Abstract
Complement sensitizes pathogens for phagocytosis and lysis. We use electron microscopy to examine the structural transitions in the activation of the pivotal protein in the complement pathway, C3. In the cleavage product C3b, the position of the thioester domain moves ≈100 Å, which becomes covalently coupled to antigenic surfaces. In the iC3b fragment, cleavage in an intervening domain creates a long flexible linker between the thioester domain and the macroglobulin domain ring of C3. Studies on two products of nucleophile addition to C3 reveal a structural intermediate in activation, and a final product, in which the anaphylatoxin domain has undergone a remarkable movement through the macroglobulin ring.
Keywords: electron microscopy, proteolytic, thioester
In all three pathways of complement activation, the pivotal step is conversion of the component C3 to C3b, which becomes covalently coupled to pathogens and antigen–antibody complexes through a reactive thioester bond in the thioester-containing domain (TED) (1, 2). Sequential cleavages (Fig. 1A) activate the C3 protein, allowing covalent attachment of C3b on antigen surfaces as a component of C3 and C5 convertases, and modulate the activities of the C3b, iC3b, and C3dg fragments as opsonins for five different cellular receptors (3, 4). Conformational change is evident from differences in binding to cofactors and receptors between C3, C3b, iC3b, and C3c; antibodies to neoepitopes (5, 6), chemical probes (7), H/D exchange (8), and scattering of x-rays and neutrons (9). Previous EM has provided a low-resolution view of C3b (10). Crystal structures have revealed essential atomic information about C3 and its final degradation products C3a, C3c, and C3d (11–14). However, the structures of many important intermediates in C3 activation remain unknown.
Fig. 1.
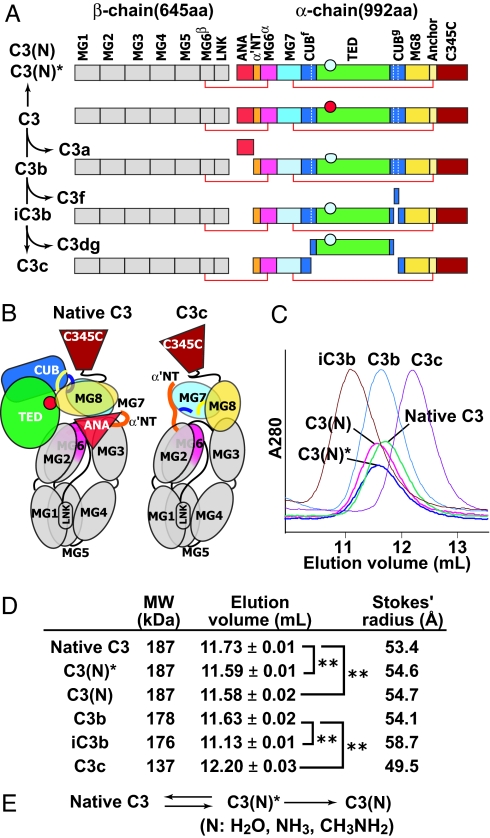
Complement C3 activation. (A) Primary structure of C3 and its products. The thioester group in the TED is shown as a circle before (red) or after (cyan) nucleophilic addition. (B) Domain architecture of C3 and C3c revealed by x-ray crystallography (12). Colors are the same as in A. (C) Overlaid elution profiles of C3 products in S200 size exclusion chromatography. (D) Hydrodynamic behavior of C3 and its products. Elution volume in S200 gel filtration was measured three times for each component. Stokes' radius was calculated from the elution positions of thyroglobulin, γ-globulin, BSA, ovalbumin, and chymotrypsinogen. ∗∗, P < 0.001. (E) The two-step reaction of C3 upon nucleophile (N) addition to the thioester.
Using EM, we now report the conformation of the biologically active species that are intermediate in the pathway between C3 and the terminal fragments. To produce an unambiguous density map, today's cryo-EM techniques require the sample to be structurally homogeneous. As described below, C3 activation is however accompanied by large structural rearrangements and conformational variability. We therefore used negative-stain EM combined with image classification because it is currently the most reliable tool to visualize and characterize heterogeneous protein samples (15). We first compare the EM projections of C3 and C3c with crystal structures. We then report the projection structures of the biologically active intermediate products, C3b and iC3b. Finally, we report the conformation of intermediate and stable products of nucleophile attack on the thioester in native C3, and present a model for C3 activation.
