Abstract
We have defined regions of the skeletal muscle ryanodine receptor (RyR1) essential for bidirectional signaling with dihydropyridine receptors (DHPRs) and for the organization of DHPR into tetrad arrays by expressing RyR1–RyR3 chimerae in dyspedic myotubes. RyR1–RyR3 constructs bearing RyR1 residues 1–1681 restored wild-type DHPR tetrad arrays and, in part, skeletal-type excitation–contraction (EC) coupling (orthograde signaling) but failed to enhance DHPR Ca2+ currents (retrograde signaling) to WT RyR1 levels. Within this region, the D2 domain (amino acids 1272–1455), although ineffective on its own, dramatically enhanced the formation of tetrads and EC coupling rescue by constructs that otherwise are only partially effective. These findings suggest that the orthograde signal and DHPR tetrad formation require the contributions of numerous RyR regions. Surprisingly, we found that RyR3, although incapable of supporting EC coupling or tetrad formation, restored a significant level of Ca2+ current, revealing a functional interaction with the skeletal muscle DHPR. Thus, our data support the hypotheses that (i) the structural/functional link between RyR1 and the skeletal muscle DHPR requires multiple interacting regions, (ii) the D2 domain of RyR1 plays a key role in stabilizing this interaction, and (iii) a form of retrograde signaling from RyR3 to the DHPR occurs in the absence of direct protein–protein interactions.
Keywords: calcium release, freeze–fracture, myotubes, voltage clamp
The basis for excitation–contraction (EC) coupling in skeletal muscle is a direct functional interaction between the dihydropyridine receptor (DHPR) in the plasmalemma/T tubules and ryanodine receptor (RyR)1 of the sarcoplasmic reticulum. The interaction requires an appropriate intermolecular link between the two skeletal muscle channels and depends on a specific positioning of the DHPR relative to the RyR. Three convenient experimental assays are available for assessing this structure–function correlation. The first approach is a functional assay that detects orthograde signaling, by which DHPRs control the functional state of RyRs (1–3) and produces depolarization-induced sarcoplasmic reticulum Ca2+ release in the absence of Ca2+ permeation through DHPRs. The second method measures the retrograde signal by which RyRs control the gating of DHPRs (4–6) as reflected in the size of DHPR Ca2+ currents via whole-cell voltage clamp. The third approach is a structural assay based on the technique of freeze–fracture for electron microscopy, which permits the study of the intermolecular interaction at the basis of the DHPR–RyR conformational coupling (7–10). In images from freeze–fracture replicas, DHPRs arrange themselves in tetrads in a RyR1-dependent manner.
In previous studies, various RyR1–RyR2 (11, 12) and several skeletal–cardiac and skeletal–insect DHPR chimerae (13–15) have been tested to assess the contributions of various domains to DHPR–RyR1 coupling. Although some details are not fully agreed on, it is clear that a specific segment of the α1S DHPR subunit II–III loop and the DHPR β1a subunit are involved in both structural and functional interactions with RyR1 (8, 16–18). A fairly large domain of RyR1, probably divided into smaller subdomains, has also been identified as a significant component in the structural coupling with DHPRs and in the reciprocal signaling between the two proteins (11, 12, 19, 20). One general conclusion from these studies is that an appropriate stereospecific DHPR–RyR positioning is absolutely necessary but not entirely sufficient for functional EC coupling. Signaling does not occur in the absence of alignment, yet it is also possible that the two channels may be appropriately aligned but incapable of reciprocal signaling. Therefore, the domain(s) involved in the structural link is not exactly the same as those involved in the functional coupling. Clearly, neither the functional nor the structural data can separately tell the whole story. In this regard, the contributions to the effectiveness of EC coupling by multiple cytosolic domains of α1S (21) and requirement for the β1a subunit of DHPR (22) may very well be due to an effect on the stability of the structural connection (8) rather than simply a direct effect on the functional coupling.
We have chosen to further explore this question by probing the contribution of RyR1–RyR3 chimerae to the structural and functional coupling with skeletal muscle DHPRs. The sections of RyR1 that were exchanged in chimeras were selected on the basis of two criteria. One criterion is past work showing that a broad central region of the cytoplasmic domain (residues 1635–3720) contains critical elements for the RyR–DHPR structural and functional interaction (11, 12). The second criterion is the fact that RyR3 uniquely lacks the D2 region, which is present (and fairly similar) in RyR1 and RyR2. Although in the absence of D2, the RyR1 central region does, in part, confer a Ca2+-free response to RyR3–RyR1 chimerae (20), the functional restoration is quite limited. Here we demonstrate that the D2 region is a critical element in stabilizing the RyR–DHPR interaction and that this domain, in addition to other domains of the receptor, may be important to intermolecular coupling and/or the structural integrity of RyR1.
Results and Discussion
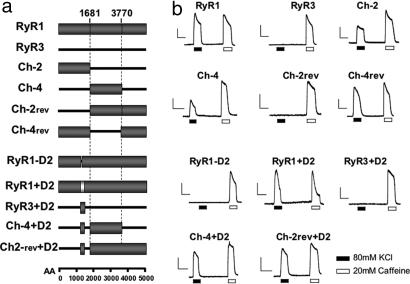
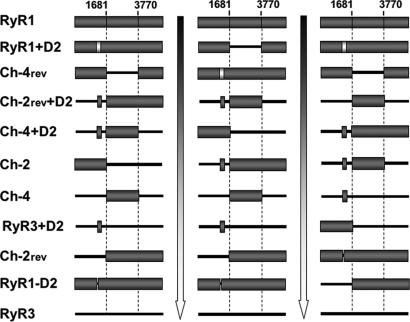
To assess the role of different domains of RyR1 on their structural/functional interaction with DHPR, two groups of chimeric RyR1–RyR3 constructs were tested in dyspedic myotubes for their effect on (i) skeletal type EC coupling, (ii) DHPR tetrad arrangement, and (iii) DHPR Ca2+ current restoration. The first group (Ch-2; Ch-4; and their respective RyR1-based reverse constructs, Ch-2rev and Ch-4rev) (Fig. 1) tested the contribution of the N-terminal and central domains of RyR1. A second group (RyR1−D2, RyR1+D2, RyR3+D2, Ch-2rev+D2, and Ch-4+D2) (Fig. 1) specifically tested the importance of the D2 region, whose critical role has been previously revealed (20, 23).
