Abstract
Sometimes infectious agents invade and become established in new geographic regions. Others may be introduced yet never become established because of the absence of suitable hosts in the new region. This phenomenon may be particularly true for the many parasites with complex life cycles, where various life stages require different host species. Homogenization of the world's biota through human-mediated invasions may reunite hosts and parasites, resulting in disease outbreaks in novel regions. Here we use molecular genetics to differentiate invasion pathways for two digenean trematode parasites and their exotic host, the Asian mud snail, Batillaria attramentaria. All of the snail haplotypes found in introduced populations in North America were identical to haplotypes common in the areas of Japan that provided oysters for cultivation in North America, supporting the hypothesis that the snails were introduced from Japan with seed oysters. Two cryptic trematode species were introduced to North American populations in high frequencies. We found a marked reduction of genetic variation in one of these species, suggesting it experienced a bottleneck or founder event comparable to that of the host snail. In contrast, no genetic variation was lost in the other parasite species. We hypothesize that this parasite was and is dispersed naturally by migratory shorebirds and was able to establish only after the host snail, B. attramentaria, was introduced to North America. Evaluation of the nature of invasion pathways and postinvasion consequences will aid mitigation of spreading diseases of humans, livestock, and wildlife in an increasingly globalized world.
Keywords: Batillaria attramentaria, Trematode parasites
Introduced species tend to lose both alleles (1, 2) and parasites (3, 4). During invasion, founder effects and population bottlenecks can reduce genetic variation (e.g., refs. 5–7) and partly drive the loss of parasites in introduced populations (8). Indeed, reduced genetic variability (2) and reduced parasitism (9) should also influence the ecology and evolution of introduced species, presumably in an opposing manner. In contrast, multiple invasions from different source locations can restore some of the missing parasites (9). Analogously, multiple invasions may integrate genetic structure from disparate source locations, resulting in an increase in gene diversity in the introduced range (e.g., ref. 10).
Although most parasites are left behind when species invade new regions, some parasites do manage to invade the new region and infect their host (3, 4). The routes by which parasites invade are scarcely studied. Logically, they are often assumed to invade with their hosts, traveling in or on infected individuals (barring deliberately introduced parasites for biological control). However, many parasites have complex life cycles, wherein different stages sequentially parasitize different host species (8, 11). For these parasites, it is possible for hosts of one stage to invade a region, leaving the parasite behind, but to be “reunified” with their parasite because of the activities of hosts for another stage of the parasite (12, 13). This occurrence is especially possible if the second host naturally has a broad range. For parasites, these disparate invasion pathways will likely affect the distribution and extent of genetic variation within and among its introduced populations.
The marine mud snail, Batillaria attramentaria (= Batillaria cumingi) is a widespread and often common intertidal gastropod in its native range in northeastern Asia (14). B. attramentaria was presumably introduced to the West coast of North America with the importation of Pacific oysters, Crassostrea gigas, from Japan in the early 1900s (15, 16). This snail now occurs in five disjunct populations from Boundary Bay in British Columbia south to Elkhorn Slough, Monterey, CA (17). In its native range, B. attramentaria is geographically genetically structured (18). This local genetic differentiation allowed us to identify the source of the introduced populations in North America. We predicted that the introduced snails would have originated from areas in Japan (identified in ref. 19) that provided high quantities of seed oysters for cultivation in North America.
B. attramentaria is infected by parasites in both its native and introduced ranges. In Japan, B. attramentaria is infected, as first intermediate host, by a suite of eight morphologically distinct digenean trematode species (see ref. 20 and refs. therein). These trematode species generally use fishes or invertebrates as second intermediate hosts and birds as final hosts. Of these eight morphologically distinct trematode species, only the most common species, Cercaria batillariae (20), has invaded North America with its snail host (21). An interesting caveat is that, within its native range, this single morphologically recognized parasite species is actually a complex of eight genetically distinct cryptic species (morphologically similar but genetically distinct species) (20). Further, these cryptic species of parasite vary geographically in distribution and abundance, some being common in the potential source areas of the snails introduced to North America (20).
There are two possible routes for the trematode parasites to have invaded North America: (i) with infected host snails or (ii) as adult stages traveling in migratory final host birds after their snail hosts became established. These routes are not mutually exclusive. However, the primary pathway taken should leave distinctive genetic evidence. As Miura et al. (20) predicted, if the parasites came with snails, the introduced parasite in North America should be a subset of the cryptic species found in snails in source areas of oysters exported to North America. We would also expect intraspecific parallels if genetic structure among populations of the cryptic parasite species occurs within the native range. Finally, if the parasites were introduced with their snail host, they would have likely experienced a bottleneck and have decreased genetic diversity in the new range. Alternatively, if migrating birds are the vector for parasite invasion, a genetic match between the native populations in the snails from oystering regions should be obscured, because birds would transport a greater genetic diversity of cryptic species and alleles within cryptic species to North America.
