling molecules are needed in certain localized areas in a short period. Further, the constitutive synthesis, followed by the delivery of these proteins to distant axonal areas, can become energy-inefficient if these proteins are not needed constitutively. Recently, it has been demonstrated that a rapid supply of newly synthesized protein occurs at distinct subcellular sites, which can influence neuronal functions (20, 45). There also exists solid evidence for axonal translation of mRNAs, especially for the structural proteins (20, 25, 30). With regard to axon guidance and regeneration, it is known that local protein synthesis plays an important role (27). Furthermore, the axon guidance signaling Ephs can be locally synthesized, and the binding by ephrin triggers a Vav-dependent endocytosis of receptor–ligand complexes (46). Our study demonstrates the presence of a category of mRNAs that code for neither structural nor growth-related proteins in the axons of sensory neurons. Our result also expands the repertoire of axonal proteins in sensory neurons that can be synthesized locally. More importantly, this study demonstrates the regulation of localized translation in axons after local depolarization, which may implicate a previously ignored, intricate coordination of RNA transport and translational regulation in the axons. However, it is also possible that stimulated translation may involve, partially, changes in mRNA stability and remains to be further investigated.
It is intriguing that in Campenot chambers, no GFP could be detected in the side compartment at 20 h after mRNA was introduced into the central compartment and vice versa. Because GFP can presumably freely diffuse, our results would suggest that the diffusion of GFP from the soma into axons, or from axons to soma, is not readily efficient for delivering a sufficient amount of GFP proteins that can be detected at 20 h after transfection. This hypothesis is supported by our observation that GFP proteins can be detected in axons of DRG neurons transfected with free GFP usually after 2–3 days after transfection (data not shown). However, a possibility of efficient, free diffusion of a small amount of proteins into axons has not been ruled out. To this end, it is generally believed that proteins, if destined for specific areas of the axons, need to be carried by specific cargos for efficient transport. Free diffusion of most proteins from the soma to the extended axons is probably not efficient to meet the local need in a distant axonal area. The mechanism of mRNA transport coupled to local regulation of its translation could provide an effective way of delivering and regulating certain proteins that are needed in specialized areas of extended axons at a specific time. These mRNAs can be stored locally until that particular site is stimulated. This process would greatly conserve the energy that may otherwise be wasted in synthesizing and transporting unneeded proteins. Our results provide the evidence supporting such a hypothetical mechanism that could regulate local expression of certain neuronal proteins that are probably needed at a higher level only under certain conditions. It would be interesting to investigate whether this mechanism can be generalized for many other axonal proteins that have to be regulated for localized expression. It is also critical in the future to determine the regulatory mechanisms underlying axonal transport of these mRNAs.
Methods
The experiments involving animals have been approved by the University's Institutional Animal Care and Use Committee. All of the experiments conform to the regulatory standards.
Plasmid Constructs.
The plasmids for the single-stranded RNA phage capsid protein MS2-fused nuclear GFP (MS2-GFP-nls), the MS2-binding site tagged kor mRNA (5′K3′K), and the MS2 site-tagged negative control mRNA (5′tk3′SV40) are as described (17). The KOR-GFP translation cassettes were engineered by replacing the entire coding region of kor of the intact kor cDNA with the GFP coding region at the NcoI site, and with 5′ or 3′ UTR replaced with a generic counterpart derived from pGL, which were made in pSp73 (Promega, Madison, WI) for in vitro transcription.
In Situ Hybridization.
The mouse kor cDNA in pGEM-T Easy vector (Promega) was used to prepare kor-specific riboprobes. Linearized DNA was used for the synthesis of DIG-labeled riboprobes by in vitro transcription. Slides were fixed in 4% paraformaldehyde at room temperature for 30 min and dehydrated by incubating successively in 70%, 90%, and 100% ethanol, followed by 100% xylene. Slides were incubated with 2 μg/ml proteinase K at 37°C for 5 min, washed with 1× PBS, and postfixed in 4% paraformaldehyde for 10 min. After washing, slides were incubated in a prehybridization buffer (50% deionized formamide/4× SSC/1× Denhardt's solution/10% dextran sulfate/0.5 mg/ml ssDNA/0.25 mg/ml yeast tRNA) and hybridized with the DIG-labeled riboprobes (10 ng/μl) overnight. Washing and detection procedures were conducted according to the manufacturer's protocol (Roche, Indianapolis, IN).