Fig. 1.
EC coupling restoration. (a) Schematic representation of chimeric RyR1–RyR3 constructs. The N-terminal and central regions of RyR1 (amino acids 1–1681 and 1682–3769, respectively) were inserted into the corresponding RyR3 sequence (Ch-2 and Ch-4). In the reverse chimeric constructs (Ch-2rev and Ch-4rev) the same residues of RyR1were replaced with RyR3 residues 1–1576 and 1577–3620. The D2 domain from RyR1 (amino acids 1272–1455) was inserted in-frame into the corresponding region of RyR3, Ch-4, and Ch-2rev (RyR3+D2, Ch-4+D2, and Ch-2rev+D2). RyR1+D2 contains RyR2 D2 (gray box, amino acids 1285–1448) in the context of the RyR1 background. (b) Representative fluorescent records of dyspedic myotubes loaded with Fluo-4. Cells were infected with different constructs and subjected to K+-depolarization and caffeine stimulation in the absence of extracellular Ca2+ plus Cd2+ and La3+. [Scale bars: vertical, 1,000 a.u. (RyR1 and RyR3) and 300 a.u. (all other graphs); horizontal, 10 s.]
Restoration of EC Coupling.
Fig. 1 illustrates representative Ca2+ transients recorded from control and chimerae-transfected myotubes in response to depolarization with 80 mM KCl in Ca2+-free solution. A large fraction of WT RyR1-expressing cells responded to KCl depolarization, showing Ca2+ transients of similar and often larger amplitude than transients induced by caffeine stimulation. As previously reported, WT RyR3 was unable to restore any depolarization-induced Ca2+ release (19, 24). The chimeric constructs had diverse responses. Constructs that included the N-terminal domain of RyR1 (Ch-4rev and Ch-2) reliably produced skeletal-type EC coupling. Replacement of the N-terminal domain of RyR1 with the corresponding RyR3 domain (Ch-2rev) resulted in a total loss of depolarization-induced Ca2+ release (Fig. 1). Exchange of the central domain of RyR1, however, seems to be less disruptive, because Ch-4rev retained a significant level of skeletal-type EC coupling. Interestingly, Ch-4, which contains the N and C termini of RyR3 and the central domain of RyR1, restored small Ca2+ transients as compared with WT RyR1. This finding suggests that there may be both positive and negative RyR–DHPR interactions.
Substitution of the RyR1 D2 region with the corresponding sequence of RyR3 (chimera RyR1−D2) (Fig. 1) resulted in the complete abolition of EC coupling ability. Substitution of the RyR1 D2 region with the corresponding domain of RyR2 (chimera RyR1+D2) produced near full restoration of the EC coupling capability of the myotubes. However, insertion of the RyR1 D2 domain into RyR3 (RyR3+D2) did not rescue the WT phenotype, showing that, although this domain is necessary, it is not sufficient to restore the EC coupling signal. This was confirmed by the fact that the addition of the RyR1 D2 region into the less efficient or silent chimeras Ch-4 and Ch-2rev (Ch-4+D2 and Ch-2rev+D2) resulted in a significant enhancement of the Ca2+ transient induced by depolarization (Fig. 1).
To determine the efficiency of all constructs to mediate skeletal type EC coupling, we measured the magnitude of the K+-induced Ca2+ transient (area under the curve). The Ca2+ transients were normalized by the caffeine response as a way of correcting for variations in the level of RyR expression, the relative volume of junctional sarcoplasmic reticulum, the extent of sarcoplasmic reticulum Ca2+ loading, and the loading of Fluo-4 (Table 1). The chimerae fall into three groups: (i) RyR1+D2, Ch-4rev, and Ch-2rev+D2 have a large WT RyR1-like response; (ii) Ch-4+D2 and Ch-2 have smaller but substantial responses; and (iii) RyR3+D2; Ch-2rev; RyR1-D2, like RyR3, have no response. Ch-4 is an outlier and produced much smaller responses than the second group, yet much larger than the third group. Whereas the insertion of the key domain of RyR1 is likely to be responsible for most of the change in EC coupling efficiency, a subtle contribution to the magnitude of the Ca2+ transient by potential changes in the Ca2+ channel properties of the chimeric receptors cannot be totally ruled out.
Table 1.
Effect of chimeric RyR1–RyR3 constructs on Ca2+-independent, K+-induced Ca2+ release in dyspedic 1B5 myotubes
| Construct | No. of cells | Cells responding to 80 mM K+, % | (F − Fo)K+/(F − Fo)Caff, % ± SE |
|---|---|---|---|
| RyR1 | 100 | 100 | 102 ± 4 |
| RyR1+D2 | 37 | 72.3 | 87 ± 5* |
| Ch-4rev | 33 | 75.8 | 85 ± 9* |
| Ch-2rev+D2 | 28 | 92.8 | 78 ± 5† |
| Ch-4+D2 | 36 | 91.7 | 66 ± 5‡ |
| Ch-2 | 39 | 82.0 | 45 ± 4‡ |
| Ch-4 | 106 | 39.0 | 29 ± 4‡ |
| RyR3+D2 | 36 | 0 | <1‡ |
| Ch-2rev | 38 | 0 | <1‡ |
| RyR1−D2 | 28 | 0 | <1‡ |
| RyR3 | 32 | 0 | <1‡ |
Signficance values were in comparison to RyR1 and were as follows:,
*, P > 0.05;
†, P < 0.01;
‡, P < 0.001.
