Abstract
Multiple phosphorylation of β-catenin by glycogen synthase kinase 3 (GSK3) in the Wnt pathway is primed by CK1 through phosphorylation of Ser-45, which lacks a typical CK1 canonical sequence. Synthetic peptides encompassing amino acids 38–64 of β-catenin are phosphorylated by CK1 on Ser-45 with low affinity (Km ≈1 mM), whereas intact β-catenin is phosphorylated at Ser-45 with very high affinity (Km ≈200 nM). Peptides extended to include a putative CK1 docking motif (FXXXF) at 70–74 positions or a F74AA mutation in full-length β-catenin had no significant effect on CK1 phosphorylation efficiency. β-Catenin C-terminal deletion mutants up to residue 181 maintained their high affinity, whereas removal of the 131–181 fragment, corresponding to the first armadillo repeat, was deleterious, resulting in a 50-fold increase in Km value. Implication of the first armadillo repeat in β-catenin targeting by CK1 is supported in that the Y142E mutation, which mimics phosphorylation of Tyr-142 by tyrosine kinases and promotes dissociation of β-catenin from α-catenin, further improves CK1 phosphorylation efficiency, lowering the Km value to <50 nM, approximating the physiological concentration of β-catenin. In contrast, α-catenin, which interacts with the N-terminal region of β-catenin, prevents Ser-45 phosphorylation of CK1 in a dose-dependent manner. Our data show that the integrity of the N-terminal region and the first armadillo repeat are necessary and sufficient for high-affinity phosphorylation by CK1 of Ser-45. They also suggest that β-catenin association with α-catenin and β-catenin phosphorylation by CK1 at Ser-45 are mutually exclusive.
Keywords: casein kinase 1, Wnt pathway, substrate recruitment, α-catenin
Protein kinase CK1 (formerly casein kinase 1) makes up a separate subfamily within the superfamily of eukaryotic protein kinases (1). In vertebrates, there are genes encoding for seven CK1 isoforms (α, β, γ1, γ2, γ3, δ, and ε), which also generate different proteins through alternative splicing (2–5). All CK1 isoforms display a high degree of sequence identity within their kinase domains (>50%), but they differ significantly in their N- and C-terminal extensions.
CK1 has been implicated in a wide variety of cellular processes, including chromosome segregation (6, 7), spindle formation (8–10), circadian rhythm (11), nuclear import (12), Wnt pathway (13–18), and apoptosis (19, 20). Deregulation of CK1 isoforms has been observed in neurodegenerative and sleeping disorders (21–23) and in cancer (24–27).
Implication in so many functions implies the ability to recognize specifically the physiologically relevant phosphoacceptor sites in its protein targets. Pertinent to target site recognition is the perplexing question concerning the consensus sequence(s) recognized by CK1. Early studies, mostly performed with artificial substrates (casein and synthetic peptides), revealed that CK1 is a “phosphate-directed” protein kinase able to phosphorylate with high efficiency Ser/Thr residues specified by a prephosphorylated side chain (either pS or pT) at position n–3 (or less effectively n–4) (28–31). This observation led to the concept of “hierarchical phosphorylation” (32) and the term “primed phosphorylation” to indicate the ability to phosphorylate residue(s) specified by another phosphorylated residue at a predetermined (critical) position. This feature is shared by a small number of acidophilic Ser/Thr kinases, notably CK1 and CK2, glycogen synthase kinase 3 (GSK3), and the Golgi apparatus casein kinase (G-CK) (33). However, primed phosphorylation by CK2 and G-CK is optional because the phosphorylated determinant can be effectively replaced by a carboxylic side chain. In the case of CK1 and GSK3, such a hierarchical mechanism was initially held as a rule. It soon became clear, however, that CK1 can also phosphorylate nonprimed substrates in which the phosphorylated determinant is surrogated by a cluster of acidic residues at positions upstream from n–2 (e.g., DARP-P32) (34, 35) and the peptides routinely used to assay CK1 contain nonprimed acidic consensus sequences (36). Recently, however, the incontrovertible identification of CK1 phosphorylation sites in its physiological targets brought an additional level of complexity to the issue because several residues phosphorylated by CK1 neither are primed by prephosphorylation nor specified by acidic residues upstream. Systematic studies with peptide substrates derived from NF-AT4, β-catenin, and APC, combined with mutational analyses of full-length β-catenin and CK1α (37, 38) have provided evidence that the phosphorylation of these noncanonical sequences is dictated by atypical specificity determinants, notably a hydrophobic side chain at position n+1 and an acidic cluster 5–11 residues downstream. These atypical determinants are equally important in the peptide and in the protein substrate because their alteration also compromises the phosphorylation of full-length β-catenin by CK1 (37).