Results
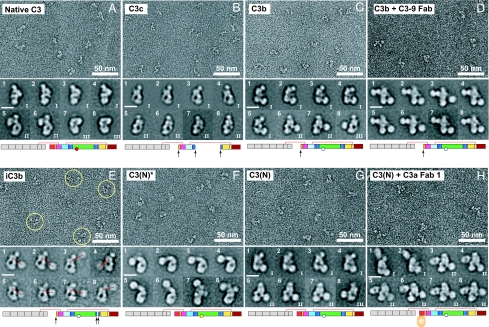
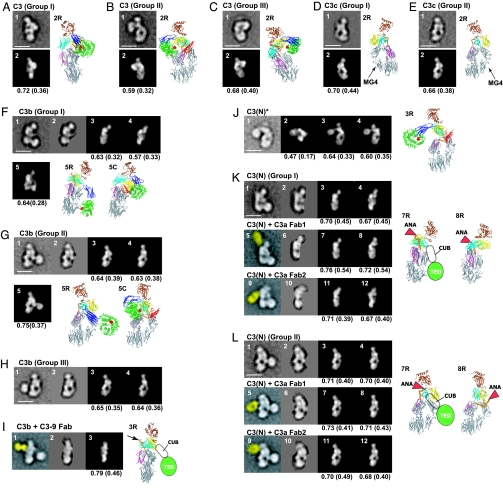
Proteins were purified by Mono-S cation exchange [supporting information (SI) Fig. 6A] and Superdex 200 chromatography (Fig. 1C), and immediately negatively stained with uranyl formate (Fig. 2 and SI Fig. 7). Five thousand to 9,000 particles were subjected to multivariate statistical analysis and multireference alignment into 100–200 classes (SI Table 1 and SI Fig. 8). The eight averages shown for each preparation in Fig. 2 represent the most populous classes, the range of conformations, and most structural detail. For cross-correlation with EM projection averages, crystal structures or models were resolution-filtered to 25 Å, and projections of the 3D map were calculated at 2° angular intervals (Fig. 3).
Fig. 2.
Negative stain EM. Shown are EM raw micrographs and, below each, representative class averages of the indicated C3 products or Fab complexes. (Scale bars in class averages: 100 Å.) In E, some iC3b particles are circled.
Fig. 3.
Cross-correlation between EM class averages and crystal structure projections. (A–E) C3 and C3c. The images represent EM averages (1) and the best correlating C3 or C3c projection (2). Ribbon diagrams in same orientation as a given image have R suffixes. (F–I) C3b and C3b complexed with C3–9 antibody. The images represent EM averages without (1) or with (2) masking, the best correlating C3c (3) or C3Δ (4) projections to the masked averages, two different C3b models made from C3c and C3 CUB domain and TED that correlate best with unmasked averages (5), the corresponding ribbon diagram (5R), and for comparison C3 in the same orientation (5C) superimposed by using MG domains 1, 2, 4, and 5. In I, the C3–9 Fab is highlighted in yellow (1), and the Fab binding site is indicated by an arrow in the corresponding ribbon diagram (3R). (J) C3(N)*. The images represent EM average (1), best correlating projections of C3 (2), model of C3 with TED and CUB domains manually repositioned (3), model of C3c with TED and CUB domains added (4), and ribbon diagram corresponding to image 3 (3R). (K and L) C3(N) and C3(N) complexed with C3a Fab 1 (from Quidel) and Fab 2 [H13 (30)]. The images represent EM averages without (1, 5, and 9) or with (2, 6, and 10) TED (and Fab) masked, the best correlating C3c (3, 7, and 11) or C3Δ (4, 8, and 12) projections to the masked averages, and corresponding C3c (7R) or C3Δ (8R) ribbon diagrams with positions of ANA and for C3c the CUB domain and TED schematically indicated. Corresponding ribbon diagrams for the Fab 2 complex are essentially identical in orientation to those in 7R and 8R. The Fab density in images 5 and 9 is highlighted in yellow.
Native C3 itself is biologically inactive. C3 is derived by proteolytic processing of a single-chain proC3 precursor to β- and α-chains (Fig. 1A). C3 contains eight MG (macroglobulin) domains that form a key ring, with MG1–4 completing one ring, MG5 and MG6 forming a second, overlapping half-ring, and MG7 and MG8 extending the second half-ring (12, 14) (Fig. 1B). The C345C domain forms a knob-like protrusion (Fig. 1B). The TED is inserted in the CUB domain, which in turn is inserted between MG domains 7 and 8 (Fig. 1 A and B). The reactive thioester bond is shielded from solvent within the interface of the TED with the MG8 domain (Fig. 1B). EM revealed three different groups (I–III) of C3 class averages with different orientations on the grid (Fig. 2 A Lower), which show excellent cross-correlation with projections from the crystal structure (Fig. 3 A–C). The domains in C3 are most clearly visible in the group II orientation (Fig. 3B, compare with Fig. 1B).
