Abstract
Neurons, with their long axons and elaborate dendritic arbour, establish the complex circuitry that is essential for the proper functioning of the nervous system. Whereas a catalogue of structural, molecular, and functional differences between axons and dendrites is accumulating, the mechanisms involved in early events of neuronal differentiation, such as neurite initiation and elongation, are less well understood, mainly because the key molecules involved remain elusive. Here we describe the establishment and application of a microscopy-based approach designed to identify novel proteins involved in neurite initiation and/or elongation. We identified 21 proteins that affected neurite outgrowth when ectopically expressed in cells. Complementary time-lapse microscopy allowed us to discriminate between early and late effector proteins. Localization experiments with GFP-tagged proteins in fixed and living cells revealed a further 14 proteins that associated with neurite tips either early or late during neurite outgrowth. Coexpression experiments of the new effector proteins provide a first glimpse on a possible functional relationship of these proteins during neurite outgrowth. Altogether, we demonstrate the potential of the systematic microscope-based screening approaches described here to tackle the complex biological process of neurite outgrowth regulation.
INTRODUCTION
Proper functioning of the nervous system requires connections between neurons and their targets. Undifferentiated cells have to expand cylindrical extensions with a growth cone at a distal tip in a process called neurite outgrowth. This process, for all neurons, can be seen as a three step event. First, the round shape of the cell is broken down and a filopodia-like extension is generated. Second, the extension elongates and it is transformed into a proper neurite. Finally, the neurite differentiates into an axon or a dendrite (reviewed in da Silva and Dotti, 2002). Understanding how the neurite initiation-site forms and what discriminates it from the rest of the cell on the molecular level is a major challenge, not only because it is an important event during nervous system development but also because the establishment of subcellular domains with distinct molecular components and properties is a fundamental problem in cell biology. Another important question is how neurite elongation is controlled? Genetic and biochemical approaches have been applied to identify and characterize single molecular components involved in neurite outgrowth. For example, a wealth of evidence in recent years suggests that actin and microtubules dynamics and membrane traffic play a central role in neurite outgrowth initiation and neurite elongation. One of the first events that happens during initiation of neurite outgrowth is actin cytoskeleton rearrangement, mediated by a plethora of actin remodeling proteins, in particular by the Rho family of GTPases and their associated regulators (reviewed in da Silva and Dotti, 2002). Cdc42, RhoA, and Rac1 are the best characterized Rho GTPases with a role in neurite outgrowth, and, although results differ from one neuronal model to the other, it is believed that Cdc42 and Rac1 promote neurite outgrowth, whereas RhoA suppresses it (reviewed in Gallo and Letourneau, 1998; da Silva and Dotti, 2002). Microtubules are equally essential for neurite outgrowth (Solomon, 1980; Rochlin et al., 1996), and thus proteins that regulate microtubule dynamics are also potential regulators of neurite outgrowth (for reviews see Dehmelt and Halpain, 2004; Grenningloh et al., 2004). It has been proposed that microtubules have a role in neurite elongation (Rochlin et al., 1996); however, microtubules have also been observed to enter the lamellipodia of morphologically undifferentiated neuroblastoma cells before neurite initiation, suggesting they might have a role in neurite initiation as well (Dehmelt et al., 2003). Membrane traffic regulators such as SNAREs (Osen-Sand et al., 1993; Shirasu et al., 2000), Rab proteins (Huber et al., 1995), and the exocyst complex (Vega and Hsu, 2001) have also been implicated in neurite outgrowth regulation. Such role of membrane traffic regulation becomes apparent considering that the growth of neurite processes from the cell body involves a massive increase in cell surface area (Futerman and Banker, 1996). Regulation of neurite outgrowth is an important aspect not only for proper development of the nervous system but also for tissue regeneration after nerve injury and the treatment of neuropathological conditions (Jones et al., 2001).
Despite this wealth of available information on neurite outgrowth regulation, it is well recognized in the field that more systematic approaches to identify new molecules involved are needed in order to obtain a more complete molecular description underlying this complex process (Grant, 2003). Recently, a large-scale RNAi screen was used to identify genes involved in synapse structure and function (Sieburth et al., 2005) underlining the importance of such systematic approaches.
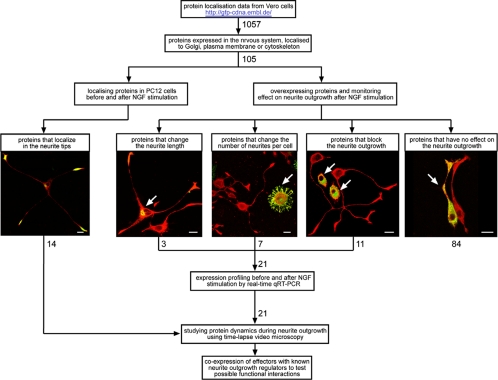
Here we describe the establishment and application of a microscope-based screening approach to identify novel human proteins involved in neurite outgrowth. The rational of our experimental strategy is shown in Figure 1. Candidate proteins that are expressed in nervous tissue are preselected based on their subcellular localization in Vero cells. Cells are transfected with chosen GFP-tagged open reading frames (ORFs) and subsequently their localization during neurite outgrowth and the effect of their overexpression on this process are investigated. The assumption of this is that those proteins localizing to the site of neurite outgrowth and/or interfere with the process due to their overexpression are highly likely to be involved in this process. This identified 21 proteins that effected neurite outgrowth when overexpressed in cells. Time-lapse microscopy was then used to determine when these effector proteins affect neurite outgrowth. Finally, effectors are coexpressed with known neurite outgrowth regulators to reveal possible functional interactions.
Figure 1.
Strategy to identify proteins involved in neurite outgrowth in PC12 cells. Proteins were pre-selected, from a pool of 1057 GFP-tagged human proteins, according to their subcellular localization in Vero cells. Only proteins known to be expressed in brain tissues were selected. The subcellular localization of the proteins before and after NGF stimulation and their effect on neurite outgrowth when overexpressed were then determined. For those proteins that showed an effect on neurite outgrowth when overexpressed, potential changes in their expression levels in response to NGF stimulation were determined by real-time qRT-PCR. Protein dynamics during neurite outgrowth was then monitored in living cells by time-lapse microscopy. Next, the identified effector proteins were coexpressed with known neurite outgrowth regulators to test the possible functional interactions. All experimental data can be seen at: http://neurite.embl.de.