In the course of these studies, it became clear that although primed peptide substrates display Km values in the low micromolar range, noncanonical peptides, albeit phosphorylated with a similar rate, invariably display high Km values close to or higher than 1 mM (37, 38), levels incompatible with physiological conditions. A reasonable explanation could have been that the phosphorylation of noncanonical sequences does not occur in vivo unless driven by the formation of supramolecular complexes of the substrate and CK1, such as the “degradation complex” generated in the Wnt pathway where axin bridges CK1 and GSK3 to β-catenin. Although this mechanism of substrate/CK1 recruitment may play a role in the phosphorylation of β-catenin (15), it does not account for the observation that even in vitro and in the absence of interacting proteins full-length β-catenin is phosphorylated by CK1 at its noncanonical site (Ser-45) with an affinity 3–4 orders of magnitude higher than found for a 28-aa peptide that includes the same site (37). This result means that β-catenin possesses functional element(s) committed to its high-affinity recognition by CK1 independent of the sequence encompassing the phosphoacceptor site (Ser-45). A first choice for this element is a sequence (FSQSF) conforming the CK1 putative docking motif identified in NFAT1 whose mutation disrupts NFAT1–CK1 interaction (39) and that is present in proteins of the Wnt, Hedgehog, and circadian-rhythm pathways.
These considerations prompted us to undertake a study aimed at assessing the relevance of this motif and other structural elements of β-catenin in relation to the high-affinity phosphorylation of Ser-45, which marks β-catenin for degradation.
Results
The structures of β-catenin and fragments used in this study are shown schematically in Fig. 1. The central core of β-caternin contains 12 armadillo repeats that form a highly ordered structural domain committed to interaction with a variety of proteins including axin, APC, α-catenin, and E-cadherin. In contrast, the N- and C-terminal domains are disordered: the C-terminal is implicated in transcriptional functions of β-catenin, whereas the N-terminal region includes residues that commit β-catenin to ubiquitination and degradation through a process initiated by phosphorylation of Ser-45 by CK1, which primes hierarchical phosphorylation of Thr-41, Ser-37, and Ser-33 by GSK3 (14, 15).
Fig. 1.
Schematic representation of full-length β-catenin, and deletion and point mutants that were used in this work.
Phosphorylation of Ser-45 by CK1 also occurs in vitro, by using either full-length β-catenin or synthetic peptides derived from the native sequence surrounding Ser-45 (37). The kinetic constants for CK1 phosphorylation reactions, however, are sharply different when using either the protein β-catenin or synthetic peptides as substrate. In Table 1, a difference of 3–4 orders of magnitude higher affinity is seen with β-catenin substrate compared with the Ser-45 peptide encompassing residues 38–65 (Km 0.19 vs. 1,240 μM).
Table 1.
Kinetic constants for the phosphorylation of β-catenin and its fragments by CK1α
| β-Catenin and derivatives | Km, μM | Vmax* | Vmax/Km |
|---|---|---|---|
| β-CatWT | 0.19 | 3.2 | 16.8 |
| β-CatF74A | 0.11 | 2.2 | 20.0 |
| β-Cat1–422 | 0.30 | 0.72 | 2.4 |
| β-Cat1–181 | 0.10 | 0.54 | 5.4 |
| β-Cat1–131 | 5.00 | 11.8 | 2.4 |
| β-Cat1–80 | 50.0 | 18.4 | 0.37 |
| Pept. 38–80 | 460.0 | 13.2 | 0.028 |
| Pept. 38–64 | 1,240 | 26.1 | 0.021 |
Kinetic constants were determined as described in Methods. The values represent the means obtained from at least three independent experiments with SEM never exceeding 15%.
*Vmax is expressed as picomoles of phosphate transferred per minute per microgram of enzyme.
A possible explanation for this high-affinity phosphorylation of the intact protein compared with the peptide could be provided by an FSQSF sequence at positions 70–74, which is not present in the peptide 38–64. Sequences characterized by the motif FXXXF have been proposed to represent interaction modules specifically recognized in substrates of CK1 (39). The possible implication of the F70SQSF motif in the high-affinity phosphorylation of β-catenin was studied with two substrates: a synthetic peptide encompassing amino acids 38–80 of β-catenin, which includes the FSQSF motif, and full-length β-catenin, in which the second phenylalanine residue is mutated to alanine (β-cat F74A). As shown in Table 1, both modifications had only marginal effects on phosphorylation efficiency by CK1. The C-terminal extension of the peptide to include FSQSF induced a <3-fold decrease of Km that remained >3 orders of magnitude higher than full-length β-catenin. On the other hand, the β-cat F74A mutant did not increase but rather slightly decreased the Km value, ruling out the relevance of the FXXXF motif in determining high-affinity phosphorylation by CK1.
To determine the minimum size of β-catenin fragments still undergoing high-affinity phosphorylation by CK1, C-terminally truncated β-catenin derivatives were generated. These fragments (Fig. 1) were assayed as substrates of CK1. The kinetic constants indicate that removal of the entire C-terminal segment and all but one of the 12 repeated armadillo domains results in a product still compatible with Km values in the nanomolar range, as seen with the full-length protein. However, a dramatic 50-fold rise in Km occurs on deletion of the 131–181 segment containing the single remaining armadillo repeat. Thus, the shifting of the kinetic behavior from “protein-like” to “peptide-like” depends on the presence of a single armadillo repeat, manifest by this very high increase in Km (Table 1).
