Abstract
Chromatin assembly factor-1 (CAF-1), a complex consisting of