Abstract
We studied the spatiotemporal regulation of Akt (also called protein kinase B), phosphatidylinositol-3,4-bisphosphate [PtdIns(3,4)P2], and phosphatidylinositol-3,4,5-trisphosphate [PtdIns(3,4,5)P3] by using probes based on the principle of fluorescence resonance energy transfer. On epidermal growth factor (EGF) stimulation, the amount of PtdIns(3,4,5)P3 was increased diffusely in the plasma membrane, whereas that of PtdIns(3,4)P2 was increased more in the nascent lamellipodia than in the plasma membrane of the central region. The distribution and time course of Akt activation were similar to that of increased PtdIns(3,4)P2 levels, which were most prominent in the nascent lamellipodia. Moreover, we found that upon EGF stimulation 3-phosphoinositide–dependent protein kinase-1 (PDK1) was also recruited to nascent lamellipodia in an Akt-dependent manner. Because PDK1 is known to activate Ral GTPase and because Ral is required for EGF-induced lamellipodial protrusion, we speculated that the PDK1–Akt complex may be indispensable for the induction of lamellipodia. In agreement with this idea, EGF-induced lamellipodia formation was promoted by the overexpression of Akt and inhibited by an Akt inhibitor or a Ral-binding domain of Sec5. These results identified the Akt–PDK1 complex as an upstream positive regulator of Ral GTPase in the induction of lamellipodial protrusion.
INTRODUCTION
Class I phosphoinositide 3-kinase (PI3K) is a key mediator of intracellular signaling pathways that regulate actin cytoskeletal reorganization and polarized cell migration. Activated PI3K phosphorylates phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] to generate phosphatidylinositol-3,4,5-tris-phosphate [PtdIns(3,4,5)P3], which, in turn, activates a variety of pleckstrin homology (PH) domain-containing proteins such as Akt (also called protein kinase B) (Saltiel and Pessin, 2002; Sulis and Parsons, 2003). PtdIns(3,4,5)P3 is dephosphorylated to yield PtdIns(4,5)P2 by a tumor suppressor protein, phosphatase and tensin homolog deleted in chromosome 10 (PTEN), which is frequently mutated or deleted in various human cancers (Sulis and Parsons, 2003). The resulting PTEN deficiency causes accumulation of PtdIns(3,4,5)P3 in cells, and, as a consequence, an increase in cell motility in fibroblasts (Liliental et al., 2000; Higuchi et al., 2001; Hafizi et al., 2005). Another dephosphorylated derivative of PtdIns(3,4,5)P3 is PtdIns(3,4)P2 produced by phosphoinositide 5-phosphatases, including Src homology 2-containing inositol-5-phosphatase (SHIP). This PtdIns(3,4)P2 also binds to various PH domain-containing proteins, which overlap significantly to those bound to PtdIns(3,4,5)P3 (Lemmon and Ferguson, 2000; Maffucci and Falasca, 2001).
Among many PH domain-containing proteins, a serine-threonine kinase, Akt, has been studied most extensively. Akt has been implicated in the control of diverse cellular functions, including glucose metabolism, gene transcription, cell proliferation, and apoptosis (Brazil et al., 2004; Fayard et al., 2005). Structurally, Akt is composed of three functionally distinct regions: an N-terminal PH domain that provides a lipid-binding module to direct Akt to PtdIns(3,4)P2 and PtdIns(3,4,5)P3, a central catalytic domain, and a C-terminal hydrophobic motif (Brazil et al., 2004). Activation of Akt occurs when growth factors bind to receptor tyrosine kinases and activate PI3K, resulting in an increase in PtdIns(3,4,5)P3 at the plasma membrane. The binding of PtdIns(3,4,5)P3 to the PH domain anchors Akt to the plasma membrane and allows its phosphorylation and activation by 3-phosphoinositide–dependent protein kinase-1 (PDK1), another PH domain-containing protein. PDK1 phosphorylates Akt at Thr308, which causes a charge-induced conformational change, allowing Akt for substrate binding and an increased rate of catalysis. Akt is further activated by the phosphorylation of Ser473, which can be catalyzed by various kinases, including PDK2, DNA-PK, mammalian target of rapamysin, rictor complex, integrin linked kinase, and protein kinase CβII (Feng et al., 2004; Kawakami et al., 2004; Fayard et al., 2005; Sarbassov et al., 2005).
It has been demonstrated that PI3K plays an essential role in the epidermal growth factor (EGF)-induced membrane protrusion by regulating not only Akt (Higuchi et al., 2001; Nishita et al., 2004; Hafizi et al., 2005) but also Rac and Ral GTPases (Tian et al., 2002; Gavard et al., 2004; Nishita et al., 2004; Takaya et al., 2004). However, the mechanism by which Akt regulates the cytoskeletal remodeling and the relationship to Ral and Rac has been still remains elusive.
Ral GTPase has been shown to regulate exocytosis, endocytosis, and the actin cytoskeleton (for reviews, see Bos, 1998; Feig, 2003). Furthermore, essential roles for Ral have been demonstrated in cell migration (Suzuki et al., 2000; Takaya et al., 2004; Oxford et al., 2005; Rosse et al., 2006), and, more locally, in the induction of lamellipodial protrusion at the front of migrating cells (Takaya et al., 2004). We have shown that PI3K, Cdc42, and Rac play important role in such localized activation of Ral (Takaya et al., 2004). However, the mechanism by which spatial regulation of Ral activity is achieved is unknown.
Ral GTPase activity is regulated positively by Ral guanine nucleotide exchange factors (Ral GEFs) and negatively by GTPase-activating protein(s) (Feig et al., 1996; Camonis and White, 2005). Three Ral GEFs, RalGDS, Rgl, and Rlf/Rgl2, are activated by Ras (Bos, 1998). However, Ral may be also activated by calcium signaling pathways (Hofer et al., 1998). In C2C12 myoblasts and some other cell types, growth factors activate Ral in a manner dependent not only on Ras but also on calcium or PI3K signaling pathway (Suzuki et al., 2000; Tian et al., 2002; Takaya et al., 2004). In contrast to GEFs, our knowledge of GAP is extremely limited and to date, proteins exhibiting Ral GAP activity are yet to be identified.
To understand the involvement of these signaling molecules in actin cytoskeleton reorganization, their spatiotemporal regulation has to be determined in living cells. To this end, in vivo probes have been developed based on the principle of fluorescence resonance energy transfer (FRET) (Zhang et al., 2002; Jares-Erijman and Jovin, 2003). FRET is a nonradiative transfer of energy between two fluophores that are placed in proximity and in a proper relative angular orientation (Zhang et al., 2002). Variants of green fluorescent protein (GFP) have provided genetically encoded fluophores that serve as donor and/or acceptor in FRET (Heim and Tsien, 1996; Mizuno et al., 2001). Using these GFP variants and FRET technology, we previously developed genetically encoded probes for low-molecular-weight GTPases (Mochizuki et al., 2001; Itoh et al., 2005). Here, we introduce two FRET probes for the monitoring of Akt activity and the concentration of PtdIns(3,4,5)P3 and PtdIns(3,4)P2. With these newly developed FRET-based probes, we demonstrate that activation of Akt correlates more with the accumulation of PtdIns(3,4)P2 than PtdIns(3,4,5)P3. Furthermore, we have found that Akt forms a complex with PDK1 on the nascent lamellipodia, which leads to Ral activation.