Using molecular genetics, we disentangled the invasion pathway of a host and its parasites. We (i) determined the source region for the introduced snail, (ii) identified which cryptic parasite species invaded North America, and (iii) provide evidence suggesting two distinct invasion pathways for the snail and its parasites. To our knowledge, no previous work has simultaneously investigated the population genetics of an invasion by both a host and its parasites.
Results
Host Genetics.
We obtained sequence data from 180 individuals of the snail, B. attramentaria, from 18 populations (140 from 14 sites in Japan and 40 from 4 sites North America). Based on the 857-bp sequences of the CO1 gene, 23 haplotypes were found (GenBank accession nos. DQ366356–DQ366378). There was significant genetic structure among snail populations in the native range (ΦST = 0.774, P < 0.001, Table 1), whereas no significant genetic structure was found among populations in the introduced range (ΦST = 0.001, P = 1.0, Table 1). Genetic structure between the native range, and the introduced range was not significantly different (ΦCT = 0.145, P = 0.09), because of shared haplotypes between the regions (Fig. 1A) and high variation among populations in the native range (Table 1).
Table 1.
Analysis of molecular variance of B. attramentaria (snail host) and HL1 and HL6 (trematode parasites) in the native and introduced ranges
| Region | Species | Source of variation | Df | Percent of variation | ΦST | P value |
|---|---|---|---|---|---|---|
| Native range | B. attramentaria | Among | 13 | 77.45 | 0.77 | <0.0001 |
| Within | 126 | 22.55 | ||||
| Total | 139 | |||||
| HL6 | Among | 12 | 1.23 | 0.01 | 0.12 | |
| Within | 163 | 98.77 | ||||
| Total | 175 | |||||
| HL1 | Among | 12 | 2.29 | 0.02 | 0.21 | |
| Within | 86 | 97.71 | ||||
| Total | 98 | |||||
| Introduced range | B. attramentaria | Among | 3 | 0 | 0.00 | 1.00 |
| Within | 36 | 100 | ||||
| Total | 39 | |||||
| HL6 | Among | 3 | 1.04 | 0.01 | 0.32 | |
| Within | 248 | 98.96 | ||||
| Total | 251 | |||||
| HL1 | Among | 3 | −0.90 | −0.01 | 0.73 | |
| Within | 127 | 100.9 | ||||
| Total | 130 |
Because variance estimates are based on measures of covariance, percent variation can be zero or negative when the actual values are small (37).
Fig. 1.
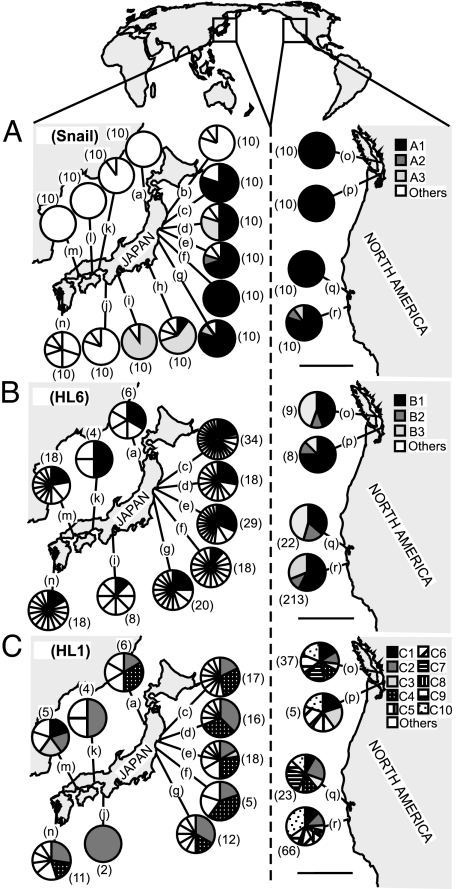
The distributions of the introduced haplotypes of B. attramentaria and its parasites (HL6 and HL1) observed in the native and introduced range. (A) B. attramentaria. (B) HL6. (C) HL1. (Scale bar, 500 km.) Because there were a total of 159 haplotypes, we identify and name only those introduced from Japan to North America. Most of the haplotypes were found only in Japan and are grouped into the category “others” (which is divided to reflect the number of haplotypes in the sample). Sample size is in parentheses. Letters indicate sampling sites: (a) Toga Bay, (b) Yamada Bay, (c) Nagazura Bay, (d) Mangoku Bay, (e) Matsushima Bay, (f) Torinoumi, (g) Matsukawa Bay, (h) Obitsu River, (i) Kumode River, (j) Tanabe Bay, (k) Ibo River, (l) Kasuga River, (m) Hiroshima Bay, (n) Ariakekai, (o) Boundary Bay, (p) Padilla Bay, (q) Bolinas Lagoon, and (r) Elkhorn Slough. c–e are the oyster culturing sites in Miyagi Prefecture.
Only three of the native haplotypes from Japan were found in introduced populations of B. attramentaria snails in North America. The numerically dominant introduced haplotype, A1, (95% of all introduced host snails examined), was also the most common haplotype in northeastern Japan (including the postulated source region, Miyagi Prefecture) (Fig. 1A). The other two less common introduced haplotypes were also found in northeastern Japan. A2 was found only in northeastern Japan, whereas A3 had a more widespread range (Fig. 1A). The phylogenetic relationships of each haplotypes are shown in Fig. 2A.