MATERIALS AND METHODS
Materials
GFP-tagged ORFs were generated and prepared as previously described (Simpson et al., 2000). Full-length cDNA for SNAP-25, VAMP-2, and RhoA were obtained from the RZPD in pCMV-SPORT6, pOTB7, and pDNR-LIB vectors, respectively and subcloned into the pEGFP-C2 vector at the EcoRI/BamHI sites. EB1-GFP and EB3-GFP constructs are kind gifts from Niels Galjart (Erasmus University, Rotterdam, The Netherlands), AnnexinA4-YFP and CaMKIIβ-GFP are kind gifts from Carsten Schultz (European Molecular Biology Laboratory [EMBL], Heidelberg, Germany), Cdc42-GFP is kind gifts from Panos Kouklis (University of Ioannina, Greece). Inhibitors used are: ROCK inhibitor Y-27632 (Calbiochem, San Diego, CA), Gsk3β inhibitor CHIR99021 (kind gift from Rudi Marquez and Natalia Shapiro from University of Dundee, United Kingdom).
Cell Culture
PC12 cells (clone 6-15), a kind gift from Dionisio Martin Zanca (University of Salamanca, Spain) were grown in medium containing DMEM (Life Technologies, Rockville, MD) supplemented with 5% fetal calf serum (PAA, Coelbe, Germany), 10% heat inactivated horse serum (Life Technologies), 1% l-glutamine (Life Technologies), and 1% penicillin/streptomycin (Life Technologies) at 37°C in 5% CO2. To facilitate otherwise poorly attaching PC12 cells all plasticware used was coated with type I collagen from rat tail (Sigma, Steinheim, Germany).
Time-Lapse Microscopy
PC12 cells were plated in 3- mm glass-bottom dishes (Willco, Amsterdam, The Netherlands) that were previously coated with poly-l-lysine (Sigma) to facilitate cell attachment to the glass. They were incubated at 37°C in 5% CO2 for 16 h, after which they where transfected with 0.5 μg of appropriate cDNA and 1.5 μl of Lipofectamine2000 (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. After 4 h the transfection medium was replaced with fresh medium and the cells were left for another 16 h. After this the medium was replaced with CO2-dependent imaging medium (2.2 g/l NaHCO3, pH 7.4, 115 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgCl2, 1.2 mM K2HPO4, 2 g/l d-glucose) supplemented with 1% horse serum and 2 mM of free radical scavenger TROLOX (Sigma), and the cells were mounted on Leica ASMDW or Leica AF6000 LX multiposition microscopes (Wetzlar, Germany) equipped with environment control boxes. Next, neurite outgrowth was stimulated with 100 ng/ml NGF (Promega, Madison, WI), and an image was acquired in fluorescence and transmission (DIC) channels every 3 min over 12–16 h. For sequence management and analysis, NIH open source software ImageJ was used.
Real-Time Quantitative RT-PCR
Total RNA was prepared from PC12 cells treated with NGF (100 ng/ml) for 24 h and nontreated cells using the Invisorb Spin Cell RNA Mini Kit (Invitek, Berlin, Germany). cDNA synthesis was achieved with 2 μg of total RNA using the SuperScript II Reverse Transcriptase (Invitrogen). A master mix containing a single gene-specific primer set, cDNA, ddH2O, and 2x-SYBR-Green PCR mastermix (Applied Biosystems, Foster City, CA) was prepared and distributed in a 96-well qPCR plate. The reaction was run on ABI Prism 7500 real-time PCR machine (Advanced Biosystems, Foster City, CA). For each experiment the reactions were done in triplicate. An actin-specific primer set was used as an internal standard. Negative controls lacking the cDNA template were run with every assay to assess specificity. Sequences of the primers used can be found in Supplementary Table 3. A threshold cycle (Ct) was determined for each sample using the exponential-growth phase and the baseline signal from fluorescence versus cycle number plots. ΔCt values were obtained by subtracting the Ct value of the gene from nontreated sample from the Ct value of the gene from NGF-treated sample. ΔCt value was also calculated for the control actin gene. ΔCt value of actin was subtracted from ΔCt value of the gene of interest to get ΔΔCt value. Positive ΔΔCt values indicate more PCR cycles and therefore less mRNA after NGF treatment, which would indicate gene repression. Conversely, negative ΔΔCt values indicate more mRNA after NGF treatment, which means gene up-regulation. Because of the exponential nature of PCR the gene's “fold change” value was calculated by 2−ΔΔCt. Each gene's “fold change” value represents the mean ± SD of three independent experiments.
Neurite Outgrowth Assay
Cells were plated on poly-l-lysine–coated glass coverslips in 12-well plates and left for 16 h at 37°C in 5% CO2. Next, they were transfected with 0.5 μg of appropriate cDNA and 1.5 μl of Lipofectamine2000 (Invitrogen) according to manufacturer's protocol. Two wells were transfected with the same cDNA of interest, and subsequently one was treated with NGF and the other not. Two wells on each plate were transfected with AnnexinA4, which served as a negative control because it has a role in nervous system function but has no role in neurite initiation or elongation. Two wells on each plate were transfected with a GTP-restricted Sar1 mutant that blocks neurite outgrowth and therefore served as a positive control. After 4 h the medium was replaced with fresh medium, and the cells were left for another 12 h to express the transfected proteins. After this the medium was replaced with low serum medium (DMEM supplemented with 1% horse serum, 0.5% fetal calf serum, 1% l-glutamine, and 1% penicillin/streptomycin), and the cells were either stimulated with 100 ng/ml nerve growth factor (Promega) or not. After 24 h the cells were fixed and permeabilized in methanol on −20°C for 4 min, plasma membrane was stained with ConA-Alexa647 (Molecular Probes, Eugene, OR) to highlight the outline of the cells, and coverslips were mounted in Mowiol and imaged using a Leica SP2 AOBS confocal microscope. Neurite length and the number of neurites per cell were measured manually using ImageJ software. Extensions longer than 15 μm (longer that one cell diameter) were considered a neurite. For every protein tested, both transfected cells and surrounding nontransfected cells were measured and compared. The distributions of the neurite length and the distributions of the number of neurites per cell in nontransfected and transfected cells, average neurite length, and average number of neurites per cell ± SD were calculated. A minimum of 60 cells were counted. Statistical significance was tested with Student's t test (p < 0.0001).
Databases
All the data generated in this project are deposited in the following website: http://neurite.embl.de.
RESULTS
Establishment and Application of the Screen for Proteins Involved in Neurite Outgrowth
Rat PC12 cells were chosen as the model cell culture system as they are round shaped in their undifferentiated state but extend long neurites and acquire a sympathetic neuron-like phenotype in response to nerve growth factor (NGF; Greene and Tischler, 1976). A PC12 cell line stably transfected with human NGF receptor (PC12 6-15) was chosen, as this line shows a rapid response to NGF stimulation (Hempstead et al., 1992), and thus is suitable for the screening experiments.