MATERIALS AND METHODS
FRET Probes
Plasmids for FRET-based monitors were constructed essentially as described previously (Mochizuki et al., 2001; Sato et al., 2003). Akt Indicator (Akind) consisted of PH domain of Akt (a.a. 1-144), a spacer (Phe-Gly), yellow fluorescent protein (YFP), a spacer (Leu-Asp), catalytic domain of Akt (a.a. 133-480), a spacer (Gly-Gly-Arg), and cyan fluorescent protein (CFP). A FRET probe for PtdIns(3,4)P2, Pippi-PI(3,4)P2, consisted of CFP, a spacer (Glu-Ala-Ala-Ala-Arg-Asp), PH domain of TAPP1 (a.a. 175-300), a spacer (Glu-Ala-Ala-Ala-Arg-Asp-Gly-Gly-Glu-Ala-Ala-Ala-Arg-Asp), YFP (a.a. 1-237), a spacer (Glu-Ala-Ala-Ala-Arg-Asp), and the CAAX box of Ki-Ras4B (a.a. 169-188). Other FRET probes, Raichu-Ras, Raichu-RalA, and Pippi-PI(3,4,5)P3, have been described previously (Mochizuki et al., 2001; Sato et al., 2003; Takaya et al., 2004; Aoki et al., 2005).
Plasmids
cDNAs of Akt, Akt3A, and mDPH-Akt were subcloned into pERedNES, pCAGGS-Flag-C-mCFP, and/or pCAGGS-C-mVenus: pERedNES is for the internal ribosomal entry site-mediated expression of red fluorescent protein (RFP) with the nuclear export signal, allowing identification of the transfected cells under fluorescence microscope. pCAGGS-Flag-mCFP and pCAGGS-Flag-mVenus are expression vectors encoding a monomeric CFP and monomeric Venus, a YFP variant, respectively (Nagai et al., 2002). cDNA of PDK1 was obtained from Alex Toker (Harvard Medical School, Boston, MA) and subcloned into pCXN2-mCFP, pCXN2-mVenus, pCXN2-HA, and pERedNLS. pERedNLS is similar to pERedNES except that RFP is tagged with nuclear localization signal (Yoshizaki et al., 2004). pCXN2 vector is derived from pCAGGS and carries a neomycin resistance gene. cDNA of RalGDS was obtained from A. Wittinghofer (Max-Planck-Institut für Molekulare Physiologie, Dortmund, Germany) and subcloned into pCXN2–5myc vector. cDNA of Sec5 was obtained from Y. Ohta (Harvard Medical School). A DNA fragment of Sec5 (a.a. 1-99) was polymerase chain reaction (PCR) amplified and subcloned into pGEX-4T3.
Cell Culture, Transfection, and Immunoblotting
NIH 3T3 cells were purchased from the RIKEN Gene Bank (Wako-shi, Japan). Cos7 cells used in this study were Cos7/E3, a subclone of Cos7 cells established by Y. Fukui (Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, University of Tokyo, Tokyo, Japan). NIH 3T3 and Cos7 cells were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal calf serum. For transient expression studies, cells were transfected using Polyfect (QIAGEN, Valencia, CA). Cells were analyzed at 24 h after transfection. Mock transfection was performed using the empty pCAGGS expression vector. For immunoblotting, proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane, followed by detection with antibodies described below. The bound antibodies were detected by an enhanced chemiluminescence (ECL) detection system (GE Healthcare, Little Chalfont, Buckinghamhsire, United Kingdom) and binding was quantified with the aid of an LAS-1000 image analyzer (Fuji-Film, Tokyo, Japan). Anti-GFP rabbit serum was prepared in our laboratory. Anti-RalA antibody was purchased from BD Biosciences (San Jose, CA). Anti-FLAG M2 antibody was purchased from Sigma-Aldrich. Anti-Myc 9E10 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-GFP antibody was purchased from Takara Bio (Otsu, Japan). Anti-phospho-glycogen synthase kinase (GSK)3β (Ser21/9), Anti-PDK1, anti-Akt, and anti-phospho Akt (Thr308) antibodies were purchased from Cell Signaling Technology (Beverly, MA). Akt inhibitor IV and LY294002 were purchased from Calbiochem (San Diego, CA).
Akt Kinase Assay
Akind-expressing cells were lysed for 10 min on ice in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μM phenylmethylsulfonyl fluoride [PMSF], 10 μg/ml leupeptin, and 10 μg/ml aprotinin). Akind was immunoprecipitated with high-affinity rat monoclonal anti-(hemagglutinin [HA]) A antibody (Roche Diagnostics, Indianapolis, IN) for 1 h at 4°C on protein A-Sepharose beads. Immunoprecipitates of Akind were washed twice with the lysis buffer and twice with the kinase buffer (25 mM Tris-HCl, pH 7.5, 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, and 10 mM MgCl2). Akind on beads was incubated for 30 min at 30°C in 40 μl of kinase buffer supplemented with 200 μM ATP and 1 μg of GSK3 fusion protein (Cell Signaling Technology). The GSK3 proteins were separated by SDS-PAGE followed by immunoblotting with anti-phospho-GSK Ser21/9 antibodies. Bound antibodies were detected by an ECL chemiluminescence detection system (GE Healthcare).
Coimmunoprecipitation Analysis among Akt, PDK1, and RalGDS
Akt-, HA-PDK1–, and Myc-RalGDS–expressing cells were lysed in lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM Na3VO4, 1 μM PMSF, and 10 μg/ml aprotinin) and clarified by centrifugation. The supernatant was incubated with anti-anti-(HA A) antibody, anti c-Myc antibody (Sigma-Aldrich), or anti-Akt antibody (Cell Signaling Technology) for 30 min at 4°C. The lysate were incubated with protein A-Sepharose beads (GE Healthcare) for 1 h at 4°C, and the bound proteins and cell lysates were separated by SDS-PAGE, followed by immunoblotting with anti-anti-(HA A) antibody, anti-c-Myc antibody, or anti Akt antibody. Bound antibodies were detected by an ECL chemiluminescence detection system (GE Healthcare) and quantified with an LAS-1000 image analyzer (Fuji-Film).
Detection of RalA-GTP by Bos' Pull-Down Assay
Bos' pull-down assay for Ral proteins was performed essentially as described previously (Wolthuis et al., 1998; Takaya et al., 2004). Briefly, cells were lysed in Ral buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 1% NP-40, 10% glycerol, 1 mM Na3VO4, 1 mM phenylmethylsulfunyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) and clarified by centrifugation. The supernatant was incubated with GST-Sec5-RBD fusion proteins for 30 min at 4°C. The resulting complexes of Ral-GTP and GST-Sec5-RBD were incubated with glutathione-Sepharose beads (GE Healthcare) for 1 h at 4°C, and the bound proteins and cell lysates were separated by SDS-PAGE, followed by immunoblotting with anti-RalA antibodies. Bound antibodies were detected by an ECL chemiluminescence detection system (GE Healthcare) and quantified with an LAS-1000 image analyzer (Fuji-Film).