To reinforce this conclusion, we reasoned that a peptide reproducing the first armadillo repeat should prove able to counteract in a competitive manner the high-affinity phosphorylation of β-catenin. Such experiments, however, performed with either a recombinant GST-β-Cat131–181 fragment or with a synthetic peptide reproducing the 131–181 sequence of the first armadillo repeat were unable to affect β-catenin phosphorylation by CK1 significantly if added in up to a 10-fold molar excess over β-catenin (data not shown).
A possible explanation for the failure could be that the isolated armadillo repeat is unable to attain a correct conformation for tight interaction with CK1. Indeed, neither recombinant nor the synthetic 131–181 fragments show any secondary structure if analyzed by CD (Fig. 2), whereas a larger fragment (131–422), including the first armadillo repeat together with the following five, is obviously folded.
Fig. 2.
CD spectrum of the β-catenin fragments used for competition assays. The percentage of α-helix was calculated as described in Methods.
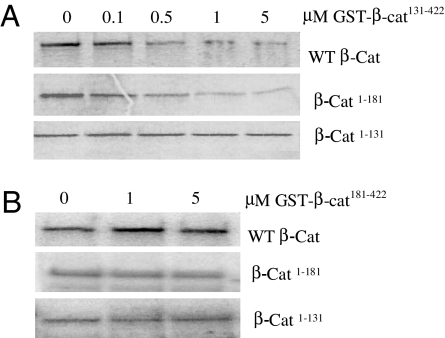
It was observed (Fig. 3A) that the 131–422 fragment inhibits β-catenin phosphorylation by CK1 in a dose-dependent manner. Two other observations are pertinent. First, the inhibitory efficacy of the 131–422 fragment is quite evident if the β-catenin1–181 fragment is used as substrate; however, little or no inhibition is seen with the shorter fragment substrate β-catenin1–131 whose low-affinity phosphorylation by CK1 does not depend on the integrity of the first armadillo repeat. Second, if the 131–422 fragment is replaced by 181–422, still folded as judged from the CD spectra (Fig. 2) but deprived of the first armadillo repeat, the inhibitory potency against β-catenin phosphorylation almost disappears (Fig. 3B).
Fig. 3.
Inhibition of β-catenin phosphorylation by nonphosphorylatable β-catenin fragments depends on the first armadillo repeat. Phosphorylation of β-cateninWT and deletion mutants (100 nM) by CK1α was determined in the presence of increasing concentrations of GST-β-Cat131–422 (A) or fragment GST-β-Cat181–422 (B), as described in Methods.
It can be concluded therefore that high-affinity phosphorylation of β-catenin Ser-45 by CK1 relies on the integrity of an N-terminal region including the first armadillo repeat. This repeat segment directly contributes to the binding of the kinase, as shown by the competition experiments (Fig. 3). However, the formation of stable complexes could not be detected by a number of approaches: BIAcore X10 surface plasmon resonance, pulldown, and far Western blots (data not shown).
Additional evidence that the first armadillo repeat plays a crucial role in the phosphorylation of Ser-45 was provided by mutating Tyr-142, present in this repeat and shown to be critical in determining the dissociation of β-catenin from α-catenin. Notably, dissociation is promoted by Tyr-142 phosphorylation by a number of tyrosine kinases, notably Fer and Fyn (40), an effect that can be mimicked by mutating Tyr-142 to glutamic acid (40). The β-catenin Y142E mutant is in fact constitutively unable to bind α-catenin. Interestingly, the same mutation significantly improves the efficiency of Ser-45 phosphorylation in intact β-catenin by inducing a 5-fold drop in the Km value (Fig. 4A), which is in the range of the physiological concentration of β-catenin. The effect of Y142E on the phosphorylation of Ser-45 by the protein kinase CK1 is also demonstrated by using an antibody specific for phosphoserine-45 (Fig. 4B). This finding provides an additional argument to support the concept that the first armadillo repeat is functionally linked to phosphorylation of Ser-45 by CK1.
Fig. 4.
Phosphorylation efficiency of β-catenin Ser-45 by CK1 is increased by the Y142E mutation. (A) β-CateninWT and the Y142E mutant were subjected to phosphorylation with CK1α as described in Methods. (B) Twenty micromolar GST-β-CatWT and the Y142E mutant were phosphorylated by CK1α. Phosphorylated products were detected by Western blotting with an anti-phosphoserine-45 (anti-pS45) antibody, and β-catenin was detected by using an antibody that recognizes full-length β-catenin. *, Vmax is expressed as picomoles of phosphate transferred per minute per microgram of enzyme.
The phosphorylation of Ser-45 is prevented in a dose-dependent manner by α-catenin (Fig. 5A), which interacts with a region of β-catenin partially encompassing its N-terminal domain and its first armadillo repeat (41, 42). As seen in Fig. 5B, such an inhibition is even more pronounced if phosphorylation of Ser-45 is monitored by using the β-catenin 1–181 fragment, whereas the low-affinity phosphorylation occurring with the 1–131 fragment (lacking the first armadillo repeat) is unaffected by α-catenin.
Fig. 5.
α-Catenin prevents β-catenin phosphorylation by CK1 in a dose-dependent manner. (A) Phosphorylation of β-cateninWT by CK1α was tested in the presence of increasing concentrations of α-catenin. (B) The phosphorylation by CK1 of either β-cateninWT and deletion mutants (all 100 nM) was assayed in the absence or presence of 1 μM α-catenin. In both A and B, radioactive products were subjected to SDS/PAGE and autoradiography.