We took advantage of a collection of human full-length cDNAs (Wiemann et al., 2001), the ORFs of which have previously been GFP-tagged at their N and C termini, and have been classified according to their subcellular localization in Vero cells (Simpson et al., 2000). From the 1057 available GFP-tagged ORFs only those were considered that were derived from a fetal brain or hypothalamus cDNA library (Wiemann et al., 2001) or SOURCE database information indicates their expression in the brain. From these, only the GFP-tagged proteins localizing to the plasma membrane, cytoskeleton, or Golgi complex (Simpson et al., 2000; see web page http://gfp-cdna.embl.de) were chosen for further functional experiments. This resulted in a total of 105 preselected proteins of which 35 localize to the cytoskeleton, 31 to the Golgi complex, 22 to the plasma membrane, and 17 to both the plasma membrane and Golgi complex (see Supplementary Table 1).
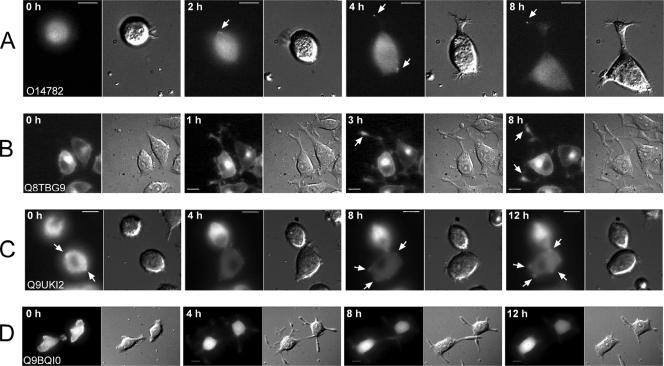
First, all the 105 preselected GFP-tagged proteins were expressed and localized in PC12 cells, and the results were compared with their localization in Vero cells (see Supplementary Table 1). Only those cells showing low expression levels of the GFP-tagged candidate proteins were analyzed. Fourteen proteins localized to neurite tips after NGF stimulation (see Table 1 and Supplementary Figure 1). To resolve the moment of their association with neurite tips, they were further analyzed by time-lapse microscopy. Depending on the time of their association with the neurite tip after NGF stimulation, they were classified as “early” or “late” (see summary in Table 1, selected examples in Figure 4, A and B, and Supplementary Videos 1 and 2).
Table 1.
List of GFP-tagged proteins localizing to neurite tips after NGF stimulation
| Early* | Late* |
|---|---|
| Kinesin-like protein Kif3C (O14872) Sorbin, SH3P12 (Q9UFT2) Gamma-BAR (Q9H0V0) Hypothetical protein, Gamma-BAR splice variant (Q96GG6) Coiled-coil domain containing 8, CCDC8 (Q9H0W5) Hypothetical protein (Q8N3S9) |
Neuronal specific septin 3 (Q8N3P3) Calcium-independent phospholipase A2 gamma (Q8N3I3) Kinesin family member 23, Kif23 (Q8WVP0) Synaptoporin (Q8TBG9) Hypothetical protein (Q9Y4S1) Hypothetical protein (Q8NFW0) Serine protease 23 (O95084) Secreted phosphoprotein 1, SPP-1 (Q96IZ1) |
* The 14 proteins localizing to neurite tips after NGF stimulation were analyzed in more detail by time-lapse microscopy and classified as “early,” when association occurred before any neurites were visible or coincidental with the beginning of neurite outgrowth. They were classified as “late” when association was first observed after neurites had already clearly formed. SwissPROT IDs are given in parentheses.
Figure 4.
Examples of protein dynamics during neurite outgrowth and overexpression effects on neurite growth. PC12 cells were transfected with GFP-tagged ORFs and incubated for 12 h at 37°C. After that they were analyzed by time lapse microscopy as described in Materials and Methods. Images were acquired in fluorescence and transmission (DIC) channels every three minutes for 12 h. Cells expressing very low levels of fluorescently labeled protein were selected for time-lapse imaging. Proteins localizing to neurite tips were classified according to the time of the protein association with the neurite tip or site of neurite growth (Table 1). (A) O14782 (Kif3C) is an example of very early protein association with the sites of neurite outgrowth. (B) Q8TBG9 (synaptoporin) is an example of later association with the neurite tip. Proteins that blocked neurite outgrowth were classified regarding the dynamics of neurite growth (Table 3) (C) Q9UKI2 (Borg2) is an example where neurites never develop. (D) Q9BQI0 (hypothetical protein) is an example of neurite growth followed by neurite retraction. Arrows point to site of protein accumulation. Time, top left corner in each image, indicates the time after addition of NGF. Scale bars, 10 μm.
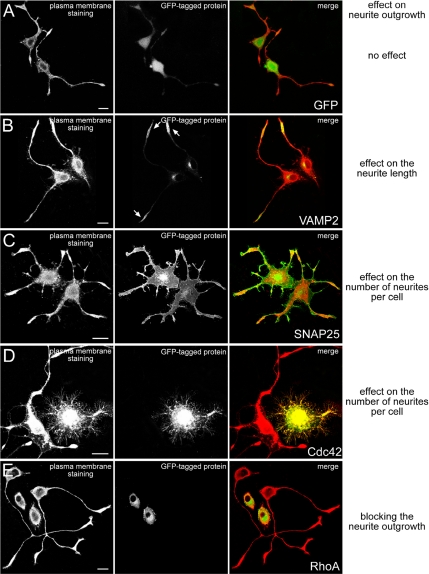
Next the 105 candidate proteins were overexpressed in PC12 cells, and their effect on neurite outgrowth was determined. To characterize parameters such as reproducibility and sensitivity of this approach, several control experiments were conducted first. Negative controls such as GFP alone, several major cytoskeletal and membrane traffic components, and proteins with a clear role in neuronal function but not involved in neurite outgrowth all showed little or no effect on neurite outgrowth when overexpressed (Supplementary Table 2, see also examples in Figure 2 and quantification in Supplementary Figure 2). Several proteins, known from the literature to influence neurite number or length or to block neurite outgrowth when overexpressed were used as positive controls (Supplementary Table 2, see also examples in Figure 2 and quantification in Supplementary Figure 2). Overexpression of the positive controls influenced neurite outgrowth consistent with their involvement in neurite outgrowth proposed in the literature (see citations in Supplementary Table 2), demonstrating the potential of our experimental approach to identify effectors of neurite outgrowth.
Figure 2.