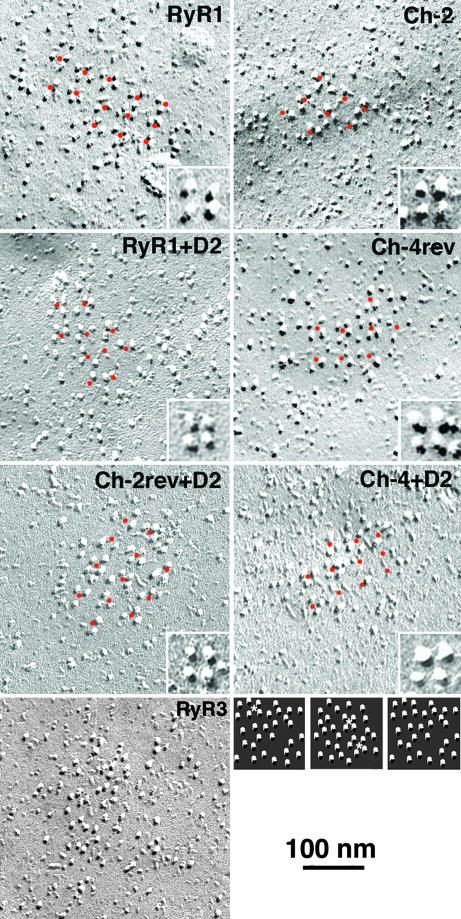
Freeze–Fracture.
As shown previously (9), the expression of RyR1 in 1B5 myotubes restores the arrays of RyR-associated DHPR tetrads (Fig. 2). RyR3, on the other hand, also associates with DHPRs at peripheral couplings, but RyR3-associated DHPRs are not arrayed into tetrads. We assessed the freeze–fracture results by determining the frequency of particles in each cluster apparently linked to an orthogonal arrangement of expected dimensions from association with an RyR lattice (11). Dots were placed in the centers of the most obvious tetrads and then were extended at equal intervals of ≈40 nm over the adjacent areas (Fig. 2). For particle clusters that did not show tetrads or any apparent order, a comparable array of dots with the appropriate spacing was superimposed on the image. Each particle within the dotted region was then classified as either belonging to the array (if appropriately clustered around the dots) or having a random disposition (if located either too close or too far from the dots). Note that in the arrays the space between ordered tetrads is mostly empty. The constructs fall into two separate groups. One group restores tetrads (Fig. 2 and Table 2) with 74–89% of the particles having an appropriate position within an orthogonal array. The remaining particles (at least 11% even in the case of WT RyR1) may represent DHPRs that were distorted during fracturing or were temporarily detached from RyRs. Within this group, Ch-4rev, RyR1+D2, and Ch-4+D2 are not significantly different in their efficacy from WT RyR1, and Ch-2 and Ch-2rev+D2 are significantly different, but the difference is very small in amplitude. A second group of constructs (RyR3+D2; Ch-2rev; and RyR1-D2, like WT RyR3) do not rescue tetrads and differ significantly from those in the first group [Table 2; see supporting information (SI) Fig. 6 and SI Methods]. Interestingly, the structural link with RyRs, as indicated by tetrad formation, seems to be either close to the level of WT RyR1 (full association) or to that of WT RyR3 (no specific association). Ch-4 is intermediate in the sense that some of its clusters have a pattern and others do not; therefore, the average clustering is low (Table 2). This may be related to the fact that this construct does not seem to effectively cluster at sites of peripheral couplings (SI Figs. 6 and 7 and SI Methods).
Fig. 2.
Freeze–fracture replicas of plasmalemma from RyRs-expressing myotubes. Like WT RyR1, Ch-4rev, Ch-4+D2, Ch-2, Ch-2rev+D2, and RyR1+D2 rescued arrays of tetrads, although some tetrads are incomplete. Orange dots marking the center of complete and incomplete tetrads appear equally spaced at ≈40-nm intervals from each other, following an orthogonal array. Presence of short-range order in the tetrad arrays for Ch-2, RyR1+D2, Ch-4rev, Ch-2rev, and Ch-4+D2 are consistent with the reduced skeletal muscle EC coupling efficiency presented by these constructs. (Insets) Higher-magnification views of representative tetrad array from each picture. Expression of WT RyR3 shows that DHPR particles do not form tetrads but are randomly arranged. The small images at the bottom right are examples of randomly dispersed particle clusters illustrating the frequency of three- to four-particle, tetrad-like arrangements (asterisks) due to random chance.
Table 2.
