Abstract
Control of the establishment of cell polarity is an essential function in tissue morphogenesis and renewal that depends on spatial cues provided by the extracellular environment. The molecular role of cell–cell or cell–extracellular matrix (ECM) contacts on the establishment of cell polarity has been well characterized. It has been hypothesized that the geometry of the cell adhesive microenvironment was directing cell surface polarization and internal organization. To define how the extracellular environment affects cell polarity, we analyzed the organization of individual cells plated on defined micropatterned substrates imposing cells to spread on various combinations of adhesive and nonadhesive areas. The reproducible normalization effect on overall cell compartmentalization enabled quantification of the spatial organization of the actin network and associated proteins, the spatial distribution of microtubules, and the positioning of nucleus, centrosome, and Golgi apparatus. By using specific micropatterns and statistical analysis of cell compartment positions, we demonstrated that ECM geometry determines the orientation of cell polarity axes. The nucleus–centrosome orientations were reproducibly directed toward cell adhesive edges. The anisotropy of the cell cortex in response to the adhesive conditions did not affect the centrosome positioning at the cell centroid. Based on the quantification of microtubule plus end distribution we propose a working model that accounts for that observation. We conclude that, in addition to molecular composition and mechanical properties, ECM geometry plays a key role in developmental processes.
Keywords: image quantification, micropattern, cell standardization
Cell polarity is defined by the expression of a morphological and functional asymmetry of cell compartmentalization relative to a polar axis (1–4). Cell adhesion regulates many morphogenetic processes during the development and orderly turnover of tissues (5). The polarity of the epithelial cells reflects its cell–cell and cell–extracellular matrix (ECM) interactions (2, 6–8). Cell–cell interactions, mediated by cadherins, can initiate the segregation of proteins within the plane of the membrane (9). Cell adhesion to ECM, mediated by the super family of integrins, provides a spatial cue for the establishment of the asymmetric distribution of cell surface receptors and the orientation of cell polarity (10–13).
Extracellular contact sites induce the local assembly of cytoskeletal and signaling proteins at contacting membranes (7, 14–16). Localized actin cytoskeleton assembly serves as a scaffold for recruiting signaling proteins such as adenomatous polyposis coli (APC) that further guide microtubule (MT) growth (17–19). Then, the crosstalk between actin and tubulin cytoskeletons propagates extracellular cues from cell surface to cell interior (3, 17, 20). The isotropic astral array of MTs can reorganize into a polarized array by selective stabilization of MT plus ends at the cell surface cortex (8, 21, 22). Nucleus and centrosome reorient in the cytoplasm along an axis of polarity relative to position of the cue(s) (8, 23, 24).
Yeaman et al. (25) proposed that establishment of structural anisotropy in the plasma membrane was the first critical event in the orientation of cell polarity. According to this proposition, the extrinsic spatial cues mediated by cell adhesion, physically and molecularly, define contacting and noncontacting surfaces. The anisotropic distribution of these contacts would hence imply membrane polarity and thereby overall cell polarity. Thus far the influence of cell–ECM interactions on epithelial cell polarity has been highlighted by mutations or function-blocking antibodies affecting ECM proteins or their surface receptors (26). However, the role of the anisotropic distribution of cell adhesions in the orientation of cell polarity has never been directly demonstrated. This problem can be tackled by the use of defined concave fibronectin micropatterns, which can make individual cells spread across adhesive and nonadhesive zones and create spatial environments that can induce a structural asymmetry in the plasma membrane (27, 28).
Results
Cell Cortex Is Polarized in Response to the Anisotropy of Cell Adhesive Environment.
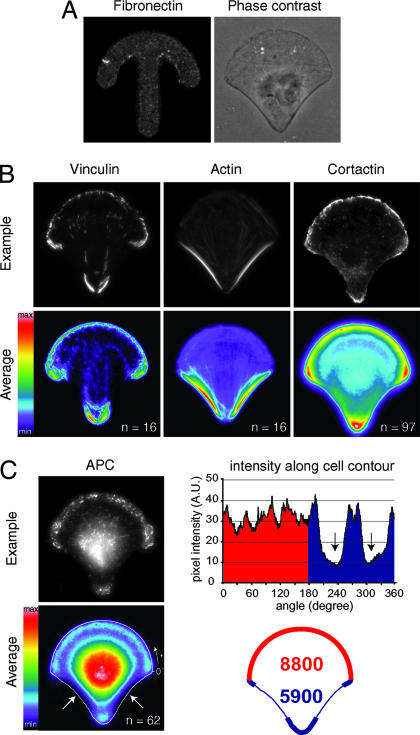
We first analyzed whether the anisotropy of a fibronectin adhesive pattern had an effect on cell surface polarity of human retinal pigment epithelial (RPE) cells. We previously observed that the presence of adhesion sites along cell edges stimulates the polymerization of actin in membrane protrusions (27). In addition, thin stress fibers are formed along straight and adhesive edges, whereas large ones are developed along straight and nonadhesive edges (28). Here, by plating human RPE1 cells on fibronectin micropatterns we observed that a curved and adhesive border prevented the development of conspicuous stress fibers. Therefore it appeared appropriate to use crossbow-shaped micropatterns, which impose a curved adhesive border to half of the cell and two nonadhesive edges to the other half, to polarize the actin cytoskeleton in protruding and contracting zones (Fig. 1A). Four hours after deposition on the micropatterns, cells were fixed for further analyses.