Fig. 2.
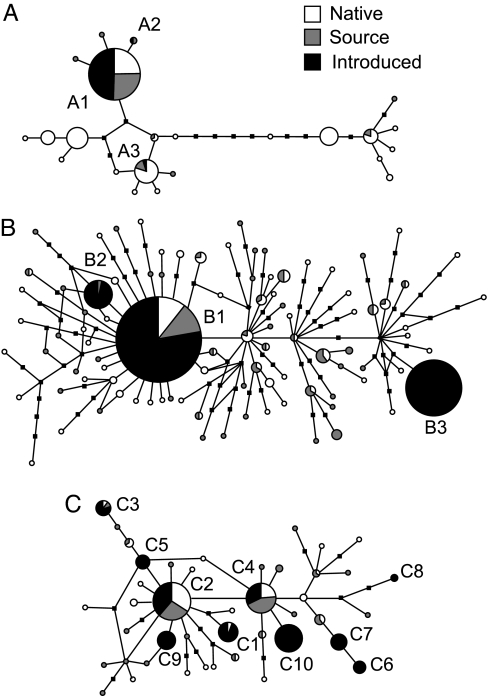
The haplotype networks of B. attramentaria and its trematode parasites (A), HL6 (B), and HL1 (C). Circle sizes are proportional to the number of individuals observed for each haplotype. The black squares represent unobserved single-nucleotide substitutions. The pie charts indicate the regions where samples were collected. Letters lie adjacent to the pie charts correspond to introduced haplotypes listed in Fig. 1A for B. attramentaria and Fig. 1 B and C for the two parasites.
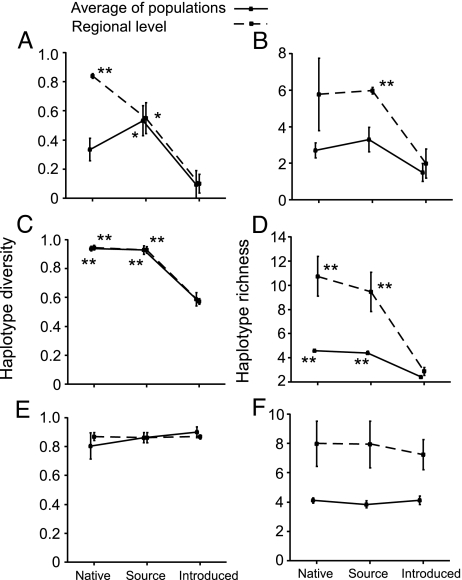
The introduced populations exhibited 72–88% less haplotype diversity than the native populations across all of Japan and compared with the native populations within Miyagi Prefecture (Fig. 3A). Rarefied haplotype richness showed a similar pattern. The haplotype richness of the introduced population was 44–67% lower than that of the native populations across all of Japan as well as just within the Miyagi Prefecture (Fig. 3B). The McDonald–Kreitman test did not reject neutral evolution of the observed molecular variation for both the native (P = 0.57) and introduced snails (P = 1.00).
Fig. 3.
Changes in genetic diversity associated with introduction. Haplotype diversity is equivalent to expected heterozygosity for diploid data, and haplotype richness is the rarefied number of haplotypes in a sample. Plots on the left are of haplotype diversity across populations and that calculated for each region as a whole. Plots on the right are of haplotype richness across populations and that calculated for each region as a whole. The top two plots, A and B, are for the snail host, B. attramentaria; the middle two, C and D, are for the HL6 trematode; and the bottom two, E and F, are for the HL1 trematode. Error bars represent ±SEM. [∗ denotes significance only within a line against the introduced population; ∗, P < 0.05; ∗∗, P < 0.01 (actual P value in each test is listed in SI Table 3)].
Parasite Genetics.
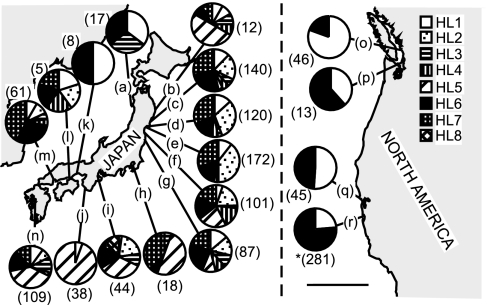
We performed PCR-based restriction fragment length polymorphism analysis on 1,315 individuals of the trematode morphospecies, C. batillariae (930 from 14 sites in Japan and 385 from 4 sites in North America). As in Miura et al. (20), we found eight genetically distinct cryptic species of C. batillariae [HL1–HL8 of Miura et al. (20)] in Japan. We found only three species introduced to North America (34% of HL1, 0.5% HL2, and 65.5% HL6; Fig. 4). Two of the introduced parasites, HL2 and HL6, were among the three most common species found in Japan (23.2% of all individuals for HL7, 20.7% for HL2, and 18.9% for HL6; Fig. 4) and in the Miyagi Prefecture (30.1% for HL7, 30.8% for HL2, and 18.8% for HL6).