Examples of control proteins affecting neurite outgrowth when overexpressed in PC12 cells. PC12 cells were transfected with GFP-tagged ORFs and incubated for 12 h at 37°C. Thereafter cells were stimulated with NGF for 24 h, fixed and stained with ConA-Alexa647 to highlight the outline of the cells. The images show phenotypes of GFP (A) or control proteins VAMP2 (B), SNAP25 (C), Cdc42 (D), and RhoA (E) known to have an effect on neurite outgrowth upon overexpression. Each panel consists of three pictures: The left one shows ConA-Alexa647 staining; the middle one shows the overexpressed GFP-tagged protein; and the right one is a merge picture with the ConA-Alexa647 staining depicted in red and the GFP-tagged protein in green. Statistical analysis for these experiments is summarized in Supplementary Figure 1. Scale bars, 10 μm.
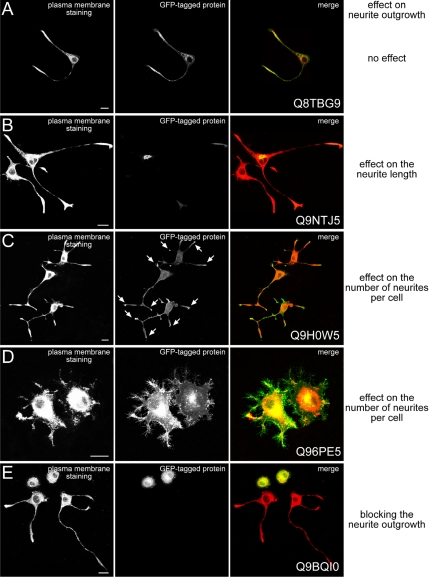
From the 105 preselected human GFP-tagged ORFs (see above) 21 were identified as effectors of neurite outgrowth and either influenced neurite length or number or completely blocked neurite outgrowth when overexpressed (Table 2; see also selected examples in Figure 3, the phenotypes off all effectors is shown in Supplementary Figure 3 and quantification in Supplementary Figure 2). Classification of the identified effector proteins according to the phenotypes obtained in the control experiments with the positive effectors VAMP2 (Figure 2B), SNAP25 (Figure 2C), Cdc42 (Figure 2D), or RhoA (Figure 2E) is summarized in Table 2 and Supplementary Figure 3, and the quantification of the phenotypes is shown in Supplementary Figure 2. From these 21 effectors, 10 cDNAs and their encoded proteins (Swissprot ID: Q9NTJ5, Q9H0W5, Q9H0H6, Q69YW2, Q9BQJ4, Q9H083, Q9BQI0, Q9Y4P9, Q9H0Q7, and Q96PE5) are completely new, and the data presented here are the first information about their putative function. Four effectors of neurite outgrowth (RP/EB family member 2, Borg2, PRC17, and γ-BAR) represent previously characterized proteins, although a role in neurite outgrowth had so far not been described for them (Renner et al., 1997; Joberty et al., 1999; Pei et al., 2002; Neubrand et al., 2005). For seven of the effector proteins identified (muscarinic receptor m2, synaptopodin, RP/EB family member 3, SH3P12, frizzled homologue 7, Kinesin2, and Kif2C) some role in neurite differentiation has already been described (Baxter and Chiba, 1999; Deller et al., 2000; Nakagawa et al., 2000; Lebre et al., 2001; Ciani and Salinas, 2005; Hirokawa and Takemura, 2005), but not specifically in the context of neurite initiation and elongation.
Table 2.
Proteins affecting neurite outgrowth upon overexpression in cells
| SNAP-25 Phenotype* | ||
| Protein name (SwissPROT ID) | Localization | Overexpression effect on neurite outgrowth |
| Coiled-coil domain containing 8, CCDC8 (Q9H0W5) | Plasma membrane, neurite tips | Increases the number of neurites per cell (from 2 to 3.5 neurites per cell on average) |
| Frizzled homologue 7, FZD7 (Q96B74) | Plasma membrane | Increases the number of neurites per cell (from 2 to 2.8 neurites per cell on average) |
| RP/EB family member 3 (Q9UPY8) | Microtubules | Increases the number of neurites per cell |
| Cdc42 Phenotype | ||
| Hypothetical protein (Q9H0H6) | Plasma membrane | Induces numerous filopodia-like extensions all around the cell cortex and increases the number of neurites per cell |
| Hypothetical protein, new isoform of m2 receptor (Q4VBK6) | Plasma membrane | Induces numerous filopodia-like extensions all around the cell cortex and short neurites |
| Hypothetical protein (Q69YW2) | Plasma membrane | Induces numerous filopodia-like extensions all around the cell cortex |
| Transmembrane protein 10, TM10 (Q96PE5) | Plasma membrane, Golgi | Induces numerous filopodia-like extensions all around the cell cortex |
| VAMP-2 Phenotype | ||
| TBC1 domain family member 3, PRC17 (Q8IZP1) | Plasma membrane | Decreases neurite length by 57% |
| γ-BAR (Q9H0V0) | Golgi, neurite tips | Decreases neurite length by 27% |
| SACM1L (Q9NTJ5) | Golgi | Increases neurite length by 25% |
| RhoA Phenotype | ||
| Transmembrane 4 superfamily member 10, TM4SF10 (Q9BQJ4) | Plasma membrane | Blocks neurite outgrowth in 50% of the cells, remaining 50% is not affected |
| Hypothetical protein, zinc and ring finger 1, ZNRF1 (Q9H083) | Plasma membrane, Golgi | Blocks neurite outgrowth in 43% of the cells |
| Hypothetical protein (Q9BQI0) | Cytoplasm, Golgi, cytoskeleton | Blocks neurite outgrowth in 81% of the cells |
| Sorbin, SH3P12 (Q9UFT2) | Cytoskeleton, neurite tips | Blocks neurite outgrowth in 70% of the cells |
| Hypothetical protein (Q9Y4P9) | Microtubules, nucleus | Blocks neurite outgrowth in 65% of the cells |
| Hypothetical protein (Q9H0Q7) | Microtubules in neurites only, not in the cell body | Blocks neurite outgrowth in 59% of the cells, remaining 41% is not affected |
| CRIB-containing BORG2 protein (Q9UKI2) | Cytoskeleton, cytoplasm | Blocks neurite outgrowth in 82% of the cells |
| Kinesin family member 2C, KIF2C (Q99661) | Cytoplasm | Blocks neurite outgrowth in 72% of the cells |
| Synaptopodin-2, SYP-2 (Q9UMS6) | Microtubules | Blocks neurite outgrowth in 72% of the cells |
| RP/EB family member 2 (Q15555) | Microtubules | Blocks neurite outgrowth in 59% of the cells |
| Kinesin-associated protein 3, KAP3 (Q5VXW0) | Cytoplasm | Blocks neurite outgrowth in 70% of the cells |
* The effects of over-expression of the GFP-tagged proteins were classified according to the phenotypes obtained with the positive control cDNAs encoding VAMP2, Cdc42, SNAP25, and RhoA as shown in Figures 2 and 3 and Supplementary Figure 3 and quantification of these phenotypes is shown in Supplementary Figure 2.