Fig. 1.
Cortical polarity. (A) RPE1 cell plated on a fibronectin crossbow micropattern (Left) and visualized in phase contrast (Right). (B) Labelings of vinculin, F-actin, and cortactin (Upper) were averaged over 16–97 cells (Lower). Actin polymerizes in membrane ruffles upon the curved adhesive edge where small dot-like focal adhesions accumulate. Actin assembles in contractile stress fibers anchored to fibronectin via large focal adhesions upon nonadhesive edges. (C) Spatial distribution of APC. Immuno-labelings of APC (Upper Left) were averaged over 62 cells (Lower Left). On the average picture, a 2-μm-wide line scan of pixel intensities along cell contour (white line) shows a reduction of APC density upon nonadhesive edges (arrows). This local reduction induces an imbalance in the spatial distribution of APC. (Right) Integration of pixel intensities along the contour shows a higher content of APC along the adhesive half of the cell (8,800 a.u., red zone) than along the nonadhesive one (5,900 a.u., blue zone).
Focal adhesions are transmembrane structures where the cell contacts extracellular environment. Their spatial distribution was quantified by averaging the intensity of the labeling of vinculin over several cells [see Materials and Methods and supporting information (SI) Fig. 7]. Vinculin-positive structures were asymmetrically distributed all along the cell contour: these accumulated at the extremities of the adhesive zones, but were regularly distributed at a lower level along the curved adhesive border. Vinculin-positive structures were absent from nonadhesive zones (Fig. 1B). The average distributions of the two exclusive states of actin dynamics were then quantified: the stress fibers revealed by the F-actin staining, and the polymerizing meshwork found in membrane ruffles revealed by the labeling of cortactin. Stress fibers were almost absent from the curved adhesive border, whereas they were enriched upon the nonadhesive edges (Fig. 1B). In contrast, cortactin was restricted to the adhesive sides (Fig. 1B). Thus, the actin network reproducibly displayed a polarized organization: a polymerizing meshwork within membrane ruffles at the adhesive border and contractile stress fibers at the nonadhesive edges.
APC is an actin-binding protein known to have an higher affinity for areas of cell protrusions (18). In addition, APC is known to participate to the connection between the actin and MT networks (18, 19, 29, 30). The average distribution of APC in cells plated on the crossbow showed a clear asymmetric accumulation at the cell periphery in addition to its cytoplasmic localization (Fig. 1C). This average peripheral localization was quantified by recording pixel intensities along a 2-μm-wide line scan over the cell contour. APC was homogeneously distributed all along the cell adhesive periphery and absent from nonadhesive edges. The actin network polarization appeared thus associated with an asymmetric cortical distribution of APC, which accumulated in membrane ruffling zones (Fig. 1C). The MT plus end-binding protein EB1 associates with APC, where this interaction ensures MT plus end capping at the cell cortex (31). Thus, the asymmetry of the spatial distribution of APC would be likely to influence MT dynamics.
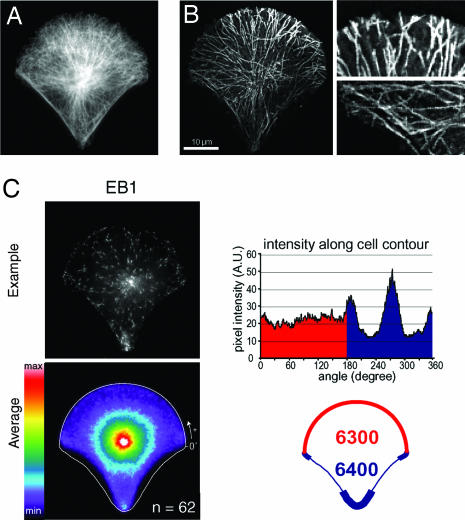
MT Growth Is Governed by Cell Cortex Composition.