FRET Imaging
FRET imaging was performed essentially as described previously (Yoshizaki et al., 2003). Briefly, cells plated on a collagen-coated 35-mm-diameter glass-base dish (Asahi Techno Glass, Tokyo, Japan) were imaged every 1 min on an Olympus IX81 inverted microscope (Olympus Optical, Tokyo, Japan) equipped with a laser-based autofocusing system, IX2-ZDC, and an automatically programmable XY stage, MD-XY30100T-Meta, which allowed us to obtain the time-lapse images of several view fields in a single experiment. For dual-emission ratio imaging of the intramolecular FRET probes, we used previously described filter sets and we obtained images for CFP and FRET. After background subtraction was carried out, the FRET/CFP ratio was depicted using MetaMorph software (Molecular Devices, Sunnyvale, CA), and this image was used to represent FRET efficiency. Filters used for the dual-emission ratio imaging were purchased from Omega Optical (Brattleboro, VT): an XF1071 (440AF21) excitation filter, an XF2034 (455DRLP) dichroic mirror, and two emission filters, XF3075 (480AF30) for CFP and XF3079 (535AF26) for FRET. Cells were illuminated with a 75-W xenon lamp through a 6% ND filter (Olympus Optical) and a 60× oil immersion objective lens. The exposure time was 0.3 s when the binning of the charge-coupled device (CCD) camera was set to 4 × 4. The ratio image of FRET/CFP was created with MetaMorph software and was used to represent the efficiency of the FRET.
Imaging with a pair of intermolecular FRET probes has been described previously (Sorkin et al., 2000). Briefly, cells expressing a pair of proteins tagged with YFP and CFP, respectively, were imaged with the fluorescence microscope as described above except that an MX510 excitation filter (Asahi Spectra, Tokyo, Japan) and a 575ALP emission filter (Omega Optical) were used for the acquisition of YFP images and a semitransparent glass was used as a dichroic mirror throughout the imaging. Exposure times were 300 ms for CFP and FRET images and 300 ms for YFP images. Fluorescence through the FRET filter set consisted of a FRET component (“corrected” FRET, FRET) and non-FRET components, spectral bleedthrough and cross-excitation. The non-FRET components were subtracted as described previously (Sorkin et al., 2000). For our experimental conditions, we used the following equation: cFRET = FRET − 0.37 × CFP − 0.71 × YFP.
Confocal FRET images were obtained by an IX51 upright fluorescence microscope (Olympus Optical) equipped with an intensified CCD camera (Hamamatsu Photonics, Hamamatsu, Japan), a CSU-10 spinning Nipkow disk confocal unit (Yokogawa Electric, Tokyo, Japan), a W-view emission image splitter (Hamamatsu Photonics), and a diode-pumped solid state 430-nm laser (Melles Griot, Carlsbad, CA).
RNA Interference
pSuper.retro.puro vector was used for short hairpin RNA (shRNA). The shRNA sequences for PTEN, SHIP2, and luciferase were 5′-GGATGGATTCGACTTAGAC-3′, 5′-GAATTATCTGGACATCCTG-3′, and 5′-GATTATGTCCGGTTATGTA-3′. NIH3T3 cells were transfected with the desired pSUPER constructs by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After recovery, the cells were selected by 2-d incubation with 4 μg/ml puromycin and then used for further analysis.
Quantification of Growth Factor-induced Plasma Membrane Protrusion
Monomeric RFP-expressing Cos7 cells were time-lapse imaged every 2 min. The cell area 10 min after stimulation was divided by the cell area before stimulation to quantify the growth factor-induced membrane protrusion.
RESULTS
Development of an Akt Indicator, Akind
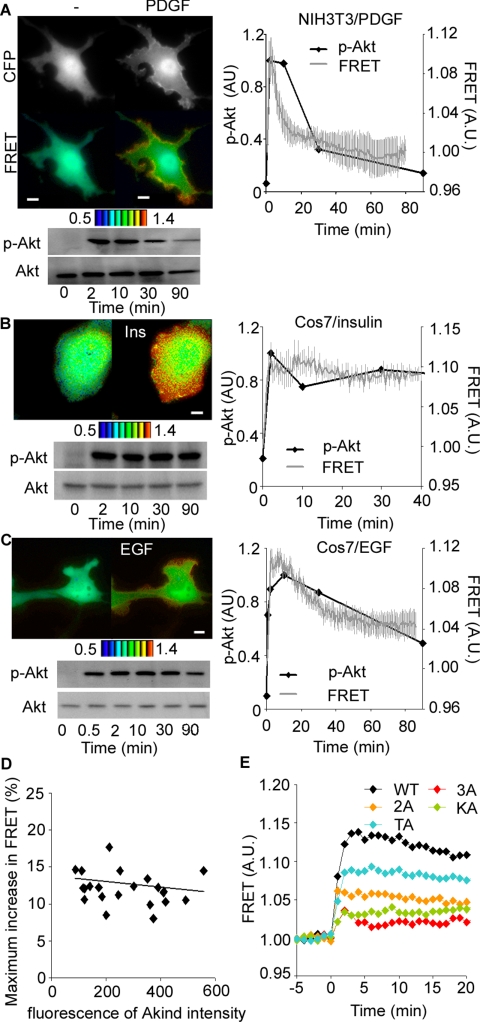
To investigate the spatiotemporal regulation of Akt, we developed a series of FRET probes for use in live cell imaging of Akt activity. For the sake of brevity, only the results obtained with the probe named Akind is described, because this probe performed best among those tested. From the amino terminus, Akind is composed of PH domain of Akt, YFP, catalytic and regulatory domains of Akt, and CFP from the amino terminus (Figure 1A). In this probe design, FRET level is low in the cytosolic inactive state and high in the membrane-recruited active state. We confirmed that Akind was regulated similarly to the endogenous Akt by showing that phosphorylation of Thr308 and in vitro kinase activity of Akind were increased upon stimulation by platelet-derived growth factor (PDGF) as efficiently as those of the endogenous Akt (Figure 1, B and C).
Figure 1.
Development of a FRET probe for Akt. (A) Schematic representation of Akt Indicator (Akind). YFP and CFP denote a yellow-emitting mutant of GFP and cyan-emitting mutant of GFP, respectively. PH and CD indicate the pleckstrin homology domain and the catalytic domain of Akt, respectively. In this probe design, the FRET level increases when the probe is activated on the plasma membrane. (B) Akind-transfected NIH3T3 cells were analyzed by Western blotting with anti-Akt or anti-phospho-Thr308 of Akt before and after PDGF stimulation. (C) Akind-HA, Flag-Akt-WT, and Flag-Akt-3A expressed in PDGF-stimulated or nonstimulated NIH3T3 cells were immunoprecipitated with an anti-HA antibody or anti-FLAG antibody. Kinase activity was measured by using recombinant GSK3 as a substrate, of which phosphorylation was detected with anti-phospho-GSK3 antibody. A small aliquot of the immunoprecipitates were also analyzed with anti-FLAG antibody.