Figure 3.
Examples of candidate proteins affecting neurite outgrowth when overexpressed in PC12 cells. PC12 cells were transfected with GFP-tagged ORFs and incubated for 12 h at 37°C. Thereafter cells were stimulated with NGF for 24 h, fixed and stained with ConA-Alexa647 to highlight the outline of the cells. The images show phenotypes of identified effector proteins, which have similar effects on neurite outgrowth as their corresponding controls shown in Figure 2. Each panel consists of three pictures: The left one shows ConA-Alexa647 staining; the middle one shows the overexpressed GFP-tagged protein; and the right one is a merge picture with the ConA-Alexa647 staining depicted in red and the GFP-tagged protein in green. Statistical analysis for these experiments is summarized in Supplementary Figure 1. Scale bars, 10 μm.
Overexpression of eight proteins was toxic to PC12 cells as judged by their low transfection efficiency (<1%) and the observed abnormal cell morphology when overexpressed. Three of the GFP-tagged proteins tested (Swissprot ID: P10636, Q8WVP0, and Q9P0W8) induced microtubule cytoskeleton abnormalities such as microtubule bundling and were not considered as effectors of neurite outgrowth and therefore were excluded from further analyses. Seventy-three proteins were classified as not interfering with neurite initiation or elongation when overexpressed.
Quantification of gene expression of the positive hits by real-time quantitative RT-PCR (qRT-PCR) strengthened our functional results. We quantified the mRNA expression in response to NGF of 15 identified effector proteins. This confirmed their expression in PC12 cells (see Supplementary Table 3 and Supplementary Figure 4). For the remaining six effector proteins we were unable to identify their rat orthologues. The relative expression levels of several genes changed during the course of neurite outgrowth. Interestingly, the changes in the gene's expression level in some of them could be correlated with the effect of protein overexpression on neurite outgrowth. Three genes that are down-regulated during neurite outgrowth (Swissprot ID: Q9BQI0, Q99661, and Q15555) encode proteins that blocked neurite outgrowth when overexpressed. Similarly, three genes that are up-regulated during neurite outgrowth (SwissprotID: Q9H0V0, Q9UPY8, and O14782) encode proteins whose overexpression induced neurite outgrowth or had no effect (Supplementary Table 3 and Supplementary Figure 4).
Next, for the proteins that blocked neurite outgrowth, we determined when the effect on neurite outgrowth occurred. For this we transfected PC12 cells with the respective GFP-tagged effectors and monitored neurite outgrowth by time-lapse microscopy for up to 16 h after NGF stimulation. This revealed two different phenotypes. One was characterized by the complete absence of neurites at any time after NGF stimulation (Table 3; also see example in Figure 4C and Supplementary Video 3). The second phenotype initially showed no apparent effect on neurite outgrowth after NGF stimulation compared with control transfected cells. However, at later time points most neurites retracted, resulting in virtually no neurites at 12 h after NGF stimulation (Table 3; also see example in Figure 4D and Supplementary Video 4). This potentially reflects on the requirements for those proteins at different stages in neurite outgrowth.
Table 3.
Time-lapse classification of proteins that block neurite outgrowth
| Initial neurite outgrowth followed by neurite retraction | No neurite outgrowth at all |
|---|---|
| Hypothetical protein (Q9BQI0) | Transmembrane 4 superfamily member 10 (Q9BQJ4) |
| Sorbin, SH3P12 (Q9UFT2) | Hypothetical protein, zinc and ring finger 1 (Q9H083) |
| Synaptopodin-2, SYP-2 (Q9UMS6) | CRIB-containing BORG2 protein (Q9UKI2) |
| Hypothetical protein (Q9Y4P9) | Kinesin family member 2C, KIF2C (Q99661) |
| RP/EB family member 2 (Q15555) | |
| Kinesin-associated protein 3, KAP3 (Q5VXW0) | |
| Hypothetical protein (Q9H0Q7) |
Experimental Testing of a Possible Functional Relationship between the Identified Effector Proteins
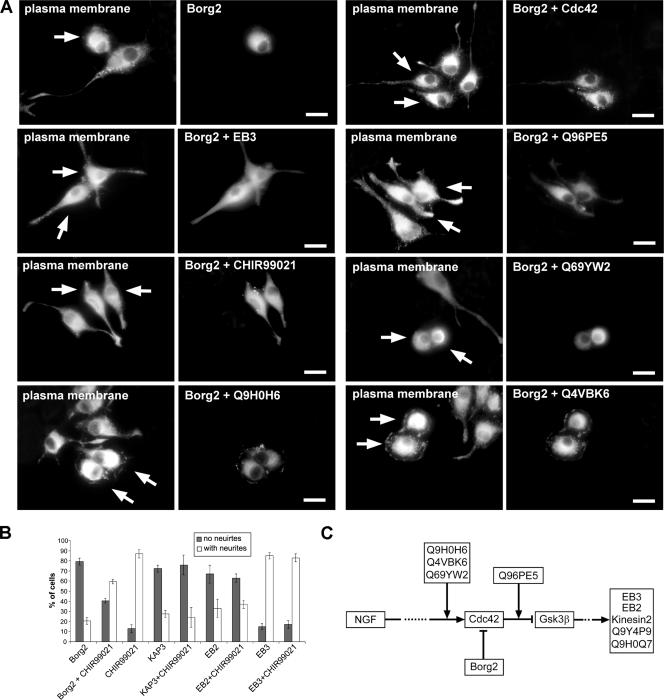
To further strengthen our screening results, we next wanted to establish a possible functional relationship between the identified effector proteins. To this end several of the identified proteins were coexpressed, and the effect on the neurite outgrowth was analyzed. Borg2, a negative regulator of Cdc42 (Joberty et al., 1999), was identified in our experiments as blocking neurite outgrowth when overexpressed (see Figure 4C and Supplementary Video 3). However, Cdc42 was able to partially overcome this neurite outgrowth block of Borg2 when both proteins were coexpressed (Figure 5). Cells with short neurites could be observed when Cdc42 and Borg2 were coexpressed, whereas those expressing Borg2 alone showed virtually no neurites (Figure 5). Expression of Cdc42 alone induced numerous filopodia-like extensions around the cell cortex (Figure 2D). Because the uncharacterized proteins Q9H0H6, Q4VBK6, Q96PE5, and Q69YW2 induced a similar phenotype as Cdc42 when overexpressed (Table 2 and Supplementary Figure 3), we next asked whether these proteins were also able to overcome the Borg2 phenotype. The coexpression of either Q9H0H6, Q4VBK6, or Q69YW2 with Borg2 was unable to rescue the Borg2-induced phenotype (Figure 5). However, Q96PE5 was able to rescue the Borg2 phenotype, and filopodia-like structures around the cell cortex were seen in the majority of cells (Figure 5), similar to when Q96PE5 or Cdc42 were overexpressed on their own (Figures 2D and 3D). Similarly, coexpression of Borg2 and EB3, a microtubule (+)-end binding protein (Nakagawa et al., 2000), whose overexpression on its own induced an increase in the number of neurites per cell (Table 2 and Supplementary Figure 3), also resulted in a rescue of the Borg2 phenotype (Figure 5). Taken together, these results show that the novel proteins Q9H0H6, Q4VBK6, and Q69YW2 cannot rescue the Borg2 phenotype, whereas Q96PE5 and EB3 are able to do so.