Fig. 4.
The distributions of genetically identified trematode parasites. The sample size is in parentheses. (Scale bar, 500 km.) Letters indicate sampling sites that are listed in Fig. 1. (We found two individuals of HL2 in Elkhorn Slough, however, representing <1% of the total sample).
We performed PCR-based single-strand conformation polymorphism (PCR-SSCP) analyses for the two common trematode species (HL1 and HL6) found in North America. We analyzed a total of 230 infections of HL1 (99 from Japan and 131 from North America) and 428 infections of HL6 (176 from Japan and 252 from North America). For HL1, we found 43 distinct SSCP CO1 patterns of the 230 sampled infections (18.6%). For HL6, we found 93 distinct SSCP CO1 patterns of the 428 sampled infections (21.7%). Sequencing an individual of each unique SSCP pattern (a total of 136 infections) resulted in a unique sequence for each SSCP pattern. Thus, each SSCP pattern represented a unique haplotype for HL1 (GenBank accession nos. AY626457–AY626466 and DQ366386–DQ366418) and for HL6 (GenBank accession nos. AY626498–AY626514 and DQ366421–DQ366497).
We found no significant genetic structure among populations in either parasite species from the native range or the introduced range (Table 1). However, for both species, there was evidence for some genetic structure between native and introduced regions. This differentiation was approximately twice as large for HL6 than for HL1 (ΦCT = 0.047, P = 0.005 for HL1; ΦCT = 0.087, P < 0.0001 for HL6).
Many of the introduced parasite haplotypes were also found in the native range (Fig. 1 B and C). Two of the three HL6 haplotypes from North America were identical to the Japanese haplotypes (Fig. 1B). Among the 10 HL1 haplotypes from North America, four were identical to the Japanese haplotypes (Fig. 1C). The phylogenetic relationships of the haplotypes are shown in Fig. 2 B and C.
Introduced HL6 populations exhibited 37–39% lower haplotype diversity than either all sampled native populations or the native populations in Miyagi Prefecture (Fig. 3C). Similarly, rarefied haplotype richness of introduced HL6 populations was 45–73% lower than that of native populations and of Miyagi Prefecture (Fig. 3D). In contrast, the haplotype diversity and rarefied haplotype richness of the other introduced parasite species (HL1) was similar to that of native HL1 populations (both throughout Japan and within thee Miyagi Prefecture) (Fig. 3 E and F). The McDonald–Kreitman test did not reject neutrality of the observed molecular variation for both native parasite species (P = 0.44 for HL1, P = 0.19 for HL6) and introduced parasite species (P = 0.49 for HL1, P = 0.60 for HL6).
Discussion
Using molecular genetics, we demonstrate that the Asian mud snail, B. attramentaria was introduced to North America from Miyagi Prefecture in Japan. Additionally, we identified three cryptic species (HL1, HL2, and HL6) of introduced trematode parasites. Introduced HL2 was very rare (<1%) and thus was not analyzed further. Our research suggests that the two common parasite species arrived in North America by different invasion pathways. One parasite was introduced to North America with its snail host, assisted by humans. In contrast, adults of the other parasite were probably continually dispersed naturally by migratory shorebirds. Before the invasion of B. attramentaria ≈100 years ago, their offspring would not have had the opportunity to recruit, because of the absence of a suitable snail intermediate host. Interestingly, oyster aquaculture likely reunited the parasite with its natural host and enabled it to establish in North America.
The genetic variation of B. attramentaria snails was highly structured in the native range in Japan (Table 1), likely because of the limited dispersal ability of the directly developing snail (18). This genetic structure enabled us to confirm the native source for the introduced North American populations. The source region historically provided seed oysters (Crassostrea gigas) for aquaculture in North America (19). The introduced haplotypes were common in or restricted to native snail populations in the postulated source region (Fig. 1A). The dominant haplotype of introduced B. attramentaria populations (A1) was identical to the dominant haplotype in Matsushima Bay, Mangoku Bay, and Nagazura Bay, sites within the source region. Perhaps more importantly, a rare haplotype (A2) in one introduced population (Elkhorn Slough) appeared only in the hypothesized source region in Japan (Matsushima Bay; Fig. 1). Further, all of the introduced snail haplotypes were phylogenetically closely related to one another (Fig. 2A), suggesting that they are not derived from large geographical range. Although it may be possible that the introduced snail haplotypes are also common in areas outside of Japan, sampling of B. attramentaria populations in Korea (18) indicates that this is likely not the case. Three haplotypes found in Korea (18) were the same or closely related to the haplotypes we found in the Japanese coast of the Sea of Japan and were not consistent with any of the introduced haplotypes (ref. 18 and data set herein). Thus, our genetic data support the hypothesis that B. attramentaria was introduced along with oysters brought to North America from Japan for aquaculture (15). Studies based on nuclear markers suggest that species invasions do not always demonstrate corresponding reductions in genetic diversity (reviewed in ref. 22). However, mitochondrial genes are particularly prone to losing diversity after invasion by a small number of founders, because mtDNA is haploid with uniparental inheritance and thus has only one-quarter the effective population size of nuclear genes (23). The low mitochondrial genetic diversity of the introduced snails is consistent with the expectation that introduced populations would have reduced genetic diversity because of founder events, bottlenecks and genetic drift (Fig. 3 A and B) (1). It is unlikely that the low genetic variation in North America was caused by selection of particular haplotypes, because the neutrality of the CO1 gene for B. attramentaria was not rejected for either the native and introduced regions.