Reconstruction efficiency of DHPR tetrads arrays in 1B5 cells transfected with cDNA for RyR1–RyR3 chimerae
| Construct | Particles in pattern, % of total | Four- and three-particle tetrads, % of total | Density of particles, no. per 104 nm2 |
|---|---|---|---|
| RyR1 | 89.5 ± 5.8 | 67.6 ± 17.0 (n + 11) | 23.0 ± 3.3 |
| Ch-4rev | 82.9 ± 11.8 | 61.2 ± 17.7 (n = 13) | 23.6 ± 5.2 |
| RyR1+D2 | 81.0 ± 13.1 | 64.4 ± 18.9 (n = 11) | 26.5 ± 3.5 |
| Ch-4+D2 | 78.8 ± 18.0 | 52.2 ± 30.4 (n = 12) | 25.1 ± 9.6 |
| Ch-2 | 74.3 ± 13.9 | 67.5 ± 14.6 (n = 28) | 29.0 ± 5.2 |
| Ch-2rev+D2 | 74.3 ± 20.3 | 57.1 ± 27.2 (n = 13) | 17.4 ± 6.3 |
| Ch-4 | 19.3 ± 3.8*† | 32.7 ± 17.4 (n = 16) | 19.3 ± 22.1 |
| RyR3+D2 | 5.0 ± 18.7* | 13.8 ± 14.0 (n = 17) | 25.0 ± 7.9 |
| Ch-2rev | 2.1 ± 5.8* | 15.9 ± 9.3 (n = 16) | 18.6 ± 2.8 |
| RyR1−D2 | 0* | 11.6 ± 5.0 (n = 12) | 19.5 ± 3.4 |
| RyR3 | 1.2 ± 4.7* | 13.9 ± 7.4 (n = 17) | 19.1 ± 3.0 |
| Dyspedic | ND | 14.5 ± 6.0 (n = 15) | ND |
Values for n in parentheses indicate the number of clusters counted for the second and third columns. Means for all constructs between RyR1 and Ch4+D2 are not significantly different from each other. Ch-2 and Ch2rev+D2 differ significantly from RyR1 (Student's t test, P < 0.01 and 0.05, respectively) but not from each other. Data are given as means ± SD. ND, no data.
*The means for these constructs are significantly different from those listed above them (P < 0.001); however, the means for all constructs between RyR3+D2 and RyR3 are not significantly different from each other.
†The mean for Ch-4 is significantly different from all the contructs listed below it (P < 0.001).
Evaluation of the relative frequency of complete (four-particle) or almost complete (three-particle) tetrads (Table 2) supports the classification of chimerae into two groups, but may predict some minor differences, possibly due to differences in affinity. Interestingly, the constructs that do not rescue DHPR arrays as well as the DHPR clusters of nontransfected 1B5 cells still show a low frequency of particles arranged in groups of four and three that mimic tetrads. This arrangement is simply due to random chance, as shown by the fact that 30 images of “particle” groupings constructed by randomly dispersing 30 particles within small areas (Fig. 2 Insets) contained an average of 14% particles within four- and three-particle “tetrads” (Table 2). Analysis of the area and the density of particles per unit of area covered by each cluster of particles in the constructs indicates that the presence and/or absence of tetrads is not linked to large differences in particle density.
Retrograde Coupling.
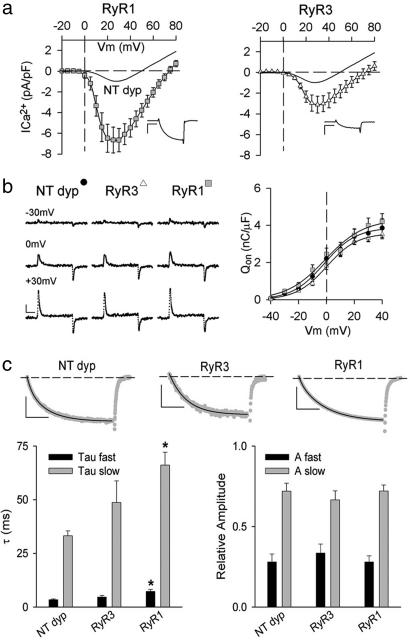
To correlate the efficiency of the structural link between RyR1 and the skeletal muscle DHPR and the degree of retrograde signaling, Ca2+ I–V relationships were obtained (Fig. 3a and Table 3). Dyspedic myotubes displayed small Ca2+ currents with peak levels no larger than 3 pA/pF and typically in the 1-pA/pF range. WT RyR1-infected cultures, on the other hand, restored Ca2+ currents similar to those seen in WT myotubes ranging between 3 and 9 pA/pF, and some ranged higher (Fig. 3a and SI Fig. 8 and SI Methods). RyR3 expression produced mixed results. RyR3 induced small Ca2+ currents in six of the 10 analyzed cells (average Ca2+ current density −1.6pA/pF). However, a significant fraction of the cells (four of 10) presented maximal Ca2+ currents at levels similar to WT RyR1 (average Ca2+ current density −5.0pA/pF). Overall, the average peak Ca2+ current of RyR3-expressing cells was smaller than that restored by RyR1, but it was also significantly higher than that observed in dyspedic myotubes (Fig. 3a and Table 3). This enhancement of thel-type Ca2+ current may be a result of the overexpression of RyR3, because double RyR1/RyR3 KO skeletal myotubes produced Ca2+ currents at levels similar to dyspedic myotubes, which include low levels of RyR3 (25, 26).
Fig. 3.