Figure 5.
Analysis of the functional relationship between Borg2/Cdc42, Gsk3β and other proteins identified in the screen. (A) Cells were cotransfected with Borg2 and other proteins identified in the screen as described in Materials and Methods. Each panel consists of two pictures: one shows ConA-Alexa647 staining of the plasma membrane to highlight the outline of the cell and the second shows the overexpression of Borg2 alone and coexpression of Borg2 with Cdc42, EB3, Q96PE5, CHIR99021, Q69YW2, Q9H0H6 and Q4VBK6, respectively. Note that Q4VBK6, Q9H0H6, Q69YW2 were unable to rescue Borg2 phenotype while Q96PE5, EB3, CHIR99021 and Cdc42 were able to partially rescue the Borg2 phenotype when coexpressed. Arrows point to cotransfected cells. Scale bars, 10 μm. (B) Cells were transfected with indicated proteins as described in Materials and Methods and treated with NGF with or without the presence of 2 μM CHIR99021. Shown is the percentage of cells with and without neurites. Error bars represent the SD of the mean of at least two independent experiments. Ten images with 20× objective were acquired and a minimum of 150 cells were counted. (C) A schematic summary of the experiments addressing a functional relationship between the effector proteins identified.
Interestingly, in our screening experiments, overexpression of the Frizzled receptor (FZD7), a well-known negative regulator of Gsk3β (for a review see Ciani and Salinas, 2005) increased the number of neurites per cell (see Table 2 and Supplementary Figure 3). Furthermore, it has been shown that Cdc42 is able to inhibit Gsk3β and thereby control microtubule stabilization and cell polarity in migrating astrocytes (Etienne-Manneville and Hall, 2003). Gsk3β was also shown to regulate microtubule stabilization in NGF induced axon growth in DRG neurons (Zhou et al., 2004). Together this suggests Gsk3β being an important regulator of neurite outgrowth.
Next we wanted to investigate the relationship of our identified effector proteins with Gsk3β function in neurite outgrowth. Cells transfected with effector proteins were treated with the specific Gsk3β inhibitor CHIR99021 (Ring et al., 2003) and analyzed for neurite outgrowth. The percentage of cells bearing four or more neurites increased in a dose-dependent manner from 11.5 ± 2% in cells treated with NGF only to 55 ± 2% in cells that were treated with NGF and 20 μM CHIR99021 (Supplementary Figure 5). Interestingly, neurite outgrowth block induced by Borg2 overexpression could be partially rescued by simultaneously inhibiting Gsk3β with CHIR99021 (Figure 5). Cells with short neurites could be observed, whereas those expressing Borg2 in the absence of the inhibitor showed virtually no neurites (Figure 5; see also Supplementary Video 3). In contrast, Gsk3β inhibition was unable to rescue the neurite outgrowth block induced by overexpression of the microtubule binding proteins EB2 and KAP3 (kinesin2 complex component; Figure 5B) or the novel proteins Q9Y4P9 and Q9H0Q7 (not shown). A summary of these experiments addressing a possible functional relationship between the effector proteins identified here is shown in Figure 5C.
DISCUSSION
Here we describe the establishment and application of a microscopy-based approach to identify new proteins involved in neurite initiation and elongation in response to NGF in PC12 cells. The candidate proteins used for the experiments were preselected according to their tissue-specific expression and subcellular localization in Vero cells (Simpson et al., 2000). On the basis of these criteria we chose from 1057 available proteins 105 that we finally tested in our experiments for a possible involvement in neurite outgrowth. This revealed 21 effector proteins with a potential role as regulators of neurite initiation or elongation. One reason for this high success rate is likely to be the preselection of the candidate proteins based on their expression in nervous tissue and subcellular localization.
We also performed cotransfection experiments, which allowed us to determine the action of new, previously uncharacterized human effector proteins with respect to Gsk3β and Cdc42-Borg2 function. These cotransfection experiments further support our screening data and provide a first glimpse on how these newly identified proteins might interact with established regulators of neurite outgrowth, such as Gsk3β and Cdc42. Further experiments on these effector proteins will be necessary in order to determine if and how they may be part of a network regulating neurite outgrowth. The experimental data presented here should provide an excellent basis for such detailed studies with the aim to obtain a more comprehensive view on the molecular networks underlying neurite outgrowth.
Although it cannot be formally ruled out, several lines of evidence exist to assure that our overexpression approach did not yield unspecific results. First, proteins with an established role in neuronal differentiation and function but not involved in neurite initiation or elongation showed no apparent effect on neurite outgrowth in our experiments. Second, 14 proteins of the 105 tested were identified as localizing to neurite tips, but 11 of them had no effect on neurite number or length when overexpressed. Third, a number of proteins we identified here as affecting neurite outgrowth are homologues or orthologues of well-characterized proteins already implicated in some aspects of neurite outgrowth in the literature. Finally, real-time qRT-PCR showed that the effector proteins identified are indeed expressed in PC12 cells and the relative expression of several of them changed during the course of neurite outgrowth in accordance with our functional data obtained by overexpression. For example, some genes that are down-regulated after NGF stimulation encode proteins whose overexpression blocked neurite outgrowth.
Our choice of time-lapse microscopy to determine when the neurite localized proteins associate with the site of neurite growth is a powerful technique to complement functional screens. Because protein localization is strongly coupled to function, the fact that proteins associating with the site of neurite growth very early could signify a function in neurite initiation or elongation, as opposed to a function for example in synaptic vesicle regulation or synapse formation when one would expect late accumulation in the neurite tip. From the 14 neurite tip proteins discovered, only 3 (Swissprot ID: Q9UFT2, Q9H0V0, and Q9H0W5) had an effect on neurite outgrowth when overexpressed, and all 3 associated with the sites of neurite growth very early (see Table 1). By extending this approach to a larger number of neurite tip–localized proteins it should be possible to generate a temporal map of protein association with neurite tips that could serve as a basis for network construction and more detailed functional studies.