With this Akind probe, the activity change of Akt was time-lapse imaged in living NIH3T3 cells stimulated with PDGF (Figure 2A and Supplemental Video 1). As an index for Akt activation, we used the FRET/CFP value, which correlates with the proportion of Akind adopting active conformation at each pixel. On PDGF stimulation, transient accumulation of Akind and an increase in the FRET level were observed at the periphery of the cells, indicating the translocation and activation of Akt to the plasma membrane. A strong correlation between the Thr308 phosphorylation of endogenous Akt and FRET level was observed when the averaged FRET/CFP values in Akind-expressing NIH3T3 cells (n = 12) were plotted against time (Figure 2A). Similarly, we observed excellent correlation between Thr308 phosphorylation and FRET level in Cos7 cells stimulated with either insulin (n = 8) or EGF (n = 24), where the Akt activation prolonged in comparison with PDGF-stimulated NIH3T3 cells (Figure 2, B and C, and Supplemental Video 2). The strong correlation between the biochemical data obtained from bulk of cells and imaging data obtained from single cells clearly demonstrated that Akt behaves as an analog, but not digital, switch even in the single cell level. It is also important to note that the maximum increase in FRET level was not affected by the expression level of Akind (Figure 2D).
Figure 2.
Akt activation in the nascent lamellipodia as monitored by Akind. (A) Akind-expressing NIH3T3 cells were serum starved for 24 h and stimulated with 50 ng/ml PDGF. CFP (excitation [ex] 440 nm/emission [em] 480 nm) and sensitized FRET (ex 440 nm/em 530 nm) images were obtained every 1 min with a time-lapse epifluorescent microscope. Ratio image of FRET/CFP was prepared to demonstrate the level of FRET (also available as Supplemental Video 1). At least 20 similar images were obtained. In the right panel, the net intensities of CFP and FRET in each cell were measured to calculate the averaged emission ratio (FRET/CFP). Then, the emission ratio values were normalized to those of the record-starting time. Data from more than 10 independent cells are shown with SE. Phosphorylation of endogenous Akt was also examined by immunoblotting and quantitated values are overlaid to the graph. Similar experiments were performed by using EGF-stimulated Cos7 cells (B and Supplemental video 2) and insulin-stimulated Cos7 cells (C). (D) Cell images used in B were used to negate the correlation between the FRET level at the zenith and the concentration of the probe. (E) Cos7 cells expressing Akt wild type or mutant (3A, 2A, KA, and TA) were serum starved for 24 h and stimulated with 50 ng/ml EGF as described in D. Bars, 10 μm.
To understand the mechanism underlying the change in FRET efficiency, we prepared various Akind mutants with impaired kinase activity by substituting alanine for the two major regulatory phosphorylation sites, Thr308 and Ser473, or for the phosphate transfer residue in the catalytic site: TA, Thr308Ala; 2A, Thr308Ala and Ser473Ala; KA, Lys179Ala; 3A, all three mutations (Aoki et al., 1998). Cos7 cells expressing these Akind mutants were stimulated with EGF and time-lapse imaged (Figure 2E). The maximum increase in the level of FRET showed an excellent correlation with the reported levels of the kinase activity of mutants: high in TA and low in 3A and KA mutants. Thus, we concluded that the activity-dependent conformational change of Akt could be monitored by the level of FRET in Akind-expressing cells.
We next examined the effect of the plasma membrane translocation of Akind on the FRET level. For this purpose, we established a semiquantitative assay for the plasma membrane translocation, which gives a translocation index as described in the Supplemental Material. With this assay, we quantitated the EGF-induced translocation of Akind to the plasma membrane and found that the increase in the translocation index correlated nicely with the increase in FRET level, indicating that the translocation and conformational change of Akt occurred almost simultaneously (Figure 3A). Importantly, however, both Akind-KA and Akind-3A mutants were found to translocate to the plasma membrane as efficiently as did the wild type (Figure 3B). This observation demonstrated that the translocation alone was not sufficient for the induction of conformational change of Akind and strongly suggested that changes in the FRET level monitored the conformational change accompanying the increased kinase activity of Akind.
Figure 3.
Semiquantitative analysis of plasma membrane translocation of Akind. Signal-dependent translocation of CFP-tagged cytosolic proteins to the plasma membrane was measured semiquantitatively by using RFP as a reference (for details, see Supplemental Material). (A) The translocation index and the FRET level of Cos7 cells expressing Akind were plotted against time. (B) Shown are time courses of the translocation index of cells expressing wild type, KA mutant, and 3A mutant of Akind.
Development of PtdIns(3,4)P2 Indicator Pippi-PI(3,4)P2
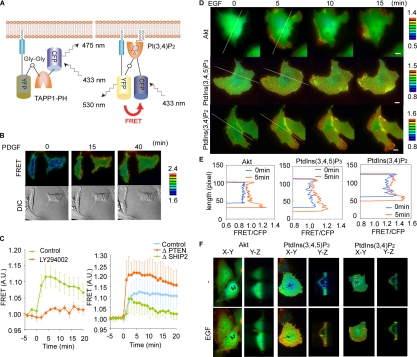
PtdIns(3,4,5)P3 and PtdIns(3,4)P2 have been shown to recruit Akt to the plasma membrane, where Akt becomes phosphorylated at Thr308 and Ser473 and is thereby activated enzymatically (Alessi et al., 1996; Fayard et al., 2005). To determine which of these two phosphoinositides determine the distribution of active Akt, we developed FRET-based monitors for the two phospholipids essentially as described previously (Figure 4A) (Sato et al., 2003). The probe for PtdIns(3,4,5)P3 named Pippi-PI(3,4,5)P3, has been already reported and the probe for PtdIns(3,4)P2 named Pippi-PI(3,4)P2 was prepared by the use of PH domain of TAPP1, which binds specifically to PtdIns(3,4)P2 (Dowler et al., 2000). Pippi-PI(3,4)P2 was found to detect PDGF-induced transient increase in PtdIns(3,4)P2 very nicely (Figure 4B). The increase in the level of FRET was markedly inhibited by LY294002 (Figure 4C, left). To further confirm the specificity of the Pippi-PI(3,4)P2 probe, we examined the effect of knockdown of SHIP2 and PTEN (Figure 4C, right). PDGF-induced increase in PtdIns(3,4)P2 was significantly enhanced in PTEN deficient cells, whereas the production of PtdIns(3,4)P2 was suppressed in SHIP2-deficient cells. Intriguingly, the initial increase in PtdIns(3,4)P2 was not impaired but the prolonged elevation was significantly suppressed in the SHIP2 knockdown cells.
Figure 4.
High Akt activity and accumulation of PtdIns(3,4)P2 in the nascent lamellipodia. (A) Schematic representation of Pippi-PI(3,4)P2. TAPP1-PH indicates the pleckstrin homology domain of TAPP1. The PH domain is sandwiched is by CFP and Venus. Venus and the PH domain are connected by means of rigid α-helical linkers consisting of repeated EAAAR sequences, between which is a single Gly-Gly motif introduced as a hinge. At the C terminus of Venus is the hypervariable region of K-Ras used as a plasma membrane-anchoring motif. Thus, the probes are designed to propagate high FRET signal when the PH domain binds to PtdIns(3,4)P2. (B) NIH3T3 cells expressing Pippi-PI(3,4)P2 were serum starved for 24 h and stimulated with 50 ng/ml PDGF. FRET imaging was performed as described in Figure 2. (C) Pippi-PI(3,4)P2-expressing NIH3T3 cells untreated or treated with 40 μM LY294002 were stimulated with 50 ng/ml PDGF. The time course of the FRET level was obtained as in Figure 2. Cells were transfected with pSuper-PTEN, SHIP2, or luciferase, an shRNA vector, and selected by puromycin. Sixty hours posttransfection, FRET imaging was performed as described above. Averaged data from five cells are shown with SD. (D) Cos7 cells expressing Akind, Pippi-PI(3,4)P2, or Pippi-PI(3,4,5)P3 were serum starved for 24 h and stimulated with 50 ng/ml EGF. FRET images were obtained by a conventional epifluorescent microscope (D and Supplemental Videos 3 and 4). The FRET/CFP ratio image was used to show the FRET level in the intensity-modulated display mode. (E) Along the white lines in D, FRET/CFP values are plotted. (F) Cos7 cells prepared as described in D were imaged with a spinning disk confocal microscope. X-Y and Y-Z sections are shown. Bar, 10 μm.