We examined whether the cortical asymmetry was transmitted to the cell interior and analyzed the spatial organization of the MT array. As anticipated, the immuno-labeling of tubulin showed that the density of the MT array was lower in angular sectors facing the two nonadhesive edges than toward the adhesive apices flanking these edges (Fig. 2A). The polarization of the MT array was further confirmed by the analysis of MT dynamics. MT plus ends trajectories were visualized by recording EB1-GFP fluorescence in time-lapse microscopy. Strikingly, MTs stopped growing when contacting the adhesive border, whereas they kept growing along nonadhesive edges up to the next adhesive site (Fig. 2B and SI Movie 1). This behavior was then quantified by calculating the average spatial distribution of EB1-positive MT plus ends for >60 fixed cells (Fig. 2C). A line scan of pixel intensities along the cell contour showed that although the total amount of EB1 was similar on the upper and lower hemi-circles of the cell contour (with respect to the orientation of the image shown), the density of EB1 was lower along nonadhesive edges. The variation of intensity along the cell contour confirmed that the low amount of MT plus ends in the vicinity of the two nonadhesive edges was balanced against an equivalent accumulation in the adhesive sites flanking these edges. These measures confirmed that the MT did not stop growing when contacting a nonadhesive edge but instead kept on growing. They were rerouted along these edges toward cell adhesive apices where they accumulated.
Fig. 2.
Polarization of the MT network. (A) MTs labeled with anti-α-tubulin in a fixed cell platted on a crossbow. (B) EB1 trajectories. (Left) Projection of 100 pictures acquired in time-lapse microscopy at two pictures per s of EB1-GFP in a cell plated on a crossbow (see SI Movie 1). (Right) Magnifications of MT plus ends trajectories show that MTs stop growing when contacting adhesive edges (Upper) and keep growing along nonadhesive edges (Lower). (C) Quantification of the spatial distribution of EB1. (Left) Immuno-labelings of EB1 (Upper) were averaged over 62 cells (Lower). (Right) Line scan of average pixel intensities along the cell contour (Upper) shows reduction of EB1 density along nonadhesive edges and accumulations of EB1 in the area flanking nonadhesive edges. Integration of pixel intensities along the cell contour shows identical amounts of EB1 along both the adhesive half border (6,300 a.u., red zone) and the nonadhesive half border (6,400 a.u., blue zone) (Lower).
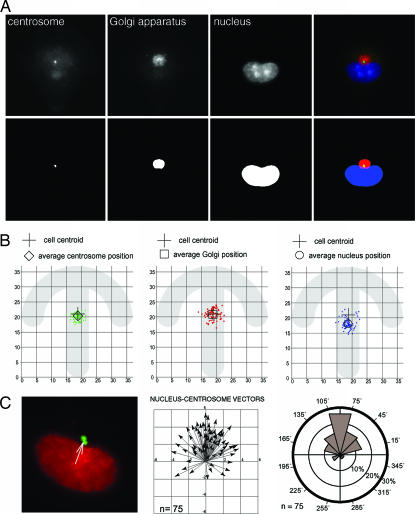
The Nucleus–Centrosome–Golgi Apparatus Is Orientated Relatively to External Cues.
We finally analyzed whether the polarity of the actin and MT networks were further propagated to the internal compartmentalization by measuring the positioning of nucleus, centrosome, and Golgi apparatus. Nucleus, centrosome, and Golgi apparatus of cells plated on crossbow micropatterns were stained after fixation. Remarkably, the Golgi apparatus was compacted around the centrosome instead of being extensively spread out in the cytoplasm as it is in classical culture conditions (SI Fig. 8). Cy3-fibronectin micropatterns were automatically detected in the acquired pictures and used as a reference for position measurements. Nucleus, centrosome, and Golgi were automatically detected by using wavelet segmentation (32). The positions of their centroids parallel to the plane of the coverslip were recorded (Fig. 3A). Interestingly, the centrosomes and Golgi were located close to the cell centroid, whereas the nuclei were off-centered toward nonadhesive edges. Such a disposition is characteristic of polarized cells (8, 23, 24, 33). The calculation of the coordinates of the nucleus–centrosome vectors and the angular distribution of their directions (Fig. 3B) showed that cells were reproducibly polarized. The nucleus–centrosome polarity axis was orientated relative to the asymmetry of the extracellular adhesive pattern. These results showed that cells were internally and cortically polarized in response to the anisotropy of the ECM.
Fig. 3.