By using high-throughput and high-content microscopy technology in fixed (Liebel et al., 2003) and living (Neumann et al., 2006) cells together with fully automated image analysis to quantify neurite outgrowth (Ramm et al., 2003), it should be possible to scale-up our approach here from low- to high-throughput and might thus become the basis for future large-scale neuronal-based proteomic studies.
It is interesting to note that there appears to be a strong correlation between the subcellular localization of the effector proteins and the phenotype generated. Six of seven proteins that induced an increase in the number of neurites per cell in our experiments localize to the plasma membrane. This finding is consistent with the view that the neurite outgrowth begins when external signals activate specific factors on the plasma membrane, which in turn triggers the formation of a neurite initiation site that should be distinct from the rest of the plasma membrane at the molecular level. Similarly, 9 of 11 proteins identified as blockers of neurite outgrowth localized either to the actin or microtubule cytoskeleton. This is in full agreement with numerous earlier studies establishing a major role of the cytoskeleton in the control of neurite outgrowth (for reviews see Gallo and Letourneau, 2000; da Silva and Dotti, 2002). Finally, effector proteins that affected neurite length upon overexpression are all localized to the Golgi or plasma membrane and are involved in membrane traffic regulation. This is consistent with the view that the delivery of new membrane to growing neurites is an important factor for their growth (for reviews see Futerman and Banker, 1996; Valtorta and Leoni, 1999).
Three proteins identified in our screen are of additional interest because they are potentially involved in human neurological disorders. Swissprot IDs: Q96PE5 (Nobile et al., 2002) and Q9BQJ4 (Christophe-Hobertus et al., 2001) have a potential role in temporal lobe epilepsy and hereditary X-linked mental retardation, respectively. However, in both cases previous research reported no mutations in these genes in families carrying the disease (Nobile et al., 2002; Christophe-Hobertus et al., 2004), and thus both genes are presently considered as “unlikely” to be involved in the respective disorders. However this earlier work did not investigate the 5′-regulatory regions of the respective genes, and therefore mutations occurring in the promoter region of the genes cannot be excluded as the cause of disease. Consistent with our data, it is therefore possible that in fact deregulation of protein expression and not loss of function is what is causing the neurological disorders related to these two genes. Therefore, based on our data here, the involvement of Q96PE5 and Q9BQJ4 in temporal lobe epilepsy and X-linked mental retardation could be possible and should thus be reevaluated with respect to their regulation and expression. The third disease-linked protein identified is SH3P12. It has been shown to interact with Ataxin-7 and huntingtin, both of which are key players in the neurodegenerative diseases Spinocerebellar ataxia and Huntington disease, respectively (Lebre et al., 2001). Because, overexpression of SH3P12 blocked neurite outgrowth in PC12 cells, nervous system dysfunction may be related to changes in SH3P12 expression.
Extending this approach to a larger number of candidate proteins and complementing it with RNAi experiments and more detailed functional experiments, such as protein-protein interaction studies will ultimately lead a more comprehensive understanding of neurite outgrowth at the molecular level.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sir Philip Cohen (University of Dundee, United Kingdom) for discussions and Gsk3β analysis and providing a number of reagents and Timo Zimmermann, Kota Miura, and Jens Rietdorf (EMBL, Heidelberg) for help with time-lapse analysis and neurite outgrowth quantification. We also thank Leica and Olympus Europe for instrument support to the Advanced Light Microscopy Facility (ALMF) at EMBL. We thank Dionisio Martin Zanca (University of Salamanca, Spain) for providing the PC12 6-15 cell line. This work was supported by Grants 01GR0420 and 1GR0423 of the National Genome Research Network, funded by the Federal Ministry for Education and Research (Bundesministerium für Bildung und Forschung). V.L. is a recipient of a Ph.D. fellowship from the Louis-Jeantet Foundation.
Abbreviations used:
- NGF
nerve growth factor
- qRT-PCR
quantitative reverse transcription PCR
- ORF
open reading frame
- Gsk3β
Glycogen synthase kinase 3β
- GFP
green fluorescent protein.
Footnotes

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0666) on November 8, 2006.
REFERENCES
- Baxter M. G., Chiba A. A. Cognitive functions of the basal forebrain. Curr. Opin. Neurobiol. 1999;9:178–183. doi: 10.1016/s0959-4388(99)80024-5. [DOI] [PubMed] [Google Scholar]
- Christophe-Hobertus C., Kooy F., Gecz J., Abramowicz M. J., Holinski-Feder E., Schwartz C., Christophe D. TM4SF10 gene sequencing in XLMR patients identifies common polymorphisms but no disease-associated mutation. BMC Med. Genet. 2004;5:22. doi: 10.1186/1471-2350-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophe-Hobertus C., Szpirer C., Guyon R., Christophe D. Identification of the gene encoding Brain Cell Membrane Protein 1 (BCMP1), a putative four-transmembrane protein distantly related to the Peripheral Myelin Protein 22/epithelial membrane proteins and the claudins. BMC Genomics. 2001;2:3. doi: 10.1186/1471-2164-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani L., Salinas P. C. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- da Silva J. S., Dotti C. G. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat. Rev. Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- Dehmelt L., Halpain S. Actin and microtubules in neurite initiation: are MAPs the missing link? J. Neurobiol. 2004;58:18–33. doi: 10.1002/neu.10284. [DOI] [PubMed] [Google Scholar]
- Dehmelt L., Smart F. M., Ozer R. S., Halpain S. The role of microtubule-associated protein 2c in the reorganization of microtubules and lamellipodia during neurite initiation. J. Neurosci. 2003;23:9479–9490. doi: 10.1523/JNEUROSCI.23-29-09479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deller T., Mundel P., Frotscher M. Potential role of synaptopodin in spine motility by coupling actin to the spine apparatus. Hippocampus. 2000;10:569–581. doi: 10.1002/1098-1063(2000)10:5<569::AID-HIPO7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- Futerman A. H., Banker G. A. The economics of neurite outgrowth—the addition of new membrane to growing axons. Trends Neurosci. 1996;19:144–149. doi: 10.1016/s0166-2236(96)80025-7. [DOI] [PubMed] [Google Scholar]