Colocalization of High Akt Activity with Accumulation of PtdIns(3,4)P2
With these two FRET probes, we monitored the dynamics of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 in EGF-stimulated Cos7 cells (Figure 4D and Supplemental Videos 3 and 4). On EGF stimulation, the amount of PtdIns(3,4,5)P3 was found to increase diffusely in the plasma membrane. This pattern of distribution was very similar to that of Ras and the EGF receptor (Mochizuki et al., 2001; Itoh et al., 2005). In contrast to PtdIns(3,4,5)P3, PtdIns(3,4)P2 concentration increased most prominently in the protruding lamellipodia and peripheral membrane ruffles. Line-scan plots of these FRET images emphasize the local accumulation of PtdIns(3,4)P2 and activation of Akt (Figure 4E). Thus, the spatial distribution of Akt activation correlated more closely to the increase in PtdIns(3,4)P2 rather than PtdIns(3,4,5)P3. Because the apparent high FRET level in the cell periphery could be caused by the relatively small height of cell body, we confirmed our observation by spinning disk confocal microscopy (Figure 4F). An increase in PtdIns(3,4,5)P3 was observed diffusely in the apical membrane. Increase in PtdIns(3,4)P2 was also observed diffusely in the dorsal membrane; however, the increase was most prominent in the lamellipodia and peripheral membrane ruffles. The distribution of increased Akt activity was similar to that of PtdIns(3,4)P2, i.e., most prominent in lamellipodia and the peripheral membrane ruffles. These results supported previous notes that Akt activation depends more on PtdIns(3,4)P2 than PtdIns(3,4,5)P3 (Franke et al., 1997; Klippel et al., 1997).
Accumulation of PDK1–Akt Complex at the Nascent Lamellipodial Protrusion
To further explore the mechanism of Akt activation in lamellipodia and peripheral membrane ruffles, we next investigated the role of PDK1, which is the major kinase that activates Akt at the plasma membrane. For this purpose, we prepared a pair of intermolecular FRET probes consisting of monomeric CFP-tagged PDK1 (mCFP-PDK1) and monomeric Venus-tagged Akt (Akt-mVenus). The intermolecular FRET level was then monitored in Cos7 cells expressing this pair of probes during EGF stimulation (Figure 5, A and B). We saw a significant increase in the FRET level in the peripheral region of cells, suggesting that the Akt–PDK1 complex was formed primarily at this location including lemellipodia. The distributions of Akt and PDK1 were similar except for nucleus and there were no significant correlation between Venus/CFP and cFRET values (Figure 5C). Thus, we could negate the possibility that the increase in FRET was caused by the accumulation of Akt-mVenus. When we used mCFP or mCFP-tagged Vav2, a PH domain-containing protein, as a FRET donor, the increase in FRET was not observed upon EGF stimulation (Figure 5B). Interestingly, we found that plasma membrane translocation of PDK1 was significantly enhanced and prolonged by the coexpression of Akt (Figure 5, D and E). Thus, the accumulation of the Akt–PDK complex at lamellipodial protrusion seemed to be caused primarily by Akt, which was recruited to the plasma membrane by PtdIns(3,4)P2 and PtdIns(3,4,5)P3.
Figure 5.
Accumulation of PDK1–Akt complex in nascent lamellipodia. (A and B) Cos7 cells expressing Akt-YFP and CFP-PDK1 were stimulated with 50 ng/ml EGF. Three images, CFP (ex 440 nm/em 480 nm), sensitized FRET (ex 440 nm/em 530 nm), and YFP (ex 510 nm/em 565 nm) were obtained every 30 s for 1 h. Corrected FRET (cFRET) images prepared as described in the text are shown in a pseudocolor mode (A). Net intensities of CFP and YFP in each cell were measured to calculate the averaged cFRET/CFP. (C) The Venus/CFP ratio values obtained from each pixel are plotted against cFRET. Pixels with YFP/CFP value over 1.0 corresponds those in the nucleus. (D) Cos7 cells expressing CFP-PDK1 in the presence or absence of Akt were stimulated with 50 ng/ml EGF and time-lapse imaged. (E) From each cell image, the translocation index of CFP-PDK1 were determined and plotted against time (n ≥ 18).
Activation of RalGDS by PDK1–Akt Complex
We speculate that the accumulation of Akt–PDK1 complex at lamellipodia might induce Ral activation, based on the following two reasons. First, RalA activation is most prominent at nascent lamellipodia as is Akt activation (Figure 4D), and RalA activation is known to be dependent on PI3K (Tian et al., 2002; Takaya et al., 2004). Second, Feig and colleagues have reported that PDK1 serves as a scaffold to recruit and activate Ral GEFs at the plasma membrane (Tian et al., 2002). In support of our model, we found that RalA activation by EGF or constitutively active mutant of PI3K was suppressed by the Akt inhibitor IV (Kau et al., 2003) and that constitutively active Akt elevated the level of RalA-GTP (Figure 6, A and B). Furthermore, EGF-induced activation of RalA was enhanced by the coexpression of PDK1 and Akt (Figure 6C). Together, these data strongly suggests that Ral activation at the nascent lamellipodia is caused by Ral GEFs activated locally by the Akt–PDK1 complex.
Figure 6.
Requirement of Akt for EGF-induced RalA activation. (A) Cos7 cells transfected with or without an expression vector for a constitutively active mutant of PI3K, p110-x. Cells were serum starved for 8 h and left untreated or treated with LY294002 or Akt inhibitor IV for 30 min. The levels of endogenous GTP-Ral were assayed by Bos' pull-down method and quantitated with an LAS-1000 image analyzer. Averaged data from two independent experiments are shown with SE. (B) Cos7 cells transfected with or without an expression vector for GFP-RalA and a constitutively active mutant of Akt mDPH-Akt were serum starved for 8 h and left untreated or treated with LY294002. Cells with or without 50 ng/ml EGF stimulation were analyzed as described in A. (C) Cos7 cells expressing GFP-RalA with or without Myc-RalGDS were serum starved for 8 h and left untreated or treated with Aki inhibitor IV for 30 min before stimulation with 50 ng/ml EGF for 10 min. The level of GTP-bound GFP-RalA was analyzed as described in text.
Requirement of Akt Activity for EGF-induced Lamellipodia Protrusion
To demonstrate the Akt–PDK1 regulation of Ral in a physiological milieu, we examined the effect of the expression of Akt or Akt inhibitor IV on the EGF-induced lamellipodial protrusion, which has been shown to be dependent on Ral (Takaya et al., 2004). The EGF-induced lamellipodia protrusion was significantly enhanced by the expression of Akt (Figure 7). This lamellipodial protrusion was markedly inhibited either by treatment with an Akt inhibitor IV or by the expression of the Ral binding domain of Sec5. Thus, our results identify the Akt–PDK complex at the cell periphery as an upstream positive regulator of Ral, which plays an essential role in the induction of lamellipodial protrusion.