Only one of eight trematode morphospecies, C. batillariae, found in the source region in Japan has invaded North America (21). Interestingly, of the eight cryptic species of this C. batillariae in Japan (20), only three species (HL1, HL6, and HL2) were found in the introduced range (Fig. 4). Several mechanisms may “filter out” parasites during the invasion process (8, 24). Parasites may be lost at the source (i.e., never get transported to the introduced area), or the parasites may be lost after the introduction. As predicted by Miura et al. (20), parasites with high prevalence in the source areas (e.g., HL6) were transported to new range. However, one cryptic parasite species (HL7) that was common in the hypothesized source region was not found in the introduced range. This result could be explained by the absence, in the introduced range, of appropriate additional hosts, or by chance.
We found some unique haplotypes in the introduced populations. This finding is most simply explained as a result of sampling error (i.e., the haplotypes went undetected in the native range) and not by mutation and fixation of new haplotypes in the introduced region (given the short time the populations have been separated (<100 years), and that neutrality was not rejected for both native and introduced populations of both parasite species).
No significant geographic genetic structure was found in either of the common introduced trematode species (HL1 and HL6) in Japanese populations, with almost all of the total diversity being distributed within populations (Table 1). Further, the introduced haplotypes were not closely related for both of these species (Fig. 2 B and C). These patterns suggest a high level of gene flow among populations in native range, likely because of high dispersal capability of adult trematodes in the final bird hosts (25). Similarly, once the parasites were introduced to North America, bird movement within the introduced range should homogenize haplotype diversity among introduced populations. Thus, it was not possible to identify the parasites' source area within Japan by comparing the distribution of haplotypes (Fig. 1 B and C), as we were able to do for their snail host. However, it was possible to infer the parasites' overall invasion pathway by assessing changes in population genetic variation. If relatively small numbers of parasites were introduced in a relatively short period (e.g., if they were introduced with the snail host by oyster transfers), we would expect a reduction in genetic diversity because of founder events, similar to what we found in the snail populations. Consistent with this hypothesis, one introduced parasite species (HL6) had populations with lower genetic diversity than the populations in the postulated source area (Fig. 3 C and D) and was likely introduced along with its host snail.
In contrast to HL6, genetic diversity of the other common introduced parasite, HL1, was not significantly different in introduced compared with native populations (Fig. 3 E and F). Also, the analysis of molecular variance (AMOVA) indicated that introduced HL1 were far more similar to native HL1 than introduced HL6 were to native HL6 (HL1 having about half the genetic differentiation across geographic regions as did HL6). This similarity suggests a limited or complete lack of a population bottleneck associated with the invasion of HL1. This high level of genetic diversity is uncommon for introduced species (1), unless they are repeatedly introduced or originate from multiple source regions (10). Importantly, the high number of introduced lineages (variation) of HL1 suggests a high level of connectivity between Japan and North America. We postulate that HL1 was repeatedly introduced to North America by migratory birds, which serve as its final hosts. Some estuarine birds (e.g., ref. 26) regularly travel from the north during alternate winter migrations down either the Asian or North American side of the Pacific (essentially connecting Asia and North America by way of their far-north breeding grounds). For individual birds, winter migration down either side of the Pacific is separated by the summer breeding season. Thus, dispersal of HL1 by these birds would require that adult parasites live a few to several months in their bird final hosts. Evidence for two confamilial (heterophyid) trematode species suggests that they can live >1 year (27, 28). Additionally, individual birds can carry 1,000s of adult trematodes in their intestines (29). Thus, it appears plausible that the relatively high genetic diversity of the introduced HL1 trematode parasite is because of recurrent introduction from its native region by the migration of its final host birds. Why would the other parasite (HL6) not be continuously dispersed along both sides of the Pacific? Although we cannot be certain, the answer may lie in differences in the biology of the adult parasites. Adult stages may differ more than one would expect based upon how similar the species appear as larvae. Perhaps adults of the HL6 species are shorter-lived than HL1 and are consequently unable to survive the breeding season of their avian final host. Thus, differential longevity of HL6 and HL1 could explain the different invasion pathways for the two parasites. Additionally, the cryptic species may differ in critical aspects of parasitism of second intermediate hosts. For instance, if HL6 infects a narrower range of second intermediate host species than HL1, it would reduce the chance both of being transported from Japan (by bird predators) and of completing its life cycle in North America. These possibilities could also explain the failure of the other trematode species in Japanese populations of B. attramentaria to invade North America.