C3c is an inactive, terminal fragment released from iC3b by the factor I protease (Fig. 1A). The C3c crystal structure demonstrates marked rearrangements compared with C3 in the MG7 and MG8 domains, lesser movements in the MG2, MG3, and MG6 domains that abut MG7 and MG8, and reorientation of the C345C domain (12) (Fig. 1B). Most remarkably, the 19-residue α′NT (α′ N-terminal) segment, which becomes N-terminal in the α-chain after removal of C3a (Fig. 1A), moves from the MG3/MG8 side of the key ring in C3 to the MG7 side in C3c, by slipping between the MG1–4 ring and MG5–6 half-ring at its C-terminal connection to MG6 (12). EM averages of C3c clearly revealed the MG domain key ring and the C345C knob (Fig. 2B) and correlated excellently with the C3c crystal structure (Fig. 3 D and E). Orientations of C3c with the MG1–4 ring in front or back can clearly be distinguished by the orientation of the C345C knob and the weak density of the key ring where it is only a single MG domain thick at MG4 (compare D and E in Fig. 3). These features enable the orientation of the C3c moiety within the fragments described below to be assessed even without reference to cross-correlations.
C3 convertases cleave C3 to generate C3b and the C3a anaphylatoxin (ANA) (2, 16). About 10% of C3b molecules become covalently linked to antigenic surfaces through ester or amide linkages to the ester moiety of the thioester, and the other 90% react with water and are released into the fluid phase (2). In contrast to C3, C3b can function as a component of C3 and C5 convertases, bind to factor H, be cleaved by factor I protease, and bind to CR1 and CRIg receptors (2, 4, 16, 17). EM averages of C3b reveal a C3c-like core, with the TED much more distal from the C345C knob than in C3 (Fig. 2C). To determine whether the conformational changes seen in the C3c crystal structure had occurred in C3b, the TED density was masked from C3b averages, and they were cross-correlated with maps prepared from (i) the C3c crystal structure and (ii) the C3 crystal structure with the domains lacking in C3c deleted (C3Δ). All 15 averages examined, including those from groups I, II, and III, showed a higher correlation with C3c than C3Δ (Fig. 3 F–H and data not shown). This highly significant result (P = 2 × 2−15 = 6 × 10−5) demonstrates that the conformation of the C3c moiety, i.e., the 8 MG domains and the C345C domain in C3b, most closely resembles that in C3c. The C3c conformation also explains the marked change in position of ≈100 Å of the TED seen in C3b [compare Fig. 2A (5 and 6) with Fig. 2C (6 and 7)]. The rearrangements in C3c bring the connections to the CUB domain in MG7 and MG8 closer to the MG1/MG4 end of the key ring and orient the connections more toward the MG1/MG4 end (12). C3b models in which CUB domain and TED were attached to C3c matched the EM 2D projections well [Fig. 3 F (1, 5, and 5R) and G (1, 5, and 5R)].
To further examine conformational change in C3b, we used the C3–9 antibody, which binds to a neoepitope present in C3b (5). The complex of the C3–9 Fab with C3b gave class averages (Fig. 2D) very similar to those of C3b group I [Fig. 3I (3R)], except for an additional density corresponding to the Fab. The Fab bound to the MG7/MG8 region of C3b, i.e., clearly in-between the C345C knob and the MG1–6 ring (Fig. 3I). Because C3–9 binds to C3c and not C3 (5) (SI Fig. 6 E, H, and I), this finding supports a C3c-like conformation of the MG7 and MG8 domains in C3b.