Figure 7.
Requirement of Akt for EGF-induced membrane protrusion. (A) Parent and Akt-expressing Cos7 cells were stimulated with 50 ng/ml EGF. Differential interference contrast images of these cells were obtained every 1 min (top). White and black lines indicated the outlines of the cells before and 10 min after EGF stimulation, respectively. Bar, 10 μm. (B) Averaged extended cell areas, which was obtained by the reduction of black-lined area from white-lined area, are shown with SE (bottom). n ≥ 12.
Coimmunoprecipitation of Atk, PDK1, and RalGDS
To demonstrate the role of PDK1 in the formation of Akt–PDK1–RalGEF complex, we expressed Akt and RalGDS in the presence or absence of PDK1 and examined the complex formation (Figure 8). In the presence of PDK1, all three complexes of Akt-RalGDS, PDK1-RalGDS, and Akt-PDK1 were readily detected. In contrast, in the absence of PDK1, the Akt–RalGDS complex could not be detected, indicating that PDK1 functions as a scaffold to Akt and RalGDS (Figure 8).
Figure 8.
Association of Akt and RalGDS in a PDK1-dependent manner. Cos7 cells expressing Akt and Myc-RalGDS with or without HA-PDK1 were serum starved for 8 h and stimulated with 50 ng/ml EGF for 10 min. Cells were lysed and immunoprecipitated with anti-Myc monoclonal antibody (mAb), anti-HA rat mAb, or anti-Akt rabbit antibody. The immunoprecipitates (IP) and total cell lysates (TLC) were analyzed by immunoblotting.
DISCUSSION
Here, we propose that Akt stabilizes the PDK1–RalGEF complex at plasma membrane and thereby enhances Ral activation. The requirement of PI3K and PDK1 for Ral activation was originally reported by Tian et al. (2002). They have shown that PI3K-activated PDK1 binds to and activates RalGEF, resulting in Ral activation. In the same study, however, requirement of Akt was negated because kinase activity of PDK1 was dispensable for Ral activation and because Ral could not be activated by Akt-CAAX, an Akt mutant that contains the C-terminal CAAX motif of Ki-Ras and thereby localizes constitutively at the plasma membrane. However, in one study, it has been reported that this Akt-CAAX mutant lacks kinase activity and inhibits insulin-induced GSK3 activation by endogenous Akt (van Weeren et al., 1998). Therefore, we used another active Akt mutant, mDPH-Akt, and found that this active Akt elevated basal and EGF-stimulated Ral activity (Figure 6). Conversely, treatment with Akt inhibitor IV decreased basal and EGF-stimulated Ral activity as did application of a PI3K inhibitor. Thus, by controlling the assembly of the PDK1–RalGEF complex, Akt serves as a critical determinant whether or not the molecular switch, Ral, should be turned on.
An important aspect of Akt regulation of Ral is in their spatial distribution. On EGF stimulation of Cos7 cells, RalGEF is recruited diffusely to the plasma membrane. Nevertheless, Ral activation is mostly observed at nascent lamellipodia, predicting the presence of another critical activator of Ral whose activity must have been elevated locally at the nascent lamellipodia (Takaya et al., 2004). Akt meets the criteria of this spatial regulator of Ral. To further explore the mechanism underlying this Akt regulation of Ral activity, an Akt substrate that is phosphorylated in or close to the Akt–PDK1–RalGEF complex must be determined in the future study.
One major controversy concerning Akt activation is the issue of which PtdIns(3,4,5)P3 and PtdIns(3,4)P2 is the critical upstream regulator of Akt in vivo (Downward, 1998; Lemmon and Ferguson, 2000; Roymans and Slegers, 2001). There are substantial in vitro data suggesting that PtdIns(3,4)P2 is more important than PtdIns(3,4,5)P3 in Akt activation (Franke et al., 1997; Frech et al., 1997; Klippel et al., 1997): Lipid vesicles containing PtdIns(3,4)P2 activate Akt, whereas those containing PtdIns(3,4,5)P3 either inhibit (Franke et al., 1997; Frech et al., 1997) or do not affect Akt activity (Klippel et al., 1997). By using cells derived from SHIP-deficient mice, an essential role of PtdIns(3,4)P2 has been demonstrated in Steel Factor-stimulated Akt activation (Scheid et al., 2002). Our observations with FRET-based probes are in favor of this model: The increase in PtdIns(3,4)P2 and activation of Akt were observed at the peripheral plasma membrane such as nascent lamellipodia, whereas an increase in PtdIns(3,4,5)P3 was observed more diffusely at the plasma membrane, including the dorsal surface of the cells (Figure 4, D–F). On the contrary, in support of PtdIns(3,4,5)P3 being the critical second messenger, PtdIns(3,4,5)P3 is reported to bind to Akt with slightly higher affinity than does PtdIns(3,4)P2 (James et al., 1996; Frech et al., 1997). Furthermore, PDK1 has a substantially higher affinity for PtdIns(3,4,5)P3 than for PtdIns(3,4)P2 (Stokoe et al., 1997) and needs to interact with the plasma membrane to phosphorylate Akt efficiently (Anderson et al., 1998). We also confirmed that the PH domain of PDK1 binds to PtdIns(3,4,5)P3 more efficiently than to PtdIns(3,4)P2 (data not shown). In vivo data in favor of PI(3,4,5)P3 as the preferential regulator of Akt is obtained by using SHIP-overexpressing cells, where Akt activity is inhibited in the presence of high amount of PtdIns(3,4)P2 (Aman et al., 1998; Liu et al., 1999; Taylor et al., 2000).
In relation to this debate, we and others have observed that growth factor-induced membrane translocation of Akt is more readily detected than that of PDK1 (Figure 5D), although the affinity of the PH domain of PDK1 to PtdIns(3,4)P2 and PtdIns(3,4,5)P3 is higher than that of the PH domain of Akt (Currie et al., 1999). This inefficient recruitment of PDK1 to the plasma membrane of growth factor-stimulated cells has been ascribed to the high-affinity binding of the PH domain of PDK1 to cytosolic Ins(1,3,4,5,6)P5 and InsP6 (Komander et al., 2004). Considering these reports and data presented in the present study, it is likely that the plasma membrane recruitment of PDK1 is mediated partly by the PH domain-mediated interaction with PtdIns(3,4,5)P3 and partly by the interaction with Akt and Ral GEF. In this context, the reason why the localization of Akt–PDK1 complex is similar to that of PtdIns(3,4)P2, but not to that of PtdIns(3,4,5)P3, should not be discussed based simply on the affinity of the PH domains to PtdIns(3,4)P2 and PtdIns(3,4,5)P3, but rather it should be considered in light of multiple factors that favors the production of PtdIns(3,4)P2 and the formation of Akt–PDK1–Ral GEF complexes at localized areas of the plasma membrane, such as nascent lamellipodia.