Although HL6 probably invaded along with its snail host, HL1 may have continually been transported by birds to North America, long before oyster aquaculture. Hence, North America may, in fact, be within the natural range of adults of this parasite. However, its infective propagules were unable to become established until the introduction of its obligate first intermediate host snail, B. attramentaria. Although HL1 may have been transported to North America for a long time, it did not adapt to use the confamilial and sympatric North American mud snail, Cerithidea californica, as an alternative first intermediate host (21). This is not surprising, given that trematodes are typically exceptionally host-specific for their first intermediate host (30, 31). Nevertheless, both of these introduced parasites are now abundant in North America, and at least one of them infects native fishes as second intermediate hosts (21). This study shows that comparative use of molecular genetic tools can be used to test hypotheses concerning mechanisms of human-mediated invasion. Further, it suggests that parasites which may have historically been “tourists” in North America became established after the introduction of a “missing” host. We suspect that these dispersal-invasion pathways are not unique and may result in future emerging infectious diseases of humans, agriculture, and wildlife.
Materials and Methods
Study Sites and Sample Collection.
Samples of B. attramentaria were collected from 14 populations in Japan and 4 populations in the U.S. (Fig. 1A). Snails were identified following Adachi and Wada (32). We dissected each snail and identified trematode species using a stereomicroscope. Both snails and parasites were fixed with 70% ethanol and stored at −20°C for molecular analysis. We modified the procedure of Doyle and Doyle (33) to isolate snail and trematode DNA. Snail and trematode tissue were separated and homogenized in a solution of 300 ml of 2× cetyltrimethylammonium bromide and 10 mg ml−1 proteinase K, incubated at 60°C for ≈1 h, extracted once with phenol/chloroform (v:v, 1:1), and precipitated with two volumes of ethanol. The DNA pellets were washed with 75% ethanol, air-dried for ≈30 min, and dissolved in 50 ml of H2O.
DNA PCR–Restriction Fragment Length Polymorphism (RFLP) Analysis.
We identified the eight cryptic species of C. batillariae [HL1–HL8, following Miura et al. (20)] using PCR-RFLP on the mitochondrial CO1 gene. PCR primers for CO1 used in this study were the regions described previously for the studies of trematodes: JB3 (34) and CO1-R trema (20) [supporting information (SI) Table 2].
PCR amplification was performed by using 35 cycles under the following conditions: denaturing at 94°C for 30 sec, annealing at 45°C for 30 sec, and extension at 72°C for 60 sec. Five microliters of unpurified PCR products was digested for 10 h with the four-base cutting restriction enzyme, endonuclease MseI (New England Biolabs, Ipswich, MA). The restricted fragments were separated by 4% Tris-acetate EDTA agarose gels for 4 h at 50 V constant voltage and detected by staining with ethidium bromide. Genotypes were identified based on the fragment patterns and scored individually.
DNA PCR-SSCP Analysis.
To assess the intraspecific genetic structure of the two common cryptic species of parasite (HL1 and HL6) that we found had invaded North America, we performed PCR-SSCP analysis. Comparative analysis of the published sequence of the CO1 gene of C. batillariae (20) enabled identification of highly variable regions that were used for PCR-SSCP analysis. Two regions (250–280 base pairs) that showed high genetic variation were selected from sequences previously published for haplotypes of the cryptic species (GenBank accession nos. AY626457–AY626466 for HL1 and AY626498–AY626514 for HL6). A total of 544 base pairs for HL1 and 542 base pairs for HL6 were investigated. We used four pairs of fluorescent-labeled primers for PCR-SSCP analysis (SI Table 2). PCR amplification was performed by using 35 cycles under the following conditions: denaturing at 94°C for 30 sec, annealing at 50°C for 30 sec, and extension at 72°C for 60 sec. Sample preparation for capillary electrophoresis involved the addition of 1 μl of diluted PCR product to the capillary electrophoresis mixture [3 μl of deionized formamide/0.5 μl of 0.1 M NaOH/0.5 μl of GeneScan-350 ROX Size Standard (PE Applied Biosystems, Foster City, CA)]. The capillary sample mixture was denatured for 2 min at 95°C and rapidly cooled on ice before loading of the instrument. Subsequent preparation, such as setup of the ABI PRISM 310 Genetic Analyzer was done in accordance with the manufacturer's instructions (PE Applied Biosystems). The nondenaturing polymer matrix used was 3% GenScan polymer with 10% glycerol. Electrophoresis conditions were set on the instrument at a 5-s injection time, a 7-kV injection voltage, a 13-kV electrophoresis voltage, a 210-s syringe pump time, a constant temperature of 30°C, and a 20-min collection time. We aligned sample peaks by size and scored them individually. As described below, we confirmed that each unique SSCP pattern represented a unique haplotype by performing DNA sequencing on an individual of each unique SSCP pattern.
DNA Sequencing.