iC3b is produced when factor I protease releases the C3f fragment (Fig. 1A). iC3b lacks convertase activity, and compared with C3b loses reactivity for CR1 and gains reactivity for CR2, CR3 (integrin αMβ2), and CR4 (integrin αXβ2) (3, 17). iC3b is the predominant C3 opsonin in vivo (17, 18). Binding of iC3b to CR2 costimulates B cell responses, and binding to CR3 and CRIg enables phagocytosis of opsonized pathogens and immune complexes (4, 18). iC3b has a markedly larger hydrodynamic radius than other C3 fragments (Fig. 1 C and D) as described (9). EM class averages of iC3b show one density that is identical to C3c, and a second, randomly oriented density that corresponds to the TED (Fig. 2E). Although in some class averages the TED or C3c moieties are averaged out (SI Fig. 8E), each is present in raw images (circled in Fig. 2E). After masking of the TED density, 10 of 10 class averages showed higher cross-correlation to C3c than C3Δ (P = 0.002). Thus iC3b, like C3b, assumes the C3c-like conformation, in agreement with reaction of C3–9 antibody with iC3b (5). Furthermore, release of the C3f fragment from the CUB domain's C-terminal portion destroys the fold of the CUB domain, and its N-terminal portion (Fig. 1A) becomes a flexible linker between the C3c-like moiety and the TED (Fig. 4D). Measuring the distance between the C terminus of the MG7 domain in the C3c moiety and the center of the TED (red lines in Fig. 2E) estimates the length of this flexible tether of 51 residues as 100 ± 16 Å (mean ± SD, n = 15), or 2 Å per residue, in agreement with expectations for an unfolded polypeptide chain.
Fig. 4.
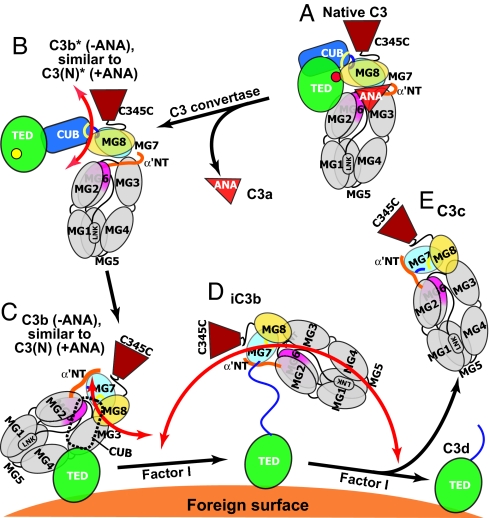
Schematic diagram of domain rearrangements and flexibility in C3 products. The positions of domains are schematically represented. The CUB domain in C3(N) and C3b is shown with a dashed outline, because it did not provide clear density in EM class averages. The unfolded N-terminal portion of the CUB segment in iC3b is shown as a blue line. The range of mobility of the key ring moiety relative to the TED is represented with orange arrows. (N) represents nucleophiles added to the thioester including H2O and CH3NH2.
The alternative pathway of complement is initiated by tick-over of C3, i.e., the spontaneous low-rate (t1/2 ∼ 200 h) hydrolysis of its thioester bond (19). Cleavage by nucleophiles (N) such as water, NH3, or CH3NH2 of the thioester bond results in formation of a conformational intermediate termed C3(N)* (Fig. 1E). Upon further incubation, the C3(N)* intermediate can yield either native C3, with reformation of the thioester, or undergo a slow (t1/2 ∼ 1–3 h) irreversible conformational transition to C3(N) (Fig. 1E) (20). C3, C3(N)*, and C3(N) can be completely isolated from one another by cation exchange chromatography (20). C3(N)* and C3(N) each show a significant increase of hydrodynamic radius compared with native C3 (Fig. 1 C and D). EM averages of the C3(N)* intermediate show that nucleophilic addition to the thioester causes the TED and CUB domain to swing away from the key ring (Fig. 2F). The TED is a well defined globular density that remains on the MG1/MG2 side of the key ring, with some variation in the angle of its connection through the CUB domain (Fig. 2F). The loss in C3(N)* of the interface between the TED and MG8 that protects the thioester bond agrees with the marked change in TED structure around the thioester when the thioester is abrogated by the C988A mutation (12, 13). Projections from native C3 coordinates correlated poorly with C3(N)* [Fig. 3J (2)]. By contrast, movement of the TED and CUB domains relative to the main body of C3 yielded a model [Fig. 3J (3R)] with a much better cross-correlation and that closely matched EM averages [Figs. 3J (3) and 4B]. A model using C3c coordinates showed lesser correlation [Fig. 3J (4)]. Importantly, the C3–9 Fab did not react with C3(N)*, suggesting that the conformation of its MG7 and MG8 domains is C3-like (SI Fig. 6F).