Two types of FRET-based monitors of Akt activity have been reported previously (Calleja et al., 2003; Sasaki et al., 2003; Ananthanarayanan et al., 2005; Kunkel et al., 2005): 1) measurement of phosphorylation of Akt substrate (Sasaki et al., 2003; Kunkel et al., 2005) and 2) measurement of the conformational change of Akt (Calleja et al., 2003). Probes belonging to the first type detect the balance between the activities of Akt and antagonizing phosphatases. For the acquisition of spatial information and high sensitivity, probes are preferentially anchored to the membrane, which may bias the interpretation of the results. Another restriction of this type of probe is in their sensitivity. Moreover, Ananthanarayanan et al. (2005) have observed a significant time-lag between the increase in the level of PtdIns(3,4,5)P3 and activation of Akt monitored by a type 1 probe. The first probe belonging to the second type of probe was reported by Calleja et al. (2003). In this probe, designed for fluorescence lifetime imaging microscopy, GFP and YFP are fused to the amino and carboxy termini of Akt. We prepared a similar probe by using CFP and YFP as a FRET pair for ratiometry with a conventional epifluorescence microscope. However, this probe did not either translocate to plasma membrane or exhibit changes in the level of FRET in NIH3T3 cells (data not shown). This failure may be ascribable to the use of CFP as a FRET donor or subtle difference in the sequence of linker peptides. Thus, we tried several different designs of FRET monitors of Akt and reached to the structure of Akind. An advantage of Akind over type 1 probes is that both the activity and translocation of Akt can be monitored simultaneously and, indeed, we could for the first time visualize the accumulation and activation of Akt at the nascent lamellipodia (Figure 2). The precise mechanism of the FRET change upon Akt activation awaits further study. At least partly, phosphorylation-induced conformational change should be responsible because amino acid substitutions of phosphorylation sites diminished the level of FRET change upon activation (Figure 2).
In conclusion, the development of Akind has enabled us to visualize the localized activation of Akt, which led to the demonstration of the Akt–PDK1–Ral GEF complex at nascent lamellipodia. This finding seems to explain the reason why EGF-induced Ral activation is limited to the nascent lamellipodia.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sam Yarwood for critical reading of this manuscript and S. Yoshiki, N. Fujimoto, and K. Fukuhara for technical assistance. This work was supported by a grant-in-aid for scientific research on priority areas entitled “Integrative Research Toward the Conquest of Cancer” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0467) on November 1, 2006.
REFERENCES
- Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Aman M. J., Lamkin T. D., Okada H., Kurosaki T., Ravichandran K. S. The inositol phosphatase SHIP inhibits Akt/PKB activation in B cells. J. Biol. Chem. 1998;273:33922–33928. doi: 10.1074/jbc.273.51.33922. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan B., Ni Q., Zhang J. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proc. Natl. Acad. Sci. USA. 2005;102:15081–15086. doi: 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. E., Coadwell J., Stephens L. R., Hawkins P. T. Translocation of PDK-1 to the plasma membrane is important in allowing PDK-1 to activate protein kinase B. Curr. Biol. 1998;8:684–691. doi: 10.1016/s0960-9822(98)70274-x. [DOI] [PubMed] [Google Scholar]
- Aoki K., Nakamura T., Fujikawa K., Matsuda M. Local phosphatidylinositol 3,4,5-trisphosphate accumulation recruits Vav2 and Vav3 to activate Rac1/Cdc42 and initiate neurite outgrowth in nerve growth factor-stimulated PC12 cells. Mol. Biol. Cell. 2005;16:2207–2217. doi: 10.1091/mbc.E04-10-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M., Batista O., Bellacosa A., Tsichlis P., Vogt P. K. The akt kinase: molecular determinants of oncogenicity. Proc. Natl. Acad. Sci. USA. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L. All in the family? New insights and questions regarding interconnectivity of Ras, Rap1 and Ral. EMBO J. 1998;17:6776–6782. doi: 10.1093/emboj/17.23.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazil D. P., Yang Z. Z., Hemmings B. A. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 2004;29:233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Calleja V., Ameer-Beg S. M., Vojnovic B., Woscholski R., Downward J., Larijani B. Monitoring conformational changes of proteins in cells by fluorescence lifetime imaging microscopy. Biochem. J. 2003;372:33–40. doi: 10.1042/BJ20030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camonis J. H., White M. A. Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol. 2005;15:327–332. doi: 10.1016/j.tcb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Currie R. A., Walker K. S., Gray A., Deak M., Casamayor A., Downes C. P., Cohen P., Alessi D. R., Lucocq J. Role of phosphatidylinositol 3,4,5-trisphosphate in regulating the activity and localization of 3-phosphoinositide-dependent protein kinase-1. Biochem. J. 1999;337:575–583. [PMC free article] [PubMed] [Google Scholar]
- Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. Mechanisms and consequences of activation of protein kinase B/Akt. Curr. Opin. Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- Fayard E., Tintignac L. A., Baudry A., Hemmings B. A. Protein kinase B/Akt at a glance. J. Cell Sci. 2005;118:5675–5678. doi: 10.1242/jcs.02724. [DOI] [PubMed] [Google Scholar]
- Feig L. A. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Feig L. A., Urano T., Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem. Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- Feng J., Park J., Cron P., Hess D., Hemmings B. A. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J. Biol. Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- Franke T. F., Kaplan D. R., Cantley L. C., Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Frech M., Andjelkovic M., Ingley E., Reddy K. K., Falck J. R., Hemmings B. A. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J. Biol. Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- Gavard J., Lambert M., Grosheva I., Marthiens V., Irinopoulou T., Riou J. F., Bershadsky A., Mege R. M. Lamellipodium extension and cadherin adhesion: two cell responses to cadherin activation relying on distinct signalling pathways. J. Cell Sci. 2004;117:257–270. doi: 10.1242/jcs.00857. [DOI] [PubMed] [Google Scholar]
- Hafizi S., Ibraimi F., Dahlback B. C1-TEN is a negative regulator of the Akt/PKB signal transduction pathway and inhibits cell survival, proliferation, and migration. FASEB J. 2005;19:971–973. doi: 10.1096/fj.04-2532fje. [DOI] [PubMed] [Google Scholar]
- Heim R., Tsien R. Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Masuyama N., Fukui Y., Suzuki A., Gotoh Y. Akt mediates Rac/Cdc42-regulated cell motility in growth factor-stimulated cells and in invasive PTEN knockout cells. Curr. Biol. 2001;11:1958–1962. doi: 10.1016/s0960-9822(01)00599-1. [DOI] [PubMed] [Google Scholar]
- Hofer F., Berdeaux R., Martin G. S. Ras-independent activation of Ral by a Ca(2+)-dependent pathway. Curr. Biol. 1998;8:839–842. doi: 10.1016/s0960-9822(98)70327-6. [DOI] [PubMed] [Google Scholar]