We sequenced the CO1 genes of 10 B. attramentaria snails (857 base pairs) from each of our 18 sites (14 in Japan and 4 in North America). We designed PCR primers (CO1-bf and CO1-br; SI Table 2) for the B. attramentaria CO1 gene based on published CO1 sequences of batillariid snails (GenBank accession nos. AB054364–AB054367). For the two common invasive cryptic species of C. batillariae (HL1 and HL6), we sequenced the CO1 genes of an individual of each unique SSCP pattern. A total of 136 infections of trematode parasites were used for the sequence analysis. The primer pairs and condition of PCR amplification for C. batillariae were the same as those used in the RCR-restriction fragment length polymorphism (described above and in SI Table 2). The PCR products of all samples were purified and sequenced by using an automated sequencer [HITACHI SQ5500 (Hitachi, Tokyo, Japan) and ABI PRISM 310 Genetic Analyzer (Applied Biosystems)]. Sequences were aligned by CLUSTALX (35). We obtained network trees based on the most parsimonious connections of haplotypes by using TCS 1.21 (36).
Estimation of Genetic Parameters and Data Analysis.
We compared haplotype variation within and among populations of the entire sampled native region, the hypothesized source region, and the introduced region, using Φ statistics from analysis of molecular variance (AMOVA) (37) using Arlequin 3.0 (38). We also compared genetic diversity of the native and introduced ranges using both haplotype diversity [gene diversity (39)] and haplotype richness [allelic richness (40)]. Haplotype diversity was calculated by Arlequin 3.0. Difference between haplotype diversity of introduced ranges and native ranges were tested by using t tests, following Nei (39). Richness measures are sensitive to sampling effort (40–42), and we sampled different numbers of populations in the different regions, and (for the parasites) different numbers of individuals within populations. Thus, to directly compare haplotype richness between regions, we used a resampling scheme to hierarchically rarify (42) measures of haplotype richness to standardized numbers of populations (sampled without replacement) and individuals within populations (sampled with replacement). To allow the most useful comparisons, we consistently resampled three populations from each region to be compared, for both snails and parasites. For snails, we resampled the 10 individuals within each population, and for parasites we resampled five individuals per population [for rarified haplotype richness of parasites, we excluded parasite populations (all in the native range) with less than five individuals sampled]. To generate the rarified richness measures, we performed 10,000 iterations, for each calculation, using the Resampling Stats Excel Add-in 3.0 (Resampling Stats, Arlington, VA). We assessed the significance of observed differences using t tests. To obtain standard errors for t tests of the rarefied richness measures, we generated the resampling distribution of differences on the null hypothesis using 10,000 iterations (resampling within each region, to allow for unequal variances) (43). All P values are two-tailed. To assess the neutrality of the quantified genetic variation at the CO1 gene in both the native and introduced regions, we applied the McDonald–Kreitman test (44) using DnaSP 4.0 (45). This test is based on a comparison of synonymous and nonsynonymous variation within and between species. To calculate the variation between species, we used sequences of Batillaria multiformis (GenBank accession no. DQ981865) as the outgroup for B. attramentaria, and HL2 (GenBank accession no. AY626472) for the two parasite species. This test is also well suited for our study because it does not assume populations are at equilibrium (44).
Supplementary Material
Acknowledgments
We thank Mamoru Fujikawa, Minoru Miura, Irit Altman, and Nadia Talhouk for assistance in collecting B. attramentaria. Steve Vollmer commented on an earlier draft of the paper. Eldredge Bermingham and two anonymous reviewers provided useful comments. This study was supported by grants from the Japan Society for the Promotion of Science (to S.C.) and by a grant from the National Institutes of Health/National Science Foundation Ecology of Infectious Disease Program (Grant DEB-0224565, to A.M.K.).
Abbreviation
- SSCP
single-strand conformation polymorphism.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY626457–AY626466, AY626472, AY626498–AY626514, DQ366356–DQ366378, DQ366386–DQ366418, DQ366421–DQ366497, and DQ981865).
This article contains supporting information online at www.pnas.org/cgi/content/full/0609603103/DC1.