Internal polarity. (A) Labeling of centrosome (green), Golgi apparatus (red), and nucleus (blue) in cells plated on crossbow. The positions of the centroids of these organelles were measured with respect to the underlying micropattern after image segmentation (see Materials and Methods). (Upper) Automated image acquisition. (Lower) Image segmentation. (B) The spatial distributions of centrosome and Golgi positions were clustered around the cell centroid. In contrast, nuclei were off-centered toward nonadhesive edges. (C) (Left) Nucleus centroid and centrosome centroid define the nucleus–centrosome vector. (Center) This vector was used as an indicator of internal cell polarity and measured on 75 cells platted on the crossbow micropattern. X and Y axes represent distances in microns. (Right) The circular histogram represents the proportions of nucleus–centrosome vectors pointing in each angular sector and highlights a clear bias of these orientations relative to the adhesive pattern geometry.
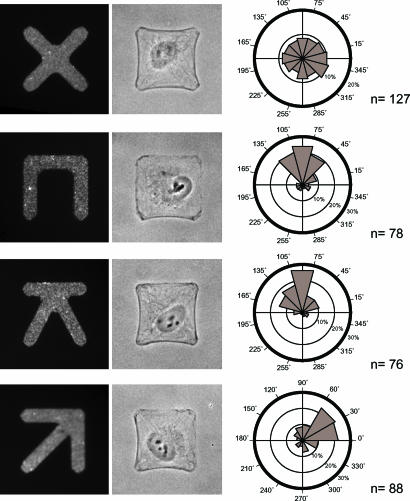
Cell Polarity Is Governed by ECM Geometry.
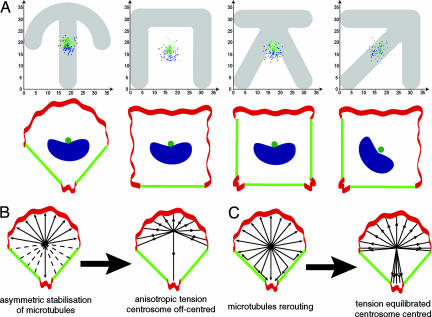
To confirm that the anisotropy of the cell adhesive environment was indeed the critical parameter for controlling cell polarity and compartmentalization, we manipulated the geometry of cell adhesion and monitored the orientation of cell polarity. Because the role of ECM asymmetry could be overridden by the influence of the elongation of cell shape (34) we designed adhesive micropatterns that imposed cells to have similar squared shapes but distinct adhesive patterns (Fig. 4). The internal polarity of RPE1 cells plated on fibronectin micropatterns having a X shape was compared with the polarity of cells plated on micropatterns with a C, K, or arrow shape (Fig. 4). The four micropatterns had the same squared envelope. The X had several symmetry axes, whereas C, K, or arrow had a single symmetry axis, polarized by nonadhesive edges opposing adhesive edges. Cells plated on X displayed no preferential orientation of the nucleus–centrosomes vectors (Fig. 4). They appeared randomly polarized in the plane of the coverslip. In contrast, on micropatterns having a single polarized symmetry axis cells were reproducibly polarized. The cell polarity axis was orientated either along a square side on C and K or along a square diagonal on arrow (Fig. 4). These results demonstrated that the internal polarity of individual nonmigrating cells, as judged by the position of the centrosome with respect to the nucleus, responds to the geometry of the cell adhesive environment.
Fig. 4.
Orientation of cell polarity depends on the adhesive pattern geometry. (Left) RPE1 cells were plated on fibronectin micropatterns having similar square convex envelops. (Center) Spread cells can be visualized in phase-contrast microscopy. (Right) The corresponding angular distributions of nucleus–centrosome vectors were measured as in Fig. 3. On X, cells displayed a random distribution of cell polarity axis. On the others, cell polarity axis was preferentially orientated from nonadhesive edges toward adhesive edges. Micropatterns are 33.5 μm wide.
Centrosome Sits at the Cell Centroid in an Anisotropic Environment.
Interestingly, the cortical anisotropy did not affect the centrosome positioning at the cell centroid (Fig. 5 A). Nuclei were off-centered toward contractile edges. Several models have been shown to account for the central positioning of the centrosome in cells having an homogeneous cortex and its off-center position when the cell cortex is anisotropic (35–37). Here the central positioning is ensured despite the anisotropy of the cell cortex. MT nucleation was isotropic (Fig. 2) and the unequal capping of MTs by APC on the upper and lower side of the cell (Fig. 1) could have led to centrosome off-centering according to only classical models of selective stabilization and pulling of MTs (22, 37) (Fig. 5B). We propose that the rerouting of MTs along nonadhesive cell edges, which guides MTs toward adhesive and protrusive areas that are probably also MT stabilization areas (30, 31, 38) where the concentration of MTs appears to be higher, counterbalances the anisotropic distribution of MT-anchoring sites (Fig. 5C). Such a mechanism implies that the centrosome only needs to support isotropic nucleation of MTs to ensure a central position to the centrosome despite any cortical heterogeneity. Hence, this rerouting of MTs and local actin contractility may contribute to the nucleus positioning toward the nonadhesive zone in the nonmigrating cell and at the cell rear in a migrating cell (24, 39).