Discussion
β-Catenin is a crucial integrator of cell–cell adhesion and gene transcription processes. It participates at the plasma membrane by bridging cadherins to α-catenin, and on release from the plasma membrane it faces two different fates: either it is ubiquitinated and destroyed, or it is free to enter the nucleus and perform a role in activating an array of genes, many of which are implicated in cell survival and proliferation. Therefore, the signaling function of β-catenin is determined by an enhancement of its stability which is dictated by the Wnt pathway through the inhibition of a multiphosphorylation step, which is a prerequisite for its subsequent ubiquitination and degradation.
As predictable, the decision of promoting cell–cell adhesion, self-destruction, or signaling is determined by a finely tuned network of interconnected regulatory mechanisms whose alterations are often causative of cancer (43, 44). Not surprisingly, several protein kinases, both Ser/Thr- or Tyr-specific, are implicated in these regulatory mechanisms. Two tyrosyl residues in the first (Tyr-142) and last (Tyr-654) armadillo repeats are essential to bind β-catenin to α-catenin and to E-cadherin, respectively (44–46), and therefore their phosphorylation switches β-catenin from its adhesive to its transcriptional function. However, phosphorylation of Tyr-142, but not of Tyr-654, also promotes the binding of β-catenin to the nuclear cofactor BCL9-2 (47), thus favoring its transcriptional role, whereas phosphorylation of Tyr-654 leaves the transcriptional and degradation options equally open. Degradation is crucially dependent on N-terminal phosphorylation at Ser/Thr residues by GSK3, which is held as the main mediator of the Wnt signaling pathway, which causes the inhibition of GSK3, thus “sparing” cytosolic β-catenin for its transcriptional functions.
Although the central role of GSK3 in Wnt signaling was soon recognized, only later did CK1 come into the scene (13). At present, there are solid arguments to implicate CK1 in the regulation of the Wnt pathway at several levels: the first indication came from its association with and phosphorylation of dishevelled (Dvl) (13), a component of the Wnt pathway also implicated in the inhibition of GSK3, followed by reports showing that CK1 phosphorylates two components of the β-catenin degradation complex, axin (48) and APC (49), and that it plays a critical role in the regulation of the LRP6 proteins, which act as coreceptors (16–18) of Wnt.
However, the most critical intervention of CK1 in determining the fate of β-catenin (or its Drosophila homolog armadillo) takes place on β-catenin itself, whose phosphorylation at Ser-45 by CK1 is required for subsequent hierarchical phosphorylation by GSK3 of 3 residues upstream, which marks β-catenin for ubiquitination and proteasomal degradation (14, 15, 50). A mechanism involving two kinases came as a surprise in two respects. First, β-catenin phosphorylation by GSK3 was believed not to be primed by previous phosphorylation (51, 52), as in the case of glycogen synthase and the majority of GSK3 targets. Second, Ser-45 does not display the typical features of canonical CK1 phosphoacceptor sites, specified by either a phosphorylated residue at position n–3 or an acidic cluster upstream from position n–2 (ref. 33 and refs. therein). Nevertheless, phosphorylation of Ser-45 β-catenin by CK1 has been well established both in vitro and in vivo, and it has been shown to take place by a mechanism that is different from that used by CK1 to recognize canonical sites (37, 38) because it is not impaired by mutation of basic residues (229KKQK232) of CK1 (38). The local determinants for such an atypical phosphorylation were defined in ref. 37 as consisting of an SLS motif followed by a downstream cluster of acidic residues. Such local determinants are important also in full-length β-catenin because its phosphorylation by CK1 is severely impaired by their alteration (37). They do not account, however, for the outstanding efficiency of β-catenin phosphorylation by CK1, displaying Km values in the nanomolar range, whereas even the best peptide substrates are phosphorylated with Km values close to 1 mM (37).
This efficiency might relate to a putative CK1 docking motif (FXXXF) found in NFAT transcription factors and also present in Wnt, Hedgehog, and circadian-rhythm proteins (39). In β-catenin, such a motif is present downstream from Ser-45, at positions 70–74 (FSQSF). However, we show here that neither inclusion of this motif in the peptide substrate nor its disruption in the full-length protein (mutant β-CatF74A) significantly affected the kinetics of phosphorylation. We conclude that in the case of β-catenin, the role of the FXXXF motif as a CK1 docking site is a marginal one. It is noteworthy that in our hands β-catenin does not make stable interactions with CK1, based on a number of experimental approaches, namely, surface plasmon resonance, far Western blotting, and pulldown assays.