RT-PCR and Southern Blot Analyses.
Axons and cell bodies were separated from mice DRG neurons cultured on two-surface culture sandwiches. RNA was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA). RT-PCRs were carried out with a one-step RT-PCR kit (Qiagen, Valencia, CA). kor cDNA was amplified by using primer pair 5′-CATCATCAGGAAACTGCA-3′ and 5′-TGGTCATGTTTGTCATC-3′. Southern blots and semiquantitative analyses were performed as described (9, 34).
DRG Neuron Culture, Transfection, and Immunohistochemistry.
DRGs were dissected from newborn rats or mice and dissociated into single neurons as described (17, 29, 47). DRGs were incubated with 50 units of collagenase for 30 min and 0.05% trypsin-EDTA for 5 min and then mechanically dissociated by triturating through a series of flame-narrowed Pasteur pipettes. The cell suspension was plated on 0.1 mg/ml poly-d-lysine-coated coverglasses, assembled two-surface culture sandwiches (31) or in the central compartments of compartmented slides (47, 48). Cells were maintained in DRG growing medium (MEM with 10% FBS, 1% N2, and 1% penicillin/streptomycin) for 3–5 days, with 5 μM arabinosylcytosine (AraC; Sigma, St. Louis, MO) added at the second day to inhibit glial proliferation. Primary rat DRG neurons were transfected by using a Lipofectamine 2000 kit (Invitrogen). Immunohistochemistry was conducted as described with anti-KOR (rabbit, 1:250; Santa Cruz Biotechnology, Santa Cruz, CA) or anti-τ antibodies (mouse, 1:1,000; Sigma) as described (17, 34).
Images were taken by using a Nikon (East Rutherford, NJ) inverted or Zeiss (Thornwood, NY) upright fluorescence microscope. To determine the mean intensity of fluorescence, several regions of axons were randomly selected, where the intensity of fluorescence was measured with the help of ImageJ software. The kor-specific signal was normalized to the τ-specific signal for the same area. A total of 7–10 individual axons were measured for each treatment, and the experiment result was analyzed by the paired Student two-tailed t test.
Compartment Culture, in Vitro Transcription, and RNA Transfection.
Sixty-millimeter plastic tissue-culture dishes were coated with 0.6 mg/ml collagen solution twice and 100 μg/ml poly-d-lysine once. On a dry coated dish, parallel scratches were made of ≈20 mm long with a sterilized Pin Rake. Campenot Chamber CAMP3 was precleaned and assembled on the coated dish by applying a rope of silicone grease (47, 48). Dissociated rat DRG neurons were plated in the central compartment in 0.3 ml of DRG culture medium. Lateral compartments were filled with 0.5 ml of DRG culture medium containing 100 μg/ml NGF and 5 μM AraC. On the second day, the medium in the central compartment was changed to DRG culture medium containing 5 μM AraC but no N2 and NGF. The neurons were fed every 2–3 days. In experiments for axons separated from soma, cuts were made along the divider to remove cell bodies or to physically separate the side from the central compartments, by using industrial razor blades.
Linear plasmid DNA was in vitro transcribed, followed by capping and poly(A) tailing by using Ambion's message Machine T7 RNA polymerase kit (Ambion, Austin, TX). The processed RNA (0.8 mg) was complexed with 4 ml of TransMessenger transfection reagent (Qiagen). The lipid mRNA complex was applied directly into the designated compartment. After experimentation, immunohistochemistry for τ was conducted, and fluorescent signals of GFP and anti-τ were quantified by using ImageJ software as described above (DRG Neuron Culture, Transfection, and Immunohistochemistry) to obtain the normalized GFP/τ ratio for each treatment. A total of 7–10 individual axons were measured for each treatment, and the experiment result was analyzed by the paired Student two-tailed t test.
Polyribosome and Postpolyribosome Fractionations.