Fig. 5.
Centrosome positions at the cell center in anisotropic cortical conditions. (A) (Upper) Centrosome positions (green dots) and nuclei positions (blue dots) as measured in Fig. 3A on crossbow (n = 75), C (n = 78), K (n = 76), and arrow (n = 88) are shown. (Lower) As illustrated in the cartoons, centrosomes were close to the geometrical center of the cell contour (cell centroid), whereas nuclei were off-centered toward nonadhesive and contractile edges (green edges). (B) Selective stabilization model. Actin dynamics at the cell cortex (red regions of cell periphery) have been shown to induce a selective stabilization of MTs. MTs contacting adhesive areas where ruffles take place are supposed to be stabilized (arrows) and therefore more amenable to be put under tension than MTs contacting nonruffling area (dashed lines). This selective stabilization, without MT rerouting, would induce an imbalance in the tension exerted on MTs by minus end-directed motors such as dynein. As a consequence centrosome would be off-centered. (C) MT rerouting compensates cortical anisotropy. MTs contacting a nonadhesive edge keep on growing along this edge without being capped. They are rerouted toward an adhesive site where they can be capped. Thereby the local absence of MT capping on nonadhesive part of the cell cortex is compensated by an increase of MT capping in the next adhesive area. This redistribution of MTs would contribute to pull back the centrosome toward the cell center.
Discussion
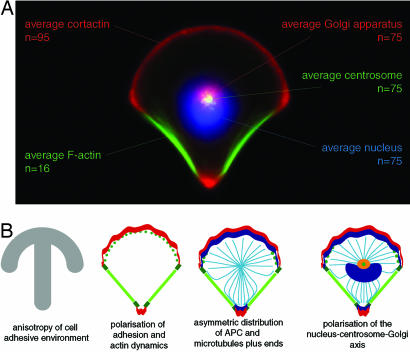
Altogether these results demonstrated that the polarity axes defined by the cortical and internal cell asymmetry were governed by the geometry of external adhesive conditions (Fig. 6). The anisotropy of the cell environment, because of the locations of adhesive and nonadhesive zones, biased the polarization process of RPE1 cells. The actin network became polarized in protruding and contracting regions depending on the presence or absence of adhesion. APC accumulated selectively upon adhesive edges. The growth of MTs appeared modulated by this asymmetric composition of the cortex. MTs reaching the cell cortex in adhesive regions stopped growing, whereas they kept growing along nonadhesive edges. MT growth along nonadhesive edges may be aided by the presence of stress fibers along these edges (40). In addition, the respective positions of the nucleus and the centrosome indicated that the internal cell polarity of these nonmigrating cells was harmonized with the polarity axis of the adhesive environment (Fig. 6A).
Fig. 6.
The polarized cell, a standard reference for normalized cell organization. (A) Cell surface polarity propagates to cell internal polarity. This map of internal cell organization on the crossbow micropattern is the combination of several average distributions of cell organelles. Nuclei, centrosomes, or Golgi stainings were averaged over 75 cells and combined to the average distribution of cortactin and F-actin (see Fig. 1). This combination highlights the coherence between cell internal polarity (orientation of the nuclei–centrosome–Golgi axis) and cell surface polarity (mutual exclusion of actin cytoskeleton protrusions and contractions). (B) From external anisotropic boundary conditions up to internal cell polarity. In response to the anisotropic distribution of fibronectin offered by the micropattern (gray) the distribution of adhesions (green) becomes uneven and concentrated at the extremities of nonadhesive edges. The actin network (red) becomes polarized in a polymerizing meshwork on adhesive edges and stress fibers on nonadhesive edges. Actin-MT connectors such as APC (blue) are segregated in membrane ruffling zones and thereby are anisotropically distributed at the cell periphery. MTs stop growing when reaching these regions, whereas they keep growing along nonadhesive edges where stress fibers are developed. The nucleus–centrosome–Golgi apparatus axis is oriented from the nonadhesive side toward the adhesive side. The Golgi apparatus is compacted around the centrosome, which sits at the cell centroid.