Retrograde signaling in WT RyRs. (a) Average I–V curves for WT RyR1- and WT RyR3-expressing myotubes. The curved line indicates the average peak current density for NT dyp myotubes (n = 6). (Insets) Representative Ca2+ current traces at +25 mV from a holding potential of −50 mV. (Scale bars: vertical, 2 pA/pF; horizontal, 75 ms.) (b) Representative transient charge movement traces from myotubes in response to a 20-ms depolarization from −50 mV to the given voltages. (Scale bars: vertical, 1 pA/pF; horizontal, 5 ms.) Graph shows average ON charge movement (Qon, mean ± SE) for NT dyp (n = 7), RyR3 (n = 7), and RyR1 (n = 6). Data were fit with Eq. 1. (c) Activation kinetics of Ca2+ currents. (Upper) Representative traces (gray) are shown in response to a 200-ms depolarization from −50 mV to + 30 mV. The Ca2+ currents were fit with a biexponential equation (black overlay). [Scale bars: vertical, 1 pA/pF (Left), 0.5 pA/pF (Center), and 4 pA/pF (Right); horizontal, 50 ms.] (Lower) (Left) Average activation kinetics for τ fast (black bar) and τ slow (gray bar) of NT dyp-, RyR3-, or RyR1-expressing cells (∗, P < 0.01 vs. NT dyp). (Right) The relative contributions of each activation component of the biexponential fit. Data presented as mean ± SE.
Table 3.
Average peak ionic current and Ca2+ conductance parameters restored by RyR1–RyR3 chimerae in dyspedic primary myotubes
| Construct | No. of cells | Average peak ICa2+, pA/pF ± SE | Gmax, nS/nF ± SE | V1/2, mV ± SE | K, mV ± SE |
|---|---|---|---|---|---|
| RyR1 | 14 | −6.6 ± 1.3 | 169 ± 21 | 16 ± 2 | 4.5 ± 0.4 |
| RyR1+D2 | 9 | −8.7 ± 1.5 | 228 ± 27 | 17 ± 2 | 5.2 ± 0.5 |
| Ch-4rev | 13 | −5.3 ± 0.8 | 167 ± 11 | 18 ± 1 | 4.8 ± 0.4 |
| Ch-4 | 10 | −4.6 ± 1.0 | 167 ± 20 | 21 ± 2 | 4.9 ± 0.4 |
| Ch-2rev+D2 | 9 | −3.5 ± 0.8 | 136 ± 12 | 19 ± 2 | 5.5 ± 0.4 |
| Ch-4+D2 | 15 | −4.2 ± 0.5 | 133 ± 12 | 17 ± 1 | 4.8 ± 0.3 |
| RyR3+D2 | 15 | −3.9 ± 0.5 | 123 ± 12 | 16 ± 1 | 4.8 ± 0.3 |
| Ch-2 | 15 | −3.4 ± 0.5* | 121 ± 14 | 21 ± 1 | 4.6 ± 0.4 |
| RyR1−D2 | 13 | −2.8 ± 0.4* | 106 ± 12* | 19 ± 1 | 4.7 ± 0.5 |
| Ch-2rev | 10 | −3.2 ± 0.8* | 91 ± 15* | 20 ± 2 | 5.0 ± 0.5 |
| RyR3 | 10 | −3.0 ± 0.8* | 94 ± 11* | 22 ± 2 | 5.2 ± 0.5 |
| Dyspedic | 6 | −1.0 ± 0.3# | 61 ± 6† | 22 ± 2 | 5.2 ± 0.2 |
Significance values are in comparison to RyR1 and are as follows:
*, P < 0.05;
†, P < 0.01.
Charge movement analysis of RyR-expressing cells (Fig. 3b) showed that (i) most of the charge movement in the voltage range critical for EC coupling can be assigned to the presence of α1S DHPRs, and (ii) the expression of either RyR1 or RyR3 did not greatly modify the average Qon values. These results suggest that the larger-than-dyspedic Ca2+ currents measured in RyR3-expressing cells are not related to increased expression levels of DHPR (Fig. 3b). Further analysis of the Gmax/Qmax ratios produced values in agreement with previously reported results [nontransfected dyspedic (NT dyp) myotubes = 15.6, WT RyR3 = 25.1, and WT RyR1 = 39.2] (27). Surprisingly, study of the Ca2+ current kinetics demonstrated that the expression of RyR3 slowed DHPR activation, although to a lesser extent than RyR1 (Fig. 3c). These unexpected results reveal that RyR3 seems to be capable of engaging in functional interactions with skeletal muscle DHPR that, although unable to trigger EC coupling, are adequate for partial restoration of l-type current amplitude.
Because tetrads were not observed with RyR3 expression, the enhancement of Ca2+ current would appear not to result from protein–protein interactions linking RyR3 to the DHPR. Although this retrograde coupling mechanism is unknown to date, the RyR3-driven Ca2+ current enhancement forces a rethinking of the nature of α1S DHPR–RyR retrograde coupling.
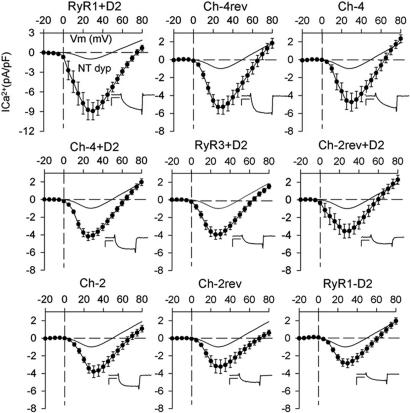
Complete I–V curves for chimeric receptors are shown in Fig. 4. Based on their maximum Ca2+ current and statistical analysis, the constructs can be classified in two groups. The first group includes RyR1+D2 and Ch-4rev, which had maximum Ca2+ currents comparable with those elicited by WT RyR1 (5 pA/pF or higher) (Table 3). RyR1+D2 showed the largest Ca2+ current recorded, which was higher than the one induced by RyR1 expression, resulting from a higher percentage of cells that had Ca2+ currents of >9 pA/pF. Interestingly, although the D2 region of RyR2 has only ≈35% similarity to the D2 region of RyR1, in the context of a RyR1 backbone, RyR2 D2 seems sufficient for superior retrograde coupling with the DHPR. The second tier of the first group, which includes Ch-4, Ch-4+D2, and RyR3+D2, have average Ca2+ currents near 4 pA/pF. Like RyR1+D2 and Ch-4rev, these chimerae seemed capable of producing large currents, but less frequently. Ch-4 proved to be more efficient at Ca2+ current restoration than at tetrad formation or EC coupling and seemed unaffected by the addition of the D2 domain (Ch-4+D2), supporting previous studies that suggest a key role for Ch-4 domain (residues 1682–3770) in retrograde signaling (12). Although the RyR3 central region in a RyR1 background (Ch-4rev) restored larger Ca2+ currents than chimera Ch-4, this does not rule out a role for this domain in Ca2+ current restoration. Because of the high degree of sequence similarity between both isoforms, it is likely that the RyR1 domain responsible for Ca2+ current restoration shares a significant structural similarity with its RyR3 counterpart. Constructs in the second group (Ch-2, Ch-2rev, Ch-2rev+D2, and RyR1-D2) have current densities very near to the RyR3 values, ≈3 pA/pF. Interestingly, with the exception of Ch-4, the Gmax values rank in close agreement with EC coupling efficiency values.