Conformational change of C3(N)* to C3(N) which occurs on an hour time scale is irreversible (20). Despite the presence of the ANA domain, C3(N) has spectroscopic and biological properties that are essentially indistinguishable from those of C3b (7, 21) enabling it to function in the alternative complement pathway. C3(N) class averages (Fig. 2G) are almost identical to those of C3b (Fig. 2C) and show the same predominant orientations on the substrate. As with C3b, the TED moves markedly toward the MG1/MG4 end of the key ring [Fig. 2G (1–8) and 4C]. When the TED was masked, 10 of 10 C3(N) class averages correlated better with C3c than C3Δ (Fig. 3 K and L) (data not shown). Furthermore, in contrast to C3(N)*, C3(N) reacts with C3–9 antibody (5) (SI Fig. 6G), showing a conformational transition in the MG7 and MG8 domains.
The above work suggests that the domains present in C3c have a similar conformation in C3c, C3b, iC3b, and C3(N), and a distinct conformation in C3 and C3(N)*. The key difference revealed between the C3 and C3c crystal structures is that after removal of the C3a fragment, i.e., the ANA domain, the newly created α′NT segment snakes through a long tunnel between the MG2, MG3, and MG6 domains, and emerges on the opposite side of the MG key ring (12) (Fig. 1B). The ANA domain acts as a stopper of this movement in C3. For a similar motion of the α′NT to occur in C3(N), the ANA stopper would have to move through the key ring along with the α′NT.
To localize the position of the small ANA domain in C3(N), we used two different C3a-specific Fabs (Fab 1 and 2). These Fabs were unreactive with C3, unreactive or weakly reactive with C3(N)*, and strongly reactive with C3(N) (SI Fig. 6 B–D and data not shown). The increased reactivity with C3(N) compared with C3(N)* is consistent with the hypothesized change in location and greater exposure of α′NT in a C3c-like conformation of C3(N), allowing better Fab access to the ANA domain. EM of the Fab–C3(N) complexes revealed similar orientations as seen with C3b and C3(N), with an additional density corresponding to the Fab bound to the MG7/MG8 end of the key ring, on the side opposite the TED (Fig. 2H and SI Fig. 8I). Complexes with the Fab and TED masked out [Fig. 3 K and L (6 and 10)] cross-correlated better with C3c [Fig. 3 K and L (7 and 11)] than C3Δ [Fig. 3 K and L (8 and 12)]. In C3c and C3Δ projections in the group II orientation, which is by far most populous for C3(N) Fab complexes, the α′NT appears on opposite sides of the key ring in C3c [Fig. 3L (7R)] and C3Δ [Fig. 3L (8R)]. Importantly, using two different Fabs, all group II class averages show binding of Fab on the left side [Fig. 2H (3–8)], i.e., the side of the key ring in which α′NT is present in C3c [Fig. 3L (5 and 9 and 7R)].
Discussion
Our findings suggest that the irreversible conformational change in C3(N) and its similarity to C3b result from movement through the key ring of the α′NT and the ANA domain, and explain long-standing observations on the similar functional properties of C3b and C3(N) (7). Whereas conversion of C3b* to C3b occurs on a time scale of <1 sec (19), the presence of the bulky ANA domain in C3(N)* slows conformational conversion to C3(N) to a time scale of ≈2 h (20). These kinetics show that an extremely rare, and thus high-energy, protein conformation is required for the transition from C3(N)* to C3(N). Insertion–deletion analysis shows that short deletions of a loop in MG6 that is surface-exposed and distal from α′NT increase C3(N) production (22). Unfolding of MG6, to which α′NT connects, could enable passage of ANA and α′NT through the water-filled cavity in the middle of the MG key ring, and is a plausible rate-limiting step for conversion to C3(N). H/D exchange shows that the MG3 and MG6 domains are more stable in C3(N) than C3 (8), consistent with the irreversibility of the conversion to C3(N).
Based on these results and the C3, C3c, and C3d crystal structures, we propose a model for C3 activation (Fig. 4). In solution, C3 is predominantly in the same conformation as in crystal structures (12, 14) (Fig. 4A). Cleavage of C3 to C3b with release of the ANA domain as C3a, makes the thioester formed by Cys-988 and Gln-991 metastable (t1/2 ∼ 60 μsec). The initial event is usually attack of His-1104 on the thioester, with rearrangement to an acyl-imidazole intermediate that is susceptible to attack by hydroxyl groups and water (2). The movements required for acyl-imidazole formation are blocked in the TED interface with MG8 (12) and are well approximated by the crystal structure of the isolated TED in C3d, which also is a good model for the TED with the thioester cleaved with nucleophile (12, 13). Therefore, we may expect a swing of the TED/CUB unit away from MG8 and flexibility in metastable C3b, i.e., C3b*, similar to that in C3(N)* (Fig. 4B). This would aid in exposing the acyl-imidazole bond of the TED to nucleophilic attack by nearby pathogenic and antigenic surfaces. Differences in orientation between the TED and MG key ring in different class averages of C3(N)* provide evidence for flexibility, summarized by orange arrows in Fig. 4, and would also facilitate reaction of the acyl-imidazole bond in C3b* with pathogenic surfaces.