These results oppose the previous statement by Jiang et al. (34) that cell shape and not cell adhesion is the critical parameter that guides the polarity of migrating cells. Noteworthy, in their study, Jiang et al. modified cell adhesion geometry while still imposing highly elongated cell shape. The effect of cell shape elongation and the limited amount of measures in the statistical analyses might have concealed the effect of cell adhesion geometry. In addition, while desorbing cell antiadhesive substrate to promote cell migration out of the patterns they allowed cells to establish new adhesions on the former nonadhesive areas and therefore could not observe the effect of the cell adhesive pattern geometry. Our results with unbiased cell shape do not contradict their conclusions on the role of cell shape elongation but brings new elements to reconsider their interpretation on the role of cell adhesion.
The morphogenetic processes of embryo development and tissue renewal are governed by the regulation of genes expression but also by physical constraints (41, 42). ECM contributes largely to the building of multicellular assemblies: on the one hand, it modulates cell signaling factors (43, 44), and on the other, it forms a structural basis for the development of cellular forces and adhesion (45). Its compliance affects cell contractility and consequently modulates cell differentiation (46, 47), division (48), and migration (49). It also serves as a scaffold transmitting and resisting intercellular and intracellular cellular forces during tissue morphogenesis (45). Our results indicate that in addition to these properties the geometry of this scaffold is a guiding cue for cell polarization. Because it also drives the orientation of cell division (27), ECM geometry appears to play a determinant role in tissue morphogenesis. Noteworthy, cell–cell and cell–ECM contacts both contribute to the establishment of cell polarity in vivo (2, 9, 13). Despite numerous similarities in their components and associated signaling pathways, these two types of contact are not identical (9, 15). What are their respective functions on cell polarity and how they cooperate are still open questions. The two types of extracellular anchorages are exclusive because cells adhere locally to either the neighboring cell or the ECM. Thus cell–cell geometry differs from cell–ECM geometry. The methodology we used in this work could be adapted to investigate the specific effect of cell–cell contact geometry and see how the two adhesive machineries cooperate to control cell polarity.
From a technological point of view, the normalization of cell internal organization should be valuable. Arrays of polarizing adhesive micropatterns, such as the one we describe, are a simple and cost-effective way to control internal organization of cultured cells with a limited intercellular variability. Current efforts for scaling up cell biology analyses are based on the parallelization and miniaturization of cell-based assays (50). However, the variability within data sets is generally a major pitfall preventing detection of genuine modifications of cell phenotypes between distinct conditions in high-throughput screens. Phenotype-based screening is challenged by the identification of characteristic features to establish a selective threshold for the detection of abnormal phenotypes (51). Improvement of image processing allows individual cell analysis, such as distinction between nuclear and cytoplasmic staining (52) or the detection of multiple nuclei (53). However, most functional analyses require more accurate subcellular quantifications, and cell-to-cell variations in the position or morphology of organelles generally prevent such fine and automated measurements. Image filtering and data processing tools have been used downstream of image acquisition to reduce the variability of raw cell data. Sophisticated statistical analyses have been performed on large amounts of data to reveal differences between data sets (54). In all events, these palliative numerical treatments can not resolve high cell-to-cell variability intrinsic to classical in vitro culture conditions. The steady-state intracellular organization of cells on anisotropic micropatterns allows a fine characterization of cell compartment positioning on fixed cells. We showed that internal compartments such as nucleus, centrosome, or the Golgi apparatus were reproducibly positioned with respect to each other (Fig. 6A). Our study is a step toward the establishment of a more complete map depicting the spatial organization of cell components. Such a map could then be used as a standard reference for all studies on these micropatterns. Specific maps would have to be established for each cell type of interest to create a database of standard cell references. The possibility of measuring the spatial distribution of a molecular marker provides key information that was lost in classical devices for automated single cell profiling (53, 55). In conclusion, the upstream reduction of cell variability by the control of the geometry of individual cell adhesive environment is an appropriate way to tackle the limitation of cell image processing and analysis caused by intercellular variability and a promising tool for quantitative cell biology.
Materials and Methods
Micropatterns fabrication, cell culture, and cell deposition on micropatterned coverslips were performed as described (28).
Cell Fixation and Labeling.
Cells were fixed for 5 min in methanol at −20°C for nucleus and centrosome positions measurements and APC labeling. Cells were fixed in 3% paraformaldehyde and 0.5% Triton X-100 in cytoskeleton buffer for 10 min for cortactin labeling. Cells were prepermealized for 15 s with 0.5% Triton X-100 in cytoskeleton buffer and fixed in 3% paraformaldehyde in cytoskeleton buffer for 10 min for cell adhesions and stress fibers labeling. Cells were fixed in 3% paraformaldehyde and 0.5% glutaraldehyde to preserve cell shape for cell centroid calculation. Cells fixed with paraformaldehyde were posttreated with 0.1 M ammonium chloride in PBS for 10 min.