Fig. 4.
Average I–V curves for all chimeric constructs expressed in dyspedic cells. Constructs are “ranked” by their average peak current density from left to right and top to bottom. Symbols and error bars represent mean ± SE. (Insets) Current traces. [Scale bars: vertical, 4 pA/pF (RyR1+D2) and 2 pA/pF (all other graphs); horizontal, 75 ms.]
Fig. 5 ranks the analyzed constructs in terms of their effectiveness in rescuing skeletal type EC coupling (Fig. 5 Left), DHPR tetrad array (Fig. 5 Center), and enhancement of DHPR channel activity (Fig. 5 Right). We find that there is a strong correlation between the ability of the construct to rescue tetrads arrays and their efficiency of skeletal-type EC coupling and Ca2+ current restoration (Gmax). Although Ca2+ current restoration seems to be more graded than the other parameters, it is clear that the constructs that show the best tetrad arrangements (WT RyR1, RyR1+D2, and Ch-4rev) also function best over the other two measures. Accordingly, constructs that show no tetrad restoration (RyR3, RyR1-D2, Ch-2rev, and RyR3+D2) fail to restore EC coupling and are least effective at restoring Ca2+ currents.
Fig. 5.
Correlation between the presence of the structural link and the efficiency of the bidirectional signaling rescued by chimeric RyRs. Chimeric constructs are classified in descending order according to their effectiveness to restore skeletal-type EC coupling (Left), produce tetrad DHPR arrays (Center), and enhance DHPR ionic current (Gmax) (Right).
A third group of constructs (Ch-2rev+D2, Ch-4+D2, Ch-2, and Ch-4) have intermediate abilities, thus revealing the importance of the N-terminal region of RyR1 for tetrad formation. Amino acids 1–1681 of RyR1 (Ch-2) seem to be sufficient to restore normal tetrad arrays. Within this region, the divergent domain D2 was shown to be essential for tetrad restoration because its addition, or addition of the corresponding RyR2 D2 sequence, to non-tetrad-restoring constructs (i.e., RyR1-D2, Ch-2rev, and Ch-4) was sufficient to rescue normal tetrad arrays. RyR3+D2 was the exception because it failed to restore tetrads, despite the presence of the RyR1 D2 region. This result suggests two possibilities: (i) the region surrounding the D2 domain, or the context of the RyR1 sequence, also may be necessary to achieve the interaction required for tetrad arrangement, and/or (ii) this region is important for the proper tertiary structure of RyR1.
A direct involvement of the D2 domain in the structural/functional linkage to the DHPR is not obvious, and a mere conformational role is not unlikely. Recent studies (28–30) using cryoelectron microscopy reconstitution located the D2 region of RyRs in the “clamp” domain, a region thought to be important for intermolecular (with DHPRs) and intramolecular (between neighboring RyRs) signaling in a right-handed lattice array (31–33). Under this model, the lack of D2 domain could result in the weakening of RyR1–RyR1 interaction and further disruption of the RyR1 arrays, which in turn could prevent DHPR–RyR1 interaction. The results from chimerae Ch-4 and Ch-2rev show that the sole addition of the D2 domain to these constructs is enough to reestablish almost normal structural linkage yet unable to restore full functional interaction with the DHPR.
Although the D2 domain was shown to be necessary for EC coupling and tetrad array formation, it is less critical for Ca2+ current restoration. Addition of the RyR1 D2 domain into chimera Ch-2rev (Ch-2rev+D2) results in a dramatic improvement of both EC coupling and tetrad restoration efficiency but has a negligible effect on Ca2+ currents. These results suggest that the determinants of RyR1 responsible for the structural linkage and orthograde signaling may be topographically close but may not be the same as those responsible for retrograde signaling. Our results also indicate that the constructs sharing both the N-terminal and C-terminal domain of RyR1 (WT RyR1, Ch-4rev, and RyR1+D2) were the most efficient in restoring Ca2+ current. These constructs also are the most efficient in restoring tetrad arrays and EC coupling. In this regard, it is noteworthy that RyR divergent domain D1 has been mapped to the “handle” domain, which is within the proximity of the D2 domain (29, 30); thus, cooperation between the handle and clamp structures (and D1 and D2) may be vital for the functional link between DHPRs and RyR1 in skeletal muscle.