Primary DRG neurons cultured on 10-cm culture dishes coated with 0.6 mg/ml collagen and 0.1 mg/ml poly-d-lysine were treated with or without 45 mM KCl for 4 h and washed with ice-cold 1× PBS. The cells were resuspended in a polysomal buffer containing 10 mM Mops (pH 7.2), 250 mM NaCl, 2.5 mM MgOAc, 0.5% Nonidet P-40, 0.1 mM phenylmethylsulfonylfluoride, 200 mg/ml heparin, and 50 mg/ml cycloheximide. The polysomes were pelleted by ultracentrifugation at 100,000 × g for 1 h at 4°C. The supernatant provided the postpolysomal fraction (49). RNA, was isolated from the polysomal and postpolysomal fractions and RT-PCR and Southern blot were conducted.
Drug Treatment.
Cultured DRG neurons were treated with 45 mM KCl, 45 mM KCl with 10 nM nifedipine, or 45 mM KCl with 2 μg of cycloheximide. All of the compounds were purchased from Sigma.
Acknowledgments
This work was supported by National Institutes of Health Grants DK54733, DK60521, K02 DA13926, and DA11190 (to L.-N.W.) and DA00564, DA01583, DA11806, and K05-DA70554 (to H.H.L.).
Abbreviations
- DRG
dorsal root ganglion
- KOR
κ-opioid receptor
- SV40
simian virus.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Goldstein A, Naidu A. Mol Pharmacol. 1989;36:256–272. [PubMed] [Google Scholar]
- 2.Pasternak GW. Neuropharmacology. 2004;47(Suppl 1):312–323. doi: 10.1016/j.neuropharm.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Law PY, Wong YH, Loh HH. Annu Rev Pharmacol Toxicol. 2000b;40:389–430. doi: 10.1146/annurev.pharmtox.40.1.389. [DOI] [PubMed] [Google Scholar]
- 4.Masabumi M, Satoh M. Neurosci Res. 1995;23:121–145. doi: 10.1016/0168-0102(95)00933-k. [DOI] [PubMed] [Google Scholar]
- 5.Kieffer BL, Gaveriaux-Ruff C. Prog Neurobiol. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 6.Law PY, Erickson LJ, El-Kouhen R, Dicker L, Solberg JM, Wang W, Miller E, Burd AL, Loh HH. Mol Pharmacol. 2000;58:388–398. doi: 10.1124/mol.58.2.388. [DOI] [PubMed] [Google Scholar]
- 7.Wei L-N, Loh HH. Curr Opin Pharmacol. 2002;2:69–75. doi: 10.1016/s1471-4892(01)00123-0. [DOI] [PubMed] [Google Scholar]
- 8.Liu HC, Shen JT, August LB, Ko JL, Loh HH. J Biol Chem. 1999;274:23617–23626. doi: 10.1074/jbc.274.33.23617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei L-N, Hu X, Bi J, Loh HH. Mol Pharmacol. 2000;57:401–408. [PubMed] [Google Scholar]
- 10.Wei L-N, Law YP, Loh HH. Frontiers Biosci. 2004;9:1665–1679. doi: 10.2741/1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drake CT, Chavkin C. In: The Pharmacology of Opioid Peptides. Tseng LF, editor. Singapore: Harwood; 1995. pp. 169–186. [Google Scholar]
- 12.Shuster SJ, Riedl M, Li XR, Vulchanova L, Elde R. J Neurosci. 1999;19:2658–2664. doi: 10.1523/JNEUROSCI.19-07-02658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoffelmeer AN, Rice KC, Jacobson AE, Van Geleren JG, Hogenboom F, Heijna MH, Mulder AH. Eur J Pharmacol. 1988;154:169–178. doi: 10.1016/0014-2999(88)90094-5. [DOI] [PubMed] [Google Scholar]
- 14.Werling LL, Frattali A, Portoghese PS, Takemori AE, Cox BM. J Pharmacol Exp Ther. 1988;246:282–286. [PubMed] [Google Scholar]
- 15.Droz B, Leblond CP. J Comp Neurol. 1963;121:325–346. doi: 10.1002/cne.901210304. [DOI] [PubMed] [Google Scholar]
- 16.Lasek RJ, Dabrowski C, Nordlander R. Nat New Biol. 1973;244:162–165. doi: 10.1038/newbio244162a0. [DOI] [PubMed] [Google Scholar]
- 17.Bi J, Hu X, Loh HH, Wei L-N. Mol Pharmacol. 2003;64:594–599. doi: 10.1124/mol.64.3.594. [DOI] [PubMed] [Google Scholar]
- 18.Bassell GJ, Kelic S. Curr Opin Neurobiol. 2004;14:574–581. doi: 10.1016/j.conb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 19.Job C, Eberwine J. Nat Rev Neurosci. 2001;2:889–898. doi: 10.1038/35104069. [DOI] [PubMed] [Google Scholar]