After convertase cleavage, conformational change to C3b is rapid (19). We find a profound, 100-Å movement of the TED toward the MG1/MG4 end of the key ring. The EM and C3–9 Fab data together with previous studies with C3–9 antibody (5) support similar rearrangements in C3b, C3(N), and C3c of the MG7 and MG8 domains, and their connections to the CUB domain. Residues in the α′NT and CUB domain are important for binding of factor B, factor H, and CR1 (23–25). Therefore, the conformational rearrangements we have described in these domains help explain the acquisition by C3b and C3(N) of convertase, cofactor, and receptor-binding activities.
It is very interesting that the orientation of the TED with respect to the MG ring differs so much between C3, the putative C3b*-like C3(N)* conformation, and C3b (Fig. 4 A–C). These changes in orientation have implications for the distance between the site of the C3 convertase and the site of covalent linkage on a foreign surface, and the radiation of foci of alternative pathway activation across surfaces (26).
Upon conversion of C3b to iC3b, the remaining portion of the CUB domain is converted to a flexible linker between the C3c and TED moieties (Fig. 4D). Thus, convertase activity and CR1 binding by C3b and C3(N) must depend on a structured CUB domain. Conversely, unfolding of the CUB domain in iC3b enhances binding to CR2, CR3, and CR4 (3, 17). Because the TED is covalently linked to antigenic surfaces, it will be partially shielded by the C3c moiety, and hence the flexibility of iC3b ensures efficient recognition by the receptors that mediate costimulation and phagocytosis.
Overall, our findings demonstrate large-scale rearrangements in C3 between four different conformational states (Fig. 4 A–D) that are governed by thioester rearrangement and release of small proteolytic fragments, and regulate the bioactivity of C3 products. Our findings with C3b and C3(N) show that C3 is in a kinetically trapped conformation, and that the ANA domain acts as a barrier to conformational change. Irreversible conformational change triggered by protease cleavage, both in C3b and iC3b, provides a clear-cut mechanism for regulating the activity of these most important C3 products.
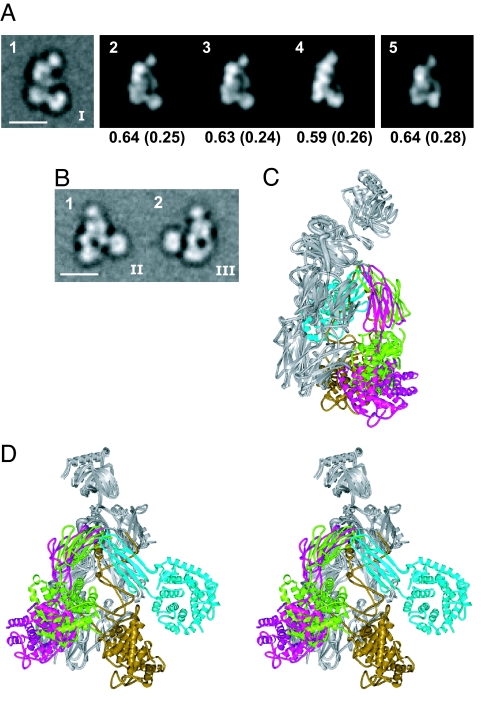
After the completion of the above study, three groups independently published C3b crystal structures (27–29). These structures fully support our conclusions on C3b by showing that C3b undergoes a rearrangement to a conformation that is essentially identical to that of C3c, and that the TED moves 75–100 Å between C3 and C3b. Moreover, the C3b crystal structures of Janssen et al. (29) and Weismann et al. (28) are essentially identical to one another and cross-correlate well with the predominant orientation on the grid we found for C3b in the group I class averages [Fig. 5A (2 and 3)]. The C3b model we made from C3c with manual rigid body movements of the CUB domain and TED from C3 shows a similarly good cross-correlation [Fig. 5A (5)], and similar arrangement of domains (Fig. 5 C and D).