The gradual truncation of β-catenin on its C-terminal side showed that the integrity of the whole molecule is not required for high-affinity phosphorylation, which is still observed by using a 181-aa N-terminal fragment that includes the first armadillo repeat. A further truncation to residue 131, which removes the first armadillo repeat, had a strongly detrimental effect on the Km value paralleled by a rise in Vmax, both reminiscent of the kinetic behavior of β-catenin-derived peptide substrates. The implication of the first armadillo repeat in the mechanism underlying efficient Ser-45 phosphorylation is corroborated by two other observations. First, a recombinant fragment of β-catenin encompassing the armadillo repeats 1–6 but lacking the first repeat is able to inhibit in a dose-dependent manner the high-affinity phosphorylation by CK1 of either full-length β-catenin or its 1–181 fragment, but it has no effect on the low-affinity phosphorylation of the 1–131 fragment. Second, in the first armadillo repeat a mutation of Tyr-142 to glutamate significantly improves the phosphorylation of Ser-45 by causing a 4-fold drop in Km. This outcome is of special interest because Tyr-142 phosphorylation is crucial for switching β-catenin from adhesion to a transcriptional role by weakening its binding to α-catenin while promoting its association with the nuclear cofactor BCL9-2 (47). Interestingly, the effects of Tyr-142 phosphorylation are mimicked by its mutation to glutamic acid (45). Thus, phosphorylation of Tyr-142, besides promoting the dissociation of β-catenin from the membrane and favoring its association with BCL9-2, could also favor the phosphorylation of Ser-45 by CK1. Note in this respect that, considering that the cellular concentration of β-catenin ranges between 30 and 50 nM (53), the decrease in Km induced by the Y142E mutation may be critical to permit cytosolic phosphorylation of β-catenin by CK1 in an axin-independent fashion.
A pool of α-catenin is also found in cytoplasm where it forms homodimers or heterodimers with β-catenin (54). The β-catenin region implicated in this interaction partially overlaps the N-terminal domain whose integrity is shown in this work to be essential for high-affinity phosphorylation of Ser-45 by CK1 (42). Our data would indicate that heterodimerized β-catenin is no longer susceptible to high-affinity phosphorylation by CK1, suggesting that phosphorylation by CK1 and association with α-catenin indicate opposite functions of β-catenin. This observation also suggests that Wnt signaling might be potentiated by factors that increase cytosolic α-catenin concentrations.
The involvement of the first armadillo repeat in the α-catenin inhibition of β-catenin phosphorylation by CK1 is supported by the fact that no inhibition is observed with fragment 1–131, which lacks this repeat.
Because the only known function of Ser-45 phosphorylation is to prime the intervention of GSK3, it is expected that the Wnt signaling pathway, besides blocking GSK3, may also prevent its priming by inhibiting CK1. A candidate for this inhibitory role could be Dvl, whose ability to interact with CK1 (13) and to prevent its phosphorylation of β-catenin Ser-45 (15) has been reported. Dvl is also implicated in the inhibition of GSK3 by the Wnt pathway (55), although, in this case, alternative mechanisms of inhibition have been proposed (56).
Independent of the above speculations, the mechanistic aspects of this work lead to the definition of a functional domain of β-catenin that encompasses the N-terminal region and the first armadillo repeat and that is instrumental to high-affinity phosphorylation of Ser-45 by CK1. The results presented also indicate that this first armadillo repeat acts as a target for the regulation of CK1 Ser-45 phosphorylation by extrinsic factors such as tyrosine kinases and α-catenin.
These observations also suggest that recognition of substrates by CK1 may differ depending on whether they contain canonical or noncanonical target sequences. Canonical sequences are efficiently recognized per se, whereas noncanonical sequences are too weak to be phosphorylated efficiently, and they require additional structure of determinants to make them functional substrates.
Methods
Materials.
Solvents, resins, and coupling reagents for peptide synthesis were from Applied Biosystems (Foster City, CA). Protected amino acids were from Novabiochem (Laufelfingen, Switzerland). Ni2+–nitrilotriacetic acid–Sepharose was from Novagen (EMD Biosciences, affiliate of Merck, Darmstadt, Germany). [γ-32P]ATP and the enhanced chemiluminescence antibody system were from Amersham (Piscataway, NJ). Monoclonal antibody against the His6 tag was from Clontech (BD Biosciences, San Diego, CA). Monoclonal antibody against β-catenin and anti-phosphoserine-45 were from Sigma (St. Louis, MO). Plasmid pGEX-2T for β-catenin from rat was a gift from A. Kikuchi, Hiroshima University, Japan.
β-Catenin Mutants.
β-Cat1–181 and β-Cat1–422 mutants were generated by cutting pGEX-2Tβ-catenin with HindIII and EcoRI, respectively. β-Cat1–131 was obtained with upstream primer 5′-GTTCCGCGTGGATCCCCGGG-3′ and downstream primer 5′-TATATAGAATTCTGACGGTTCAGCCAAGCG-3′. β-Cat1–80 was obtained with primers 5′-GTTCCGCGTGGATCCCCGGG-3′ and 5′-TATATAGAATTCAGCTACTTGCTCTTGCG-3′. β-Cat131–422 was obtained with primers TATATAGGATCCATGCTGAAACATGCAGTTGTC-3′ and 5′-TATATAGAATTCTCAGAAGAGGGAACTGGTCA-GCTC-3′. β-Cat181–422 was obtained with primers 5′-TATATAGGATCCAAGGAAGCTTCCAGACACGC-3′ and 5′-TATATAGAATTCTCAGAAGAGGGAACTGGTCA-GCTC-3′. Mutants F74A and Y142E were generated by using a QuikChange kit (Stratagene, La Jolla, CA). Mutant β-Cat-F74A used primers 5′-CCAGTCCGCCAACCAAGAGCAAGTAGCAGCAGACATCGATGGTC-3′ and 5′-GACCATCGATGTCTGCTGCTACTTGCTCTTGGTTGGCGGACTGG-3′. For mutant β-Cat-Y142E primers 5′-CAATTTGATTAACGAACAGGATGACGCG-3′ and CGCGTCATCCTGTTCGTTAATCAAATTG-3′ were used. All mutations were confirmed by DNA sequencing.