- Gallo G., Letourneau P. C. Axon guidance: GTPases help axons reach their targets. Curr. Biol. 1998;8:R80–R82. doi: 10.1016/s0960-9822(98)70051-x. [DOI] [PubMed] [Google Scholar]
- Gallo G., Letourneau P. C. Neurotrophins and the dynamic regulation of the neuronal cytoskeleton. J. Neurobiol. 2000;44:159–173. doi: 10.1002/1097-4695(200008)44:2<159::aid-neu6>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Grant S. G. Systems biology in neuroscience: bridging genes to cognition. Curr. Opin. Neurobiol. 2003;13:577–582. doi: 10.1016/j.conb.2003.09.016. [DOI] [PubMed] [Google Scholar]
- Greene L. A., Tischler A. S. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh G., Soehrman S., Bondallaz P., Ruchti E., Cadas H. Role of the microtubule destabilizing proteins SCG10 and stathmin in neuronal growth. J. Neurobiol. 2004;58:60–69. doi: 10.1002/neu.10279. [DOI] [PubMed] [Google Scholar]
- Hempstead B. L., Rabin S. J., Kaplan L., Reid S., Parada L. F., Kaplan D. R. Overexpression of the trk tyrosine kinase rapidly accelerates nerve growth factor-induced differentiation. Neuron. 1992;9:883–896. doi: 10.1016/0896-6273(92)90241-5. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Huber L. A., Dupree P., Dotti C. G. A deficiency of the small GTPase rab8 inhibits membrane traffic in developing neurons. Mol. Cell Biol. 1995;15:918–924. doi: 10.1128/mcb.15.2.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G., Perlungher R. R., Macara I. G. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol. Cell. Biol. 1999;19:6585–6597. doi: 10.1128/mcb.19.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L. L., Oudega M., Bunge M. B., Tuszynski M. H. Neurotrophic factors, cellular bridges and gene therapy for spinal cord injury. J. Physiol. 2001;533:83–89. doi: 10.1111/j.1469-7793.2001.0083b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebre A. S., et al. Ataxin-7 interacts with a Cbl-associated protein that it recruits into neuronal intranuclear inclusions. Hum. Mol. Genet. 2001;10:1201–1213. doi: 10.1093/hmg/10.11.1201. [DOI] [PubMed] [Google Scholar]
- Liebel U., Starkuviene V., Erfle H., Simpson J. C., Poustka A., Wiemann S., Pepperkok R. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 2003;554:394–398. doi: 10.1016/s0014-5793(03)01197-9. [DOI] [PubMed] [Google Scholar]
- Nakagawa H., Koyama K., Murata Y., Morito M., Akiyama T., Nakamura Y. EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene. 2000;19:210–216. doi: 10.1038/sj.onc.1203308. [DOI] [PubMed] [Google Scholar]
- Neubrand V. E., Will R. D., Mobius W., Poustka A., Wiemann S., Schu P., Dotti C. G., Pepperkok R., Simpson J. C. Gamma-BAR, a novel AP-1-interacting protein involved in post-Golgi trafficking. EMBO J. 2005;24:1122–1133. doi: 10.1038/sj.emboj.7600600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B., Held M., Liebel U., Erfle H., Rogers P., Pepperkok R., Ellenberg J. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat. Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- Nobile C., et al. Identification and characterization of a novel human brain-specific gene, homologous to S. scrofa tmp83.5, in the chromosome 10q24 critical region for temporal lobe epilepsy and spastic paraplegia. Gene. 2002;282:87–94. doi: 10.1016/s0378-1119(01)00846-0. [DOI] [PubMed] [Google Scholar]
- Osen-Sand A., Catsicas M., Staple J. K., Jones K. A., Ayala G., Knowles J., Grenningloh G., Catsicas S. Inhibition of axonal growth by SNAP-25 antisense oligonucleotides in vitro and in vivo. Nature. 1993;364:445–448. doi: 10.1038/364445a0. [DOI] [PubMed] [Google Scholar]
- Pei L., Peng Y., Yang Y., Ling X. B., Van Eyndhoven W. G., Nguyen K. C., Rubin M., Hoey T., Powers S., Li J. PRC17, a novel oncogene encoding a Rab GTPase-activating protein, is amplified in prostate cancer. Cancer Res. 2002;62:5420–5424. [PubMed] [Google Scholar]
- Ramm P., Alexandrov Y., Cholewinski A., Cybuch Y., Nadon R., Soltys B. J. Automated screening of neurite outgrowth. J. Biomol. Screen. 2003;8:7–18. doi: 10.1177/1087057102239779. [DOI] [PubMed] [Google Scholar]
- Renner C., Pfitzenmeier J. P., Gerlach K., Held G., Ohnesorge S., Sahin U., Bauer S., Pfreundschuh M. RP1, a new member of the adenomatous polyposis coli-binding EB1-like gene family, is differentially expressed in activated T cells. J. Immunol. 1997;159:1276–1283. [PubMed] [Google Scholar]
- Ring D. B., et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–595. doi: 10.2337/diabetes.52.3.588. [DOI] [PubMed] [Google Scholar]
- Rochlin M. W., Wickline K. M., Bridgman P. C. Microtubule stability decreases axon elongation but not axoplasm production. J. Neurosci. 1996;16:3236–3246. doi: 10.1523/JNEUROSCI.16-10-03236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu M., Kimura K., Kataoka M., Takahashi M., Okajima S., Kawaguchi S., Hirasawa Y., Ide C., Mizoguchi A. VAMP-2 promotes neurite elongation and SNAP-25A increases neurite sprouting in PC12 cells. Neurosci. Res. 2000;37:265–275. doi: 10.1016/s0168-0102(00)00125-5. [DOI] [PubMed] [Google Scholar]
- Sieburth D., et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Simpson J. C., Wellenreuther R., Poustka A., Pepperkok R., Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon F. Neuroblastoma cells recapitulate their detailed neurite morphologies after reversible microtubule disassembly. Cell. 1980;21:333–338. doi: 10.1016/0092-8674(80)90469-9. [DOI] [PubMed] [Google Scholar]
- Valtorta F., Leoni C. Molecular mechanisms of neurite extension. Philos. Trans. R Soc. Lond. B Biol. Sci. 1999;354:387–394. doi: 10.1098/rstb.1999.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega I. E., Hsu S. C. The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J. Neurosci. 2001;21:3839–3848. doi: 10.1523/JNEUROSCI.21-11-03839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann S., et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001;11:422–435. doi: 10.1101/gr.154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F. Q., Zhou J., Dedhar S., Wu Y. H., Snider W. D. NGF-induced axon growth is mediated by localized inactivation of GSK-3beta and functions of the microtubule plus end binding protein APC. Neuron. 2004;42:897–912. doi: 10.1016/j.neuron.2004.05.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.