- 20.Piper M, Holt C. Annu Rev Cell Dev Biol. 2004;20:505–523. doi: 10.1146/annurev.cellbio.20.010403.111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kindler S, Wang H, Richter D, Tiedge H. Ann Rev Cell Dev Biol. 2005;21:223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benn SC, Woolf CJ. Nat Rev Neurosci. 2004;5:686–700. doi: 10.1038/nrn1477. [DOI] [PubMed] [Google Scholar]
- 23.Brittis PA, Lu Q, Flanagan JG. Cell. 2002;110:223–235. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- 24.Condeelis J, Singer RH. Biol Cell. 2001;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- 25.Eng H, Lund K, Campenot RB. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glebova NO, Ginty DD. Annu Rev Neurosci. 2005;28:191–222. doi: 10.1146/annurev.neuro.28.061604.135659. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan BB, Lavina ZS, Gioio AE. Ann NY Acad Sci. 2004;1081:244–254. doi: 10.1196/annals.1296.029. [DOI] [PubMed] [Google Scholar]
- 28.Martin KC. Curr Opin Neurobiol. 2004;14:305–310. doi: 10.1016/j.conb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Q, Kelly TK, Chang B, Ryazantsev S, Rajasekaran A, Martin C, Twiss J. J Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell DS, Holt CE. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- 31.Torre ER, Steward O. J Neurosci. 1992;12:762–772. doi: 10.1523/JNEUROSCI.12-03-00762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buzas B, Rosenberger J, Cox BM. NeuroReport. 1996;7:1505–1508. doi: 10.1097/00001756-199606170-00013. [DOI] [PubMed] [Google Scholar]
- 33.Unterwald EM, Rubenfeld JM, Imai Y, Wang JB, Uhl GR, Kreek MJ. Brain Res Mol Brain Res. 1995;33:351–355. doi: 10.1016/0169-328x(95)00143-g. [DOI] [PubMed] [Google Scholar]
- 34.Bi J, Hu X, Loh HH, Wei L-N. J Neurosci. 2001;21:1590–1599. doi: 10.1523/JNEUROSCI.21-05-01590.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X, Bi J, Loh HH, Wei L-N. J Biol Chem. 2001;276:4597–4603. doi: 10.1074/jbc.M005477200. [DOI] [PubMed] [Google Scholar]
- 36.Park SW, Li J, Loh HH, Wei L-N. J Neurosci. 2002;22:7941–7947. doi: 10.1523/JNEUROSCI.22-18-07941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park SW, Huq MDM, Loh HH, Wei L-N. J Neurosci. 2005;25:3350–3357. doi: 10.1523/JNEUROSCI.0186-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tongiorgi E, Righi M, Cattaneo A. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krichevsky AM, Kosil SK. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 40.Tiruchinapalli MD, Oleynikov Y, Kelic S, Shenoy MS, Hartley A, Stanton KP, Singer RH, Bassell GJ. J Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tongiorgi E, Domenici L, Simonato M. Mol Neurobiol. 2006;33:17–32. doi: 10.1385/MN:33:1:017. [DOI] [PubMed] [Google Scholar]
- 42.Rook SM, Lu M, Kosik SK. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallee RB, Williams JC, Varma D, Barnhart LE. J Neurobiol. 2004;58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 44.Baas PW. Intern Rev Cytol. 2002;212:41–62. doi: 10.1016/s0074-7696(01)12003-6. [DOI] [PubMed] [Google Scholar]
- 45.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 46.Cowan CW, Shao YR, Sahin M, Shamah SM, Lin MZ, Greer PL, Gao S, Griffith EC, Brugge JS, Greenberg ME. Neuron. 2005;46:205–217. doi: 10.1016/j.neuron.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 47.Stucky CL, Abrahams LG, Seybold V. Neuroscience. 1998;84:1257–1265. doi: 10.1016/s0306-4522(97)00572-1. [DOI] [PubMed] [Google Scholar]
- 48.Ure RD, Campenot RB. J Neurosci. 1997;17:1282–1290. doi: 10.1523/JNEUROSCI.17-04-01282.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Mol Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]