Protein Purification and Peptide Synthesis.
CK1α from zebrafish with N-terminal His tags was expressed in Escherichia coli BL21(DE3) and purified on Ni2+–nitrilotriacetic acid–agarose as described in ref. 5. GST fusion proteins were purified by using glutathione–agarose beads and elution with 20 mM glutathione dissolved in PBS. Synthesis of peptides derived from Xenopus laevis was carried out as described in ref. 37.
35S Labeling of CK1α.
Proteins were expressed by using TNT-coupled transcription/translation reticulocyte lysate assay (Promega, Madison, WI) according to the manufacturer's recommendations by using [35S]methionine (Amersham).
Pulldown Assays.
Assays were performed by incubating 10–20 mg of GST-β-catenin fragment fusion proteins for 1 h with glutathione–Sepharose 4B beads in a buffer containing 20 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1% (vol/vol) glycerol, and 0.1% Triton X-100. The complex was washed three times with the same buffer, and beads were incubated with CK1α labeled either with 35S or with 32P by autophosphorylation. GST-axin was used as a positive control.
Measurements of Kinetic Constants.
The assay has been described in detail in ref. 5. Briefly, a range of concentrations of substrate were phosphorylated by incubation in a 30-μl volume containing 50 mM Tris·HCl buffer (pH 7.5), 10 mM MgCl2, 100 mM NaCl, and 100 μM [γ-32P]ATP (specific radioactivity, 500–1,000 cpm/pmol). The reactions were incubated for 10 min at 37°C. For peptide substrates, the reaction was stopped by absorption on phosphocellulose p81 paper. Papers were washed three times with 75 mM phosphoric acid, dried, and counted in a scintillation counter. For protein substrates, the reaction was stopped by the addition of 5× concentrated Laemmli buffer followed by SDS/PAGE and autoradiography. Phosphate incorporated was quantified by the p81 phosphocellulose filter procedure and also by SDS/PAGE, autoradiography, and measurement of the excised protein in a scintillation counter. For Km and Vmax values, initial rate data were fitted to the Michaelis–Menten equation with the program Prism (Prism Software, San Diego, CA).
Western Blot Analysis.
A 20 μM concentration of each GST-β-CatWT and the mutant Y142E was phosphorylated by CK1α and unlabeled ATP. The reaction was stopped by using 5× concentrated Laemmli buffer; SDS/PAGE was performed, and proteins were transferred to PVDF membranes. The phosphorylated product was detected by using the anti-phosphoserine-45 antibody and by enhanced chemiluminescent label. As a control of quantity of protein, the membrane was stripped and probed against anti-β-catenin antibody. For far Western blot analysis, SDS/PAGE was performed with 10 μg of β-catenin or different mutants followed by electroblotting to PVDF membranes. The membrane was blocked with 3% (wt/vol) BSA for 30 min and incubated with His-CK1α for 3 h at 4°C. After three washes with PBS, the membrane was incubated with anti-His6 antibody and detected by enhanced chemiluminescent label. The experiments were repeated three times with similar results.
Surface Plasmon Resonance Analysis.
Interactions of CK1δ protein with the mutants GST-β-catenin fragments were studied by surface plasmon resonance with a BIAcore X10 system (Biacore, Uppsala, Sweden). Purified CK1δ protein (from zebrafish) was immobilized on a certified CM5 sensor chip by using an amine-coupling kit. The HBS-EP buffer was used for diluting all of the species injected in BIAcore. Kinetic experiments were carried out at 25°C in 70 μl with a constant buffer flow rate of 10 μl/min, and no typical mass transport effect was observed. Regeneration of the sensor chip, sensorgrams normalization, and fitting of the binding curves were performed as reported in ref. 57.
Circular Dichroism.
CD spectra were recorded by using a J-810 Jasco spectropolarimeter and software provided by the manufacturer (Jasco, Easton, MD). Cuvettes with a 0.1-cm pathlength were used. Each spectrum was averaged by using nine accumulations and a scan speed of 50 nm/minute. The samples were measured in 10 mM Tris·HCl buffer (pH 7.8) with a 1.5–2 μM concentration.
Acknowledgments
We thank Dr. Catherine C. Allende for critical comments on the manuscript and Dr. Stefania Sarno for performing surface plasmon resonance experiments. The work was supported by European Commission Pro-Kinase Research Grant 503467 and an Associazione Italiana per la Ricerca sul Cancro grant (to L.A.P.) and Fondo Nacional de Desarrollo Científico y Tecnológico de Chile Project 1030462 (to J.E.A.).