Fig. 5.
Comparison between crystal structures and EM class averages of C3b. (A) Cross-correlation of a group I class average of C3b (image 1) with projections from C3b crystal structures from Janssen et al. (29) (image 2; PDB entry 2I07), Wiesmann et al. (28) (image 3; PDB entry 2ICF, the C3b moiety only), and Ajees et al. (27) (image 4; PDB entry 2HR0), and from the rigid body model described in Fig. 3 (image 5) was as described in the legend of Fig. 3. (B) Representative group II (image 1) and III (image 2) class averages of C3b from Fig. 2C. (C and D) Two crystal structures and two EM-based rigid body models of C3b from Fig. 3 F and G superimposed by using the C3c-moiety (shown in gray for all four). The CUB domain and TED are green (group I model), cyan (group II model), magenta [structure by Janssen et al. (29)], and gold (structure by Ajees et al.). (C) View in group I orientation of A. (D) Stereoview in group II orientation of image 1 of B.
Materials and Methods
Protein Purification.
Human C3 (Complement Technology, Tyler, TX) was separated into C3 and C3(H2O) [termed here C3(N)] fractions by Mono-S ion exchange chromatography (Amersham Biosciences, Piscataway, NJ) (20) (SI Fig. 6A). C3b, iC3b, and C3c were from Calbiochem (San Diego, CA). Methylamine (100 mM) treatment of C3 was in 50 mM Bicine (pH 8.3)/100 mM NaCl at room temperature for 30 min, immediately followed by purification of C3(CH3NH2)* [termed here C3(N)*] away from C3 and C3(N) by Mono-S chromatography (20) (SI Fig. 6A). Final purification of all proteins was by size-exclusion S200 chromatography (Amersham Biosciences) in 20 mM Tris·HCl (pH 7.5)/150 mM NaCl/1 mM CaCl2/1 mM MgCl2. Purified protein was immediately applied to EM grids.
C3–9 antibody (5) was from Fitzgerald Industries (Concord, MA). The C3a antibody used to make C3a Fab 1 was from Quidel (San Diego, CA). C3a H13 antibody (30), a generous gift from R. Burger (Robert Koch Institute, Berlin, Germany), was used to make C3a Fab 2. To prepare Fab fragments, IgG was digested by papain (USB, Cleveland, OH) with 1:1,000 enzyme/substrate ratio for 4 h at 37°C in 20 mM sodium phosphate (pH 7.0)/10 mM cysteine/5 mM EDTA. Reaction was terminated by adding 20 mM iodoacetamide, for 15 min at 4°C, followed by dialysis against 50 mM Tris·HCl (pH 8.0). The Fab fragment was applied to Hitrap Q FF column (Amersham Biosciences) equilibrated in 50 mM Tris·HCl (pH 8.0), and eluted with a linear 0 M to 0.5 M NaCl gradient.
Electron Microscopy and Image Processing.
Preparation of negatively stained grids, acquisition of micrographs, and image digitization, classification, and cross-correlation were performed exactly as described (31) except that 5,000–9,000 monodisperse particles were picked and windowed into 100 × 100- or 75 × 75-pixel images (4.04 Å per pixel) and aligned into 100–200 classes. Pseudoatomic models of the alternative conformation of C3b and C3(N)* were prepared by using UCSF Chimera (32).
Acknowledgments
We thank Piet Gros and Motomu Shimaoka for reviewing the manuscript. This work was supported by National Institutes of Health Grants AI72765 (to T.A.S.) and GM62580 (to T.W.), the Astellas Foundation (N.N.), and a Yoshida Scholarship (to N.N.).
Abbreviations
- ANA
anaphylatoxin
- TED
thioester-containing domain.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609791104/DC1.