- Itoh R. E., Kurokawa K., Fujioka A., Sharma A., Mayer B. J., Matsuda M. A FRET-based probe for epidermal growth factor receptor bound non-covalently to a pair of synthetic amphipathic helixes. Exp. Cell Res. 2005;307:142–152. doi: 10.1016/j.yexcr.2005.02.026. [DOI] [PubMed] [Google Scholar]
- James S. R., Downes C. P., Gigg R., Grove S. J., Holmes A. B., Alessi D. R. Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem. J. 1996;315:709–713. doi: 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares-Erijman E. A., Jovin T. M. FRET imaging. Nat. Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- Kau T. R., Schroeder F., Ramaswamy S., Wojciechowski C. L., Zhao J. J., Roberts T. M., Clardy J., Sellers W. R., Silver P. A. A chemical genetic screen identifies inhibitors of regulated nuclear export of a Forkhead transcription factor in PTEN-deficient tumor cells. Cancer Cell. 2003;4:463–476. doi: 10.1016/s1535-6108(03)00303-9. [DOI] [PubMed] [Google Scholar]
- Kawakami Y., Nishimoto H., Kitaura J., Maeda-Yamamoto M., Kato R. M., Littman D. R., Leitges M., Rawlings D. J., Kawakami T. Protein kinase C betaII regulates Akt phosphorylation on Ser-473 in a cell type- and stimulus-specific fashion. J. Biol. Chem. 2004;279:47720–47725. doi: 10.1074/jbc.M408797200. [DOI] [PubMed] [Google Scholar]
- Klippel A., Kavanaugh W. M., Pot D., Williams L. T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol. Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Fairservice A., Deak M., Kular G. S., Prescott A. R., Peter D. C., Safrany S. T., Alessi D. R., van Aalten D. M. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 2004;23:3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel M. T., Ni Q., Tsien R. Y., Zhang J., Newton A. C. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J. Biol. Chem. 2005;280:5581–5587. doi: 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M. A., Ferguson K. M. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- Liliental J., Moon S. Y., Lesche R., Mamillapalli R., Li D., Zheng Y., Sun H., Wu H. Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol. 2000;10:401–404. doi: 10.1016/s0960-9822(00)00417-6. [DOI] [PubMed] [Google Scholar]
- Liu Q., Sasaki T., Kozieradzki I., Wakeham A., Itie A., Dumont D. J., Penninger J. M. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev. 1999;13:786–791. doi: 10.1101/gad.13.7.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci T., Falasca M. Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism. FEBS Lett. 2001;506:173–179. doi: 10.1016/s0014-5793(01)02909-x. [DOI] [PubMed] [Google Scholar]
- Mizuno H., Sawano A., Eli P., Hama H., Miyawaki A. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–2510. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Yamashita S., Kurokawa K., Ohba Y., Nagai T., Miyawaki A., Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nishita M., Wang Y., Tomizawa C., Suzuki A., Niwa R., Uemura T., Mizuno K. Phosphoinositide 3-kinase-mediated activation of cofilin phosphatase Slingshot and its role for insulin-induced membrane protrusion. J. Biol. Chem. 2004;279:7193–7198. doi: 10.1074/jbc.M312591200. [DOI] [PubMed] [Google Scholar]
- Oxford G., Owens C. R., Titus B. J., Foreman T. L., Herlevsen M. C., Smith S. C., Theodorescu D. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- Rosse C., Hatzoglou A., Parrini M. C., White M. A., Chavrier P., Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol. Cell Biol. 2006;26:727–734. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roymans D., Slegers H. Phosphatidylinositol 3-kinases in tumor progression. Eur. J. Biochem. 2001;268:487–498. doi: 10.1046/j.1432-1327.2001.01936.x. [DOI] [PubMed] [Google Scholar]
- Saltiel A. R., Pessin J. E. Insulin signaling pathways in time and space. Trends Cell Biol. 2002;12:65–71. doi: 10.1016/s0962-8924(01)02207-3. [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Sato M., Umezawa Y. Fluorescent indicators for Akt/protein kinase B and dynamics of Akt activity visualized in living cells. J. Biol. Chem. 2003;278:30945–30951. doi: 10.1074/jbc.M212167200. [DOI] [PubMed] [Google Scholar]
- Sato M., Ueda Y., Takagi T., Umezawa Y. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell Biol. 2003;5:1016–1022. doi: 10.1038/ncb1054. [DOI] [PubMed] [Google Scholar]
- Scheid M. P., Huber M., Damen J. E., Hughes M., Kang V., Neilsen P., Prestwich G. D., Krystal G., Duronio V. Phosphatidylinositol (3,4,5)P3 is essential but not sufficient for protein kinase B (PKB) activation; phosphatidylinositol (3,4)P2 is required for PKB phosphorylation at Ser-473, studies using cells from SH2-containing inositol-5-phosphatase knockout mice. J. Biol. Chem. 2002;277:9027–9035. doi: 10.1074/jbc.M106755200. [DOI] [PubMed] [Google Scholar]
- Sorkin A., McClure M., Huang F., Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr. Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- Stokoe D., Stephens L. R., Copeland T., Gaffney P. R., Reese C. B., Painter G. F., Holmes A. B., McCormick F., Hawkins P. T. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- Sulis M. L., Parsons R. PTEN: from pathology to biology. Trends Cell Biol. 2003;13:478–483. doi: 10.1016/s0962-8924(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Suzuki J., Yamazaki Y., Li G., Kaziro Y., Koide H. Involvement of Ras and Ral in chemotactic migration of skeletal myoblasts. Mol. Cell Biol. 2000;20:4658–4665. doi: 10.1128/mcb.20.13.4658-4665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya A., Ohba Y., Kurokawa K., Matsuda M. RalA activation at nascent lamellipodia of epidermal growth factor-stimulated Cos7 cells and migrating Madin-Darby canine kidney cells. Mol. Biol. Cell. 2004;15:2549–2557. doi: 10.1091/mbc.E03-11-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor V., Wong M., Brandts C., Reilly L., Dean N. M., Cowsert L. M., Moodie S., Stokoe D. 5′ phospholipid phosphatase SHIP-2 causes protein kinase B inactivation and cell cycle arrest in glioblastoma cells. Mol. Cell Biol. 2000;20:6860–6871. doi: 10.1128/mcb.20.18.6860-6871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Rusanescu G., Hou W., Schaffhausen B., Feig L. A. PDK1 mediates growth factor-induced Ral-GEF activation by a kinase-independent mechanism. EMBO J. 2002;21:1327–1338. doi: 10.1093/emboj/21.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weeren P. C., de Bruyn K. M., Vries-Smits A. M., van Lint J., Burgering B. M. Essential role for protein kinase B (PKB) in insulin-induced glycogen synthase kinase 3 inactivation. Characterization of dominant-negative mutant of PKB. J. Biol. Chem. 1998;273:13150–13156. doi: 10.1074/jbc.273.21.13150. [DOI] [PubMed] [Google Scholar]
- Wolthuis R. M., Franke B., van Triest M., Bauer B., Cool R. H., Camonis J. H., Akkerman J. W., Bos J. L. Activation of the small GTPase Ral in platelets. Mol. Cell Biol. 1998;18:2486–2491. doi: 10.1128/mcb.18.5.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Kurokawa K., Itoh R. E., Nakamura T., Mochizuki N., Nagashima K., Matsuda M. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 2003;162:223–232. doi: 10.1083/jcb.200212049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Parrini M. C., Dulyaninova N. G., Bresnick A. R., Mochizuki N., Matsuda M. Cell type-specific regulation of RhoA activity during cytokinesis. J. Biol. Chem. 2004;279:44756–44762. doi: 10.1074/jbc.M402292200. [DOI] [PubMed] [Google Scholar]
- Zhang J., Campbell R. E., Ting A. Y., Tsien R. Y. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.