Abbreviations
- CK1
casein kinase 1
- CK2
casein kinase 2
- Dvl
dishevelled
- GSK3
glycogen synthase kinase 3.
Footnotes
The authors declare no conflict of interest.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Rowles J, Slaughter C, Moomaw C, Hsu J, Cobb MH. Proc Natl Acad Sci USA. 1991;88:9548–9552. doi: 10.1073/pnas.88.21.9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhai L, Graves PR, Robinson LC, Italiano M, Culbertson MR, Rowles J, Cobb MH, DePaoli-Roach AA, Roach PJ. J Biol Chem. 1995;270:12717–12724. doi: 10.1074/jbc.270.21.12717. [DOI] [PubMed] [Google Scholar]
- 4.Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM. J Biol Chem. 1995;270:14875–14883. doi: 10.1074/jbc.270.25.14875. [DOI] [PubMed] [Google Scholar]
- 5.Burzio V, Antonelli M, Allende CC, Allende JE. J Cell Biochem. 2002;86:805–814. doi: 10.1002/jcb.10263. [DOI] [PubMed] [Google Scholar]
- 6.Brockman JL, Gross SD, Sussman MR, Anderson RA. Proc Natl Acad Sci USA. 1992;89:9454–9458. doi: 10.1073/pnas.89.20.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milne DM, Looby P, Meek DW. Exp Cell Res. 2001;263:43–54. doi: 10.1006/excr.2000.5100. [DOI] [PubMed] [Google Scholar]
- 8.Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 9.Behrend L, Milne DM, Stoter M, Deppert W, Campbell LE, Meek DW, Knippschild U. Oncogene. 2000;19:5303–5313. doi: 10.1038/sj.onc.1203939. [DOI] [PubMed] [Google Scholar]
- 10.Behrend L, Stoter M, Kurth M, Rutter G, Heukeshoven J, Deppert W, Knippschild U. Eur J Cell Biol. 2000;79:240–251. doi: 10.1078/s0171-9335(04)70027-8. [DOI] [PubMed] [Google Scholar]
- 11.Camacho F, Cilio M, Guo Y, Virshup DM, Patel K, Khorkova O, Styren S, Morse B, Yao Z, Keesler GA. FEBS Lett. 2001;489:159–165. doi: 10.1016/s0014-5793(00)02434-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Shibasaki F, Price R, Guillemot JC, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- 13.Peters JM, McKay RM, McKay JP, Graff JM. Nature. 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 15.Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, Ben-Neriah Y, Alkalay I. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng X, Tamai K, Doble B, Li S, Huang H, Habas R, Okamura H, Woodgett J, He X. Nature. 2005;438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- 18.Swiatek, Kang H, Garcia BA, Shabanowitz J, Coombs GS, Hunt DF, Virshup DM. J Biol Chem. 2006;281:12233–12241. doi: 10.1074/jbc.M510580200. [DOI] [PubMed] [Google Scholar]
- 19.Beyaert R, Vanhaesebroeck B, Declercq W, Van Lint J, Vandenabele P, Agostinis P, Vandenheede JR, Fiers W. J Biol Chem. 1995;270:23293–23299. doi: 10.1074/jbc.270.40.23293. [DOI] [PubMed] [Google Scholar]
- 20.Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, Journot L, Antonsson B, Martinou JC. Mol Cell. 2001;8:601–611. doi: 10.1016/s1097-2765(01)00335-5. [DOI] [PubMed] [Google Scholar]
- 21.Schwab C, DeMaggio AJ, Ghoshal N, Binder LI, Kuret J, McGeer PL. Neurobiol Aging. 2000;21:503–510. doi: 10.1016/s0197-4580(00)00110-x. [DOI] [PubMed] [Google Scholar]
- 22.Yasojima K, Kuret J, DeMaggio AJ, McGeer E, McGeer PL. Brain Res. 2000;865:116–120. doi: 10.1016/s0006-8993(00)02200-9. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 24.Elias L, Li AP, Longmire J. Cancer Res. 1981;41:2182–2188. [PubMed] [Google Scholar]
- 25.Mishra SK, Yang Z, Mazumdar A, Talukder AH, Larose L, Kumar R. Oncogene. 2004;23:4422–4429. doi: 10.1038/sj.onc.1207569. [DOI] [PubMed] [Google Scholar]
- 26.Frierson HF, Jr, El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, Hampton GM. Am J Pathol. 2002;161:1315–1323. doi: 10.1016/S0002-9440(10)64408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuja TJ, Lin F, Osann KE, Bryant PJ. Cancer Res. 2004;64:942–951. doi: 10.1158/0008-5472.can-03-2100. [DOI] [PubMed] [Google Scholar]
- 28.Meggio F, Donella-Deana A, Pinna LA. FEBS Lett. 1979;106:76–80. doi: 10.1016/0014-5793(79)80698-5. [DOI] [PubMed] [Google Scholar]
- 29.Donella-Deana A, Grankowski N, Kudlicki W, Szyszka R, Gasior E, Pinna LA. Biochim Biophys Acta. 1985;829:180–187. doi: 10.1016/0167-4838(85)90187-6. [DOI] [PubMed] [Google Scholar]