For labeling with primary antibodies we used rabbit polyclonal anti-gamma tubulin (1:500), mouse monoclonal anti-cortactin (1:200, Upstate Biotechnology, Lake Placid, NY), rabbit polyclonal anti-APC (1:500, Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-vinculin (1:200, Sigma/Aldrich, St. Louis, MO), and a mix of his-tagged anti-Giantin TA10 (56) and mouse monoclonal anti-his (1:400, H1029, Sigma/Aldrich). For secondary antibodies we used either FITC-conjugated donkey anti-rabbit (1:200, Jackson ImmunoResearch, West Grove, PA) or Cy5-conjugated goat anti-mouse (1:500, Jackson ImmunoResearch). In some cases cells were stained with FITC-conjugated phaloidin at 1 μM (Sigma/Aldrich) to visualize F-actin and/or with DAPI to visualize the DNA. All steps were performed for 1 h at room temperature in PBS with 3% BSA and 0.1% Triton X-100. Preparations were mounted in MOWIOL solution.
Pictures Acquisitions and Processing.
Pictures of fixed cells were acquired through a ×40 PL APO oil objective with a DMRA microscope (Leica, Rveil-Malmaison, France) and a MicroMax camera (Princetown Instruments, Trenton, NJ) controlled by Metamorph software (Universal Imaging, Dowingtown, PA).
The cell array was automatically scanned. The acquired pictures were aligned by using the fluorescent Cy3-fibronectin micropattern and stacked. The “average cell,” i.e., the average spatial distribution of the labeling of interest, was obtained by calculating the average intensity of each pixel over the stack (SI Fig. 7).
Pictures of nucleus, centrosome, and Golgi were filtered by using wavelet segmentation (32). The positions of organelles centroid were automatically detected in the filtered pictures by using a homemade algorithm.
Cell centroid measurements were performed by using Metamorph morphometric analysis on the contour of average cell shape calculated with the overlay of the average actin and cortactin labeling of cells fixed in paraformaldehyde and glutaraldehyde.
Time-lapse acquisitions of EB1-GFP were performed through a ×100 UplanApo objective (NA 1.35) (Olympus, Rungis, France) on an inverse IX71 microscope (Olympus) heated in a plastic box at 37°C (Life Imaging Science, Reinach, Switzerland) using a CoolSnapHQ camera (Princetown Instruments) controlled by Metamorph acquisition software.
Supplementary Material
Acknowledgments
We thank Drs. Yohanns Bellaiche, Matthew Morgan, and Alexandra Fuchs for critical reading of the manuscript and Dr. I. S. Näthke (University of Dundee, Dundee, Scotland) for providing anti-APC antibody. This work was supported by the Centre National de la Recherche Scientifique, Institut Curie, and Human Frontier Science Program Grant Ref RGP0064/2004 (to M.B.).
Abbreviations
- ECM
extracellular matrix
- APC
adenomatous polyposis coli
- MT
microtubule
- RPE
retinal pigment epithelial.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609267103/DC1.
References
- 1.Nelson WJ. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eaton S, Simons K. Cell. 1995;82:5–8. doi: 10.1016/0092-8674(95)90045-4. [DOI] [PubMed] [Google Scholar]
- 3.Drubin DG, Nelson WJ. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 5.Gumbiner BM. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 6.Braga VM. Curr Opin Cell Biol. 2002;14:546–556. doi: 10.1016/s0955-0674(02)00373-3. [DOI] [PubMed] [Google Scholar]
- 7.Cox EA, Sastry SK, Huttenlocher A. Mol Biol Cell. 2001;12:265–277. doi: 10.1091/mbc.12.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etienne-Manneville S, Hall A. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 9.Gumbiner BM. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 10.Ojakian GK, Schwimmer R. J Cell Sci. 1994;107:561–576. [PubMed] [Google Scholar]
- 11.Ojakian GK, Ratcliffe DR, Schwimmer R. J Cell Sci. 2001;114:941–952. doi: 10.1242/jcs.114.5.941. [DOI] [PubMed] [Google Scholar]
- 12.O'Brien LE, Jou TS, Pollack AL, Zhang Q, Hansen SH, Yurchenco P, Mostov KE. Nat Cell Biol. 2001;3:831–838. doi: 10.1038/ncb0901-831. [DOI] [PubMed] [Google Scholar]
- 13.Yu W, Datta A, Leroy P, O'Brien LE, Mak G, Jou TS, Matlin KS, Mostov KE, Zegers MM. Mol Biol Cell. 2005;16:433–445. doi: 10.1091/mbc.E04-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RG, Schwartz MA. Science. 2004;303:839–842. doi: 10.1126/science.1092571. [DOI] [PubMed] [Google Scholar]