Overall, our data indicate that the D2 region of RyR1 is important for DHPR tetrad formation and voltage-dependent EC coupling, but other regions outside this N-terminal region are important for the enhancement of Ca2+ current (retrograde coupling). In fact, RyR3, which lacks the D2 region entirely, is shown to engage in functional interactions with skeletal muscle DHPR that enhances l-type Ca2+ current. This study supports our hypothesis that several independent regions of RyR1 are necessary to restore a fully functional cross-talk between DHPR and RyR1 in skeletal muscle.
Methods
Construction of Chimeric cDNAs.
Design and construction of chimeric constructs Ch-2, Ch-2rev, Ch-2rev+D2, Ch-4, Ch-4rev, Ch-4+D2, RyR3+D2, and RyR1-D2 were described previously (20). To generate chimera RyR1+D2, nucleotides 3812–4369 of RyR1 were replaced with the corresponding fragment of RyR2 (nucleotides 3852–4344) carrying divergent domain D2 (amino acids 1285–1448). cDNA constructs were cloned into an HSV-1 amplicon vector and packaged into HSV-1 virions by using a helper virus-free packaging system (34).
Cell Culturing and Ca2+ Imaging.
Ca2+ imaging and tetrad analysis were performed in dyspedic 1B5 myotubes (35) infected with viral particles containing different chimeric constructs. The cells were differentiated and imaged as described elsewhere (11, 20). The ability of the constructs to support depolarization-induced Ca2+ release was tested by exposing the cells to chemical depolarization with a 10-s exposure to high K+ buffer (50 mM NaCl/80 mM KCl/1.2 mM MgSO4/6 mM glucose/25 mM Hepes, pH 7.4) supplemented with 0.5 mM CdCl2 and 0.1 mM LaCl3 by using a multivalve perfusion system (AutoMate Scientific, Oakland, CA). In each well, the myotubes were first perfused for at least 1 min with Ca2+-free imaging buffer and then stimulated by separate 10-s exposures to Ca2+-free K+ buffer and 40 mM caffeine, with a 30-s recovery period in between. The resulting fluorescence changes were corrected for background by subtraction of the average fluorescence value in the 10 s preceding the test stimulus.
Electron Microscopy and Quantitation of Tetrad Particle Parameters.
Cells were grown and differentiated in Thermanox cover slips (Nunc, Naperville, IL). Transfected cells were washed twice in PBS at 37°C, fixed in 3.5% glutaraldehyde in 100 mM sodium cacodylate buffer (pH 7.2), infiltrated with 30% glycerol, freeze–fractured, and replicated (15, 33). Association of particles with orthogonal arrays and frequency of tetrad formation were assessed as in Protasi et al. (9, 11) and Takekura et al. (7) by using digitized images from micrographs at a magnification of ×33,900. Clusters of particles showing the best evidence of order were selected, and counts were limited to areas that had either coherent arrays of tetrads with the same orientation or an evenly distributed set of particles. For each cluster, we obtained three sets of data, which are shown in Table 2.
Measurement of Ionic Currents and Intramembrane Charge Movement.
Macroscopic Ca2+ current measurements were performed in primary cultures of dyspedic myotubes by using the whole-cell patch–clamp technique as described previously (5, 36). Myotubes were infected with 7.5 × 104 viral units per dish and examined for Ca2+ currents 24–36 h after cDNA infection. For measurement of intramembrane charge movements, ionic currents were blocked by the addition of 0.5 mM CdCl2 and 0.1 mM LaCl3 to the extracellular solution. The voltage dependence of the Ca2+ conductance and the integral of the ON transient (Qon) for each test potential (V) were fit according to Eq. 1:
where Amax is the maximal Gmax or Qmax, V1/2 is the potential causing movement of half the maximal charge, and k is a slope parameter.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant P01AR44750, National Institutes of Health Training Grant NS43115-04 (to D.C.S.), American Heart Association Grant 0530250N (to C.F.P.), and Grant-in-Aid for Scientific Research B17300210 (to H.T.).
Abbreviations
- EC
excitation–contraction
- DHPR
dihydropyridine receptor
- RyR
ryanodine receptor
- NT dyp
nontransfected dyspedic.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609473103/DC1.