- 30.Flotow H, Graves PR, Wang A, Fiol CJ, Roeske RW, Roach PJ. J Biol Chem. 1990;265:14264–14269. [PubMed] [Google Scholar]
- 31.Meggio F, Perich JW, Reynolds EC, Pinna LA. FEBS Lett. 1991;283:303–306. doi: 10.1016/0014-5793(91)80614-9. [DOI] [PubMed] [Google Scholar]
- 32.Roach PJ. J Biol Chem. 1991;266:14139–14142. [PubMed] [Google Scholar]
- 33.Pinna LA, Ruzzene M. Biochim Biophys Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 34.Desdouits F, Siciliano JC, Greengard P, Girault JA. Proc Natl Acad Sci USA. 1995;92:2682–2685. doi: 10.1073/pnas.92.7.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulgar V, Marin O, Meggio F, Allende CC, Allende JE, Pinna LA. Eur J Biochem. 1999;260:520–526. doi: 10.1046/j.1432-1327.1999.00195.x. [DOI] [PubMed] [Google Scholar]
- 36.Marin O, Meggio F, Pinna LA. Biochem Biophys Res Commun. 1994;198:898–905. doi: 10.1006/bbrc.1994.1128. [DOI] [PubMed] [Google Scholar]
- 37.Marin O, Bustos VH, Cesaro L, Meggio F, Pagano MA, Antonelli M, Allende CC, Pinna LA, Allende JE. Proc Natl Acad Sci USA. 2003;100:10193–10200. doi: 10.1073/pnas.1733909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bustos VH, Marin O, Meggio F, Cesaro L, Allende CC, Allende JE, Pinna LA. Biochem J. 2005;391:417–424. doi: 10.1042/BJ20050717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamura H, Garcia-Rodriguez C, Martinson H, Qin J, Virshup DM, Rao A. Mol Cell Biol. 2004;24:4184–4195. doi: 10.1128/MCB.24.10.4184-4195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piedra J, Miravet S, Castaõ J, Pĺmer HG, Heisterkamp N, de Herreros AG, Duãch M. Mol Cell Biol. 2003;23:2287–2297. doi: 10.1128/MCB.23.7.2287-2297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koslov ER, Maupin P, Pradhan D, Morrow JS, Rimm DL. J Biol Chem. 1997;272:27301–27306. doi: 10.1074/jbc.272.43.27301. [DOI] [PubMed] [Google Scholar]
- 42.Pokutta S, Weis WI. Mol Cell. 2000;5:533–543. doi: 10.1016/s1097-2765(00)80447-5. [DOI] [PubMed] [Google Scholar]
- 43.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 44.Liu W, Dong X, Mai M, Seelan RS, Taniguchi K, Krishnadath KK, Halling KC, Cunningham JM, Boardman LA, Qian C, et al. Nat Genet. 2000;26:146–147. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- 45.Aberle H, Schwartz H, Hoschuetzky H, Kemler R. J Biol Chem. 1996;271:1520–1526. doi: 10.1074/jbc.271.3.1520. [DOI] [PubMed] [Google Scholar]
- 46.Roura S, Miravet S, Piedra J, de Herreros AG, Duãch M. J Biol Chem. 1999;274:36734–36740. doi: 10.1074/jbc.274.51.36734. [DOI] [PubMed] [Google Scholar]
- 47.Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Genes Dev. 2004;18:2225–2230. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao ZH, Seeling JM, Hill V, Yochum A, Virshup DM. Proc Natl Acad Sci USA. 2002;99:1182–1187. doi: 10.1073/pnas.032468199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubinfeld B, Tice DA, Polakis P. J Biol Chem. 2001;276:39037–39045. doi: 10.1074/jbc.M105148200. [DOI] [PubMed] [Google Scholar]
- 50.Yanagawa S, Matsuda Y, Lee JS, Matsubayashi H, Sese S, Kadowaki T, Ishimoto A. EMBO J. 2002;21:1733–1742. doi: 10.1093/emboj/21.7.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frame S, Cohen P, Biondi RM. Mol Cell. 2001;7:1321–1327. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 52.Polakis P. Curr Biol. 2002;12:R499–R501. doi: 10.1016/s0960-9822(02)00969-7. [DOI] [PubMed] [Google Scholar]
- 53.Lee E, Salic A, Kruger R, Heinrich R, Kirschner MW. PLoS Biol. 2003;1:E10. doi: 10.1371/journal.pbio.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Yuan H, Weaver CD, Mao J, Farr GH, 3rd, Sussman DJ, Jonkers J, Kimelman D, Wu D. EMBO J. 1999;18:4233–4240. doi: 10.1093/emboj/18.15.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kikuchi A, Kishida S, Yamamoto H. Exp Mol Med. 2006;38:1–10. doi: 10.1038/emm.2006.1. [DOI] [PubMed] [Google Scholar]
- 57.Barberis M, Pagano MA, Gioia LD, Marin O, Vanoni M, Pinna LA, Alberghina L. Biochem Biophys Res Commun. 2005;336:1040–1048. doi: 10.1016/j.bbrc.2005.08.224. [DOI] [PubMed] [Google Scholar]