- 15.Geiger B, Bershadsky A, Pankov R, Yamada KM. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 16.DeMali KA, Wennerberg K, Burridge K. Curr Opin Cell Biol. 2003;15:572–582. doi: 10.1016/s0955-0674(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- 18.Nathke IS, Adams CL, Polakis P, Sellin JH, Nelson WJ. J Cell Biol. 1996;134:165–179. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reilein A, Nelson WJ. Nat Cell Biol. 2005;7:463–473. doi: 10.1038/ncb1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Etienne-Manneville S. Traffic. 2004;5:470–477. doi: 10.1111/j.1600-0854.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 21.Palazzo AF, Cook TA, Alberts AS, Gundersen GG. Nat Cell Biol. 2001;3:723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- 22.Gundersen GG, Gomes ER, Wen Y. Curr Opin Cell Biol. 2004;16:106–112. doi: 10.1016/j.ceb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Gotlieb AI, May LM, Subrahmanyan L, Kalnins VI. J Cell Biol. 1981;91:589–594. doi: 10.1083/jcb.91.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomes ER, Jani S, Gundersen GG. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 25.Yeaman C, Grindstaff KK, Nelson WJ. Physiol Rev. 1999;79:73–98. doi: 10.1152/physrev.1999.79.1.73. [DOI] [PubMed] [Google Scholar]
- 26.Matlin KS, Haus B, Zuk A. Methods. 2003;30:235–246. doi: 10.1016/s1046-2023(03)00030-6. [DOI] [PubMed] [Google Scholar]
- 27.Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. Nat Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 28.Thery M, Pepin A, Dressaire E, Chen Y, Bornens M. Cell Motil Cytoskeleton. 2006;63:341–355. doi: 10.1002/cm.20126. [DOI] [PubMed] [Google Scholar]
- 29.Zumbrunn J, Kinoshita K, Hyman AA, Nathke IS. Curr Biol. 2001;11:44–49. doi: 10.1016/s0960-9822(01)00002-1. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 32.Starck JL, Murtagh FD, Bijaoui A. Image Processing and Data Analysis: The Multiscale Approach. Cambridge, UK: Cambridge Univ Press; 1998. [Google Scholar]
- 33.Kupfer A, Louvard D, Singer SJ. Proc Natl Acad Sci USA. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang X, Bruzewicz DA, Wong AP, Piel M, Whitesides GM. Proc Natl Acad Sci USA. 2005;102:975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vallee RB, Stehman SA. Trends Cell Biol. 2005;15:288–294. doi: 10.1016/j.tcb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. J Cell Biol. 2003;162:963–969. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grill SW, Hyman AA. Dev Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Palazzo AF, Eng CH, Schlaepfer DD, Marcantonio EE, Gundersen GG. Science. 2004;303:836–839. doi: 10.1126/science.1091325. [DOI] [PubMed] [Google Scholar]
- 39.Reinsch S, Gonczy P. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 40.Small JV, Kaverina I. Curr Opin Cell Biol. 2003;15:40–47. doi: 10.1016/s0955-0674(02)00008-x. [DOI] [PubMed] [Google Scholar]
- 41.Keller R, Davidson LA, Shook DR. Differentiation. 2003;71:171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- 42.Farge E. Curr Biol. 2003;13:1365–1377. doi: 10.1016/s0960-9822(03)00576-1. [DOI] [PubMed] [Google Scholar]
- 43.Adams JC, Watt FM. Development (Cambridge, UK) 1993;117:1183–1198. doi: 10.1242/dev.117.4.1183. [DOI] [PubMed] [Google Scholar]
- 44.Thiery JP. Curr Opin Genet Dev. 2003;13:365–371. doi: 10.1016/s0959-437x(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 45.Ingber DE. Int J Dev Biol. 2006;50:255–266. doi: 10.1387/ijdb.052044di. [DOI] [PubMed] [Google Scholar]
- 46.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 47.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 48.Huang S, Chen CS, Ingber DE. Mol Biol Cell. 1998;9:3179–3193. doi: 10.1091/mbc.9.11.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lo CM, Wang HB, Dembo M, Wang YL. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 51.Lang P, Yeow K, Nichols A, Scheer A. Nat Rev Drug Discov. 2006;5:343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 52.Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 53.Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smellie A, Wilson CJ, Ng SC. J Chem Inf Model. 2006;46:201–207. doi: 10.1021/ci050404g. [DOI] [PubMed] [Google Scholar]
- 55.Krutzik PO, Nolan GP. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 56.Nizak C, Sougrat R, Jollivet F, Rambourg A, Goud B, Perez F. Traffic. 2004;5:284–299. doi: 10.1111/j.1398-9219.2004.0174.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.