References
- 1.Tanabe T, Beam KG, Adams BA, Niidome T, Numa S. Nature. 1990;346:567–569. doi: 10.1038/346567a0. [DOI] [PubMed] [Google Scholar]
- 2.Tanabe T, Mikami A, Numa S, Beam KG. Nature. 1990;344:451–453. doi: 10.1038/344451a0. [DOI] [PubMed] [Google Scholar]
- 3.Adams BA, Tanabe T, Mikami A, Numa S, Beam KG. Nature. 1990;346:569–572. doi: 10.1038/346569a0. [DOI] [PubMed] [Google Scholar]
- 4.Nakai J, Tanabe T, Konno T, Adams B, Beam KG. J Biol Chem. 1998;273:24983–24986. doi: 10.1074/jbc.273.39.24983. [DOI] [PubMed] [Google Scholar]
- 5.Nakai J, Dirksen RT, Nguyen HT, Pessah IN, Beam KG, Allen PD. Nature. 1996;380:72–75. doi: 10.1038/380072a0. [DOI] [PubMed] [Google Scholar]
- 6.Avila G, Dirksen RT. J Gen Physiol. 2000;115:467–480. doi: 10.1085/jgp.115.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takekura H, Nishi M, Noda T, Takeshima H, Franzini-Armstrong C. Proc Natl Acad Sci USA. 1995;92:3381–3385. doi: 10.1073/pnas.92.8.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schredelseker J, Di Biase V, Obermair GJ, Felder ET, Flucher BE, Franzini-Armstrong C, Grabner M. Proc Natl Acad Sci USA. 2005;102:17219–17224. doi: 10.1073/pnas.0508710102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Protasi F, Takekura H, Wang Y, Chen SR, Meissner G, Allen PD, Franzini-Armstrong C. Biophys J. 2000;79:2494–2508. doi: 10.1016/S0006-3495(00)76491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felder E, Protasi F, Hirsch R, Franzini-Armstrong C, Allen PD. Biophys J. 2002;82:3144–3149. doi: 10.1016/S0006-3495(02)75656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Protasi F, Paolini C, Nakai J, Beam KG, Franzini-Armstrong C, Allen PD. Biophys J. 2002;83:3230–3244. doi: 10.1016/S0006-3495(02)75325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakai J, Sekiguchi N, Rando TA, Allen PD, Beam KG. J Biol Chem. 1998;273:13403–13406. doi: 10.1074/jbc.273.22.13403. [DOI] [PubMed] [Google Scholar]
- 13.Franzini-Armstrong C, Protasi F, Ramesh V. Ann NY Acad Sci. 1998;853:20–30. doi: 10.1111/j.1749-6632.1998.tb08253.x. [DOI] [PubMed] [Google Scholar]
- 14.Franzini-Armstrong C, Protasi F, Ramesh V. Biophys J. 1999;77:1528–1539. doi: 10.1016/S0006-3495(99)77000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takekura H, Paolini C, Franzini-Armstrong C, Kugler G, Grabner M, Flucher BE. Mol Biol Cell. 2004;15:5408–5419. doi: 10.1091/mbc.E04-05-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregg RG, Messing A, Strube C, Beurg M, Moss R, Behan M, Sukhareva M, Haynes S, Powell JA, Coronado R, Powers PA. Proc Natl Acad Sci USA. 1996;93:13961–13966. doi: 10.1073/pnas.93.24.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beurg M, Ahern CA, Vallejo P, Conklin MW, Powers PA, Gregg RG, Coronado R. Biophys J. 1999;77:2953–2967. doi: 10.1016/S0006-3495(99)77128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheridan DC, Cheng W, Ahern CA, Mortenson L, Alsammarae D, Vallejo P, Coronado R. Biophys J. 2003;84:220–237. doi: 10.1016/S0006-3495(03)74844-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez CF, Voss A, Pessah IN, Allen PD. Biophys J. 2003;84:2655–2663. doi: 10.1016/S0006-3495(03)75071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez CF, Mukherjee S, Allen PD. J Biol Chem. 2003;278:39644–39652. doi: 10.1074/jbc.M305160200. [DOI] [PubMed] [Google Scholar]
- 21.Carbonneau L, Bhattacharya D, Sheridan DC, Coronado R. Biophys J. 2005;89:243–255. doi: 10.1529/biophysj.104.056218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coronado R, Ahern CA, Sheridan DC, Cheng W, Carbonneau L, Bhattacharya D. Biol Res. 2004;37:565–575. doi: 10.4067/s0716-97602004000400010. [DOI] [PubMed] [Google Scholar]
- 23.Yamazawa T, Takeshima H, Shimuta M, Iino M. J Biol Chem. 1997;272:8161–8164. doi: 10.1074/jbc.272.13.8161. [DOI] [PubMed] [Google Scholar]
- 24.Fessenden JD, Wang Y, Moore RA, Chen SR, Allen PD, Pessah IN. Biophys J. 2000;79:2509–2525. doi: 10.1016/S0006-3495(00)76492-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeshima H, Yamazawa T, Ikemoto T, Takekura H, Nishi M, Noda T, Iino M. EMBO J. 1995;14:2999–3006. doi: 10.1002/j.1460-2075.1995.tb07302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheridan DC, Carbonneau L, Ahern CA, Nataraj P, Coronado R. Biophys J. 2003;85:3739–3757. doi: 10.1016/S0006-3495(03)74790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avila G, O'Connell KM, Groom LA, Dirksen RT. J Biol Chem. 2001;276:17732–17738. doi: 10.1074/jbc.M009685200. [DOI] [PubMed] [Google Scholar]
- 28.Sharma MR, Jeyakumar LH, Fleischer S, Wagenknecht T. J Biol Chem. 2000;275:9485–9491. doi: 10.1074/jbc.275.13.9485. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Zhang J, Wang R, Wayne Chen SR, Wagenknecht T. J Mol Biol. 2004;338:33–45. doi: 10.1016/j.jmb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Zhang J, Li P, Chen SR, Wagenknecht T. J Biol Chem. 2002;277:46712–46719. doi: 10.1074/jbc.M208124200. [DOI] [PubMed] [Google Scholar]
- 31.Yin CC, Blayney LM, Lai FA. J Mol Biol. 2005;349:538–546. doi: 10.1016/j.jmb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Wagenknecht T, Hsieh CE, Rath BK, Fleischer S, Marko M. Biophys J. 2002;83:2491–2501. doi: 10.1016/S0006-3495(02)75260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paolini C, Protasi F, Franzini-Armstrong C. J Mol Biol. 2004;342:145–153. doi: 10.1016/j.jmb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Fraefel C, Protasi F, Moore RA, Fessenden JD, Pessah IN, DiFrancesco A, Breakefield X, Allen PD. Am J Physiol. 2000;278:C619–C626. doi: 10.1152/ajpcell.2000.278.3.C619. [DOI] [PubMed] [Google Scholar]
- 35.Moore RA, Nguyen H, Galceran J, Pessah IN, Allen PD. J Cell Biol. 1998;140:843–851. doi: 10.1083/jcb.140.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beam KG, Franzini-Armstrong C. Methods Cell Biol. 1997;52:283–306. doi: 10.1016/s0091-679x(08)60384-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.