References
- 1.Walport MJ. N Engl J Med. 2001;344:1058–1066. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 2.Law SK, Dodds AW. Protein Sci. 1997;6:263–274. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown EJ. Curr Opin Immunol. 1991;3:76–82. doi: 10.1016/0952-7915(91)90081-b. [DOI] [PubMed] [Google Scholar]
- 4.Helmy KY, Katschke KJ, Jr, Gorgani NN, Kljavin NM, Elliott JM, Diehl L, Scales SJ, Ghilardi N, van Lookeren Campagne M. Cell. 2006;124:915–927. doi: 10.1016/j.cell.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 5.Hack CE, Paardekooper J, Smeenk RJ, Abbink J, Eerenberg AJ, Nuijens JH. J Immunol. 1988;141:1602–1609. [PubMed] [Google Scholar]
- 6.Tamerius JD, Pangburn MK, Muller-Eberhard HJ. J Immunol. 1985;135:2015–2019. [PubMed] [Google Scholar]
- 7.Isenman DE, Kells DI, Cooper NR, Muller-Eberhard HJ, Pangburn MK. Biochemistry. 1981;20:4458–4467. doi: 10.1021/bi00518a034. [DOI] [PubMed] [Google Scholar]
- 8.Winters MS, Spellman DS, Lambris JD. J Immunol. 2005;174:3469–3474. doi: 10.4049/jimmunol.174.6.3469. [DOI] [PubMed] [Google Scholar]
- 9.Perkins SJ, Sim RB. Eur J Biochem. 1986;157:155–168. doi: 10.1111/j.1432-1033.1986.tb09652.x. [DOI] [PubMed] [Google Scholar]
- 10.Smith CA, Pangburn MK, Vogel CW, Muller-Eberhard HJ. J Exp Med. 1984;159:324–329. doi: 10.1084/jem.159.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber R, Scholze H, Paques EP, Deisenhofer J. Hoppe-Seyler's Z Physiol Chem. 1980;361:1389–1399. doi: 10.1515/bchm2.1980.361.2.1389. [DOI] [PubMed] [Google Scholar]
- 12.Janssen BJ, Huizinga EG, Raaijmakers HC, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 13.Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. Science. 1998;280:1277–1281. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- 14.Fredslund F, Jenner L, Husted LB, Nyborg J, Andersen GR, Sottrup-Jensen L. J Mol Biol. 2006;361:115–127. doi: 10.1016/j.jmb.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Ohi M, Li Y, Cheng Y, Walz T. Biol Proc Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reid KB, Porter RR. Annu Rev Biochem. 1981;50:433–464. doi: 10.1146/annurev.bi.50.070181.002245. [DOI] [PubMed] [Google Scholar]
- 17.Ross GD. J Immunol Methods. 1980;37:197–211. doi: 10.1016/0022-1759(80)90307-5. [DOI] [PubMed] [Google Scholar]
- 18.Yan J, Vetvicka V, Xia Y, Hanikyrova M, Mayadas TN, Ross GD. Immunopharmacology. 2000;46:39–54. doi: 10.1016/s0162-3109(99)00157-5. [DOI] [PubMed] [Google Scholar]
- 19.Muller-Eberhard HJ. Annu Rev Biochem. 1988;57:321–347. doi: 10.1146/annurev.bi.57.070188.001541. [DOI] [PubMed] [Google Scholar]
- 20.Pangburn MK. J Biol Chem. 1992;267:8584–8590. [PubMed] [Google Scholar]
- 21.Pangburn MK, Schreiber RD, Muller-Eberhard HJ. J Exp Med. 1981;154:856–867. doi: 10.1084/jem.154.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogata RT, Ai R, Low PJ. J Immunol. 1998;161:4785–4794. [PubMed] [Google Scholar]
- 23.Taniguchi-Sidle A, Isenman DE. J Immunol. 1994;153:5285–5302. [PubMed] [Google Scholar]
- 24.O'Keefe MC, Caporale LH, Vogel CW. J Biol Chem. 1988;263:12690–12697. [PubMed] [Google Scholar]
- 25.Oran AE, Isenman DE. J Biol Chem. 1999;274:5120–5130. doi: 10.1074/jbc.274.8.5120. [DOI] [PubMed] [Google Scholar]
- 26.Kozel TR, Wilson MA, Murphy JW. Infect Immun. 1991;59:3101–3110. doi: 10.1128/iai.59.9.3101-3110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ajees AA, Gunasekaran K, Volanakis JE, Narayana SV, Kotwal GJ, Krishna Murthy HM. Nature. 2006;444:221–225. doi: 10.1038/nature05258. [DOI] [PubMed] [Google Scholar]
- 28.Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, McCallum SA, Embuscado L, Deforge L, Hass PE, van Lookeren Campagne M. Nature. 2006;444:217–220. doi: 10.1038/nature05263. [DOI] [PubMed] [Google Scholar]
- 29.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 30.Burger R, Bader A, Kirschfink M, Rother U, Schrod L, Worner I, Zilow G. Clin Exp Immunol. 1987;68:703–711. [PMC free article] [PubMed] [Google Scholar]
- 31.Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA. Immunity. 2006;25:583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]