References
- 1.Nei M, Maruyama T, Chakraborty R. Evolution (Lawrence, Kans) 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 2.Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, et al. Annu Rev Ecol Syst. 2001;32:305–332. [Google Scholar]
- 3.Mitchell CE, Power AG. Nature. 2003;421:625–627. doi: 10.1038/nature01317. [DOI] [PubMed] [Google Scholar]
- 4.Torchin ME, Lafferty KD, Dobson AP, Mckenzie VJ, Kuris AM. Nature. 2003;421:628–630. doi: 10.1038/nature01346. [DOI] [PubMed] [Google Scholar]
- 5.Cristescu MEA, Hebert PD, Witt JDS, MacIsaac FJ, Grigorovich IA. Limnol Oceanogr. 2001;46:224–229. [Google Scholar]
- 6.Hanfling B, Carvalho GR, Brandl R. Mar Ecol Prog Ser. 2002;238:307–310. [Google Scholar]
- 7.Voisn M, Engel CR, Viard F. Proc Natl Acad Sci USA. 2005;102:5432–5437. doi: 10.1073/pnas.0501754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torchin ME, Lafferty KD, Kuris AM. Parasitology. 2002;124:S137–S151. doi: 10.1017/s0031182002001506. [DOI] [PubMed] [Google Scholar]
- 9.Torchin ME, Mitchell CE. Front Ecol Environ. 2004;2:183–190. [Google Scholar]
- 10.Kolbe JJ, Glor RE, Schettino LR, Lara AC, Larson A, Losos JB. Nature. 2004;431:177–181. doi: 10.1038/nature02807. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguti S. A Synoptical Review of Life Histories of Digenetic Trematodes of Vertebrates with Special Reference to the Morphology of Their Life Forms. Tokyo: Keigaku; 1975. [Google Scholar]
- 12.Warner RE. Condor. 1969;70:101–120. [Google Scholar]
- 13.van Riper C III, van Riper ML, Goff ML, Laird M. Ecol Monogr. 1986;56:327–344. [Google Scholar]
- 14.Hasegawa K. Okutani T. Marine Mollusks in Japan. Tokyo: Tokai Univ Press; 2000. pp. 130–133. [Google Scholar]
- 15.Bonnot P. Nautilus. 1935;49:1–2. [Google Scholar]
- 16.Barrett EM. Fish Bull. 1963;123:2–103. [Google Scholar]
- 17.Byers JE. Biol Invasions. 1999;1:339–352. [Google Scholar]
- 18.Kojima S, Hayashi I, Kim D, Iijima A, Furota T. Mar Ecol Prog Ser. 2004;276:161–172. [Google Scholar]
- 19.National Research Council. Nonnative Oysters in Chesapeake Bay. Washington, DC: National Acad Press; 2004. [Google Scholar]
- 20.Miura O, Kuris AM, Torchin ME, Hechinger RF, Dunhum EJ, Chiba S. Int J Parasitol. 2005;35:793–801. doi: 10.1016/j.ijpara.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Torchin ME, Byers JE, Huspeni TC. Biol Invasions. 2005;7:885–894. [Google Scholar]
- 22.Wares JP, Hughes AR, Grosberg RK. In: Species Invasions: Insights into Ecology, Evolution, and Biogeography. Sax DF, Stachowicz JJ, Gaines SD, editors. Sunderland, MA: Sinauer; 2005. pp. 229–257. [Google Scholar]
- 23.Avise JC. Molecular Markers, Natural History and Evolution. London: Chapman & Hall; 1994. [Google Scholar]
- 24.Dobson AP, May RM. In: Ecology of Biological Invasions of North America and Hawaii. Mooney HA, Drake JA, editors. New York: Springer; 1986. pp. 58–76. [Google Scholar]
- 25.Dybdahl MF, Lively CM. Evolution (Lawrence, Kans) 1996;50:2264–2275. doi: 10.1111/j.1558-5646.1996.tb03615.x. [DOI] [PubMed] [Google Scholar]
- 26.Wenink PW, Baker AJ. Auk. 1996;113:744–756. [Google Scholar]
- 27.Komiyama Y, Ito J, Yamamoto S. Jpn J Parasitol. 1958;7:7–11. [Google Scholar]
- 28.Ellis C, Williams IC. J Helminthol. 1973;47:329–338. doi: 10.1017/s0022149x00026614. [DOI] [PubMed] [Google Scholar]
- 29.Russell HT. Trematodes from Shorebirds Collected at Morro Bay, California. Los Angeles: Univ of California; 1960. [Google Scholar]
- 30.Gibson DI, Bray RA. Int J Parasitol. 1994;24:1213–1226. doi: 10.1016/0020-7519(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 31.Galaktionov KV, Dobrovolskij AA. The Biology and Evolution of Trematodes. Dordrecht, The Netherlands: Kluwer Academic; 2003. [Google Scholar]
- 32.Adachi N, Wada K. Venus. 1998;57:115–120. [Google Scholar]
- 33.Doyle JJ, Doyle JL. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 34.Bowles J, McManus DP. Mol Biochem Parasitol. 1993;57:231–240. doi: 10.1016/0166-6851(93)90199-8. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clement MP, Crandall KA. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 37.Excoffier L, Smouse PE, Quattro JM. Genetics. 1992;13:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Excoffier L, Laval G, Schneider S. Evol Bioinform Online. 2005;1:47–51. [PMC free article] [PubMed] [Google Scholar]
- 39.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ Press; 1987. [Google Scholar]
- 40.Petit RJ, El Mousadik A, Pons O. Conserv Biol. 1998;12:844–855. [Google Scholar]
- 41.Hurlbert SH. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 42.Kalinowski ST. Conserv Genet. 2004;5:539–543. [Google Scholar]
- 43.Manly BFJ. Randomization, Bootstrap and Monte Carlo Methods in Biology. 2nd Ed. London: Chapman & Hall; 1997. [Google Scholar]
- 44.McDonald JH, Kreitman M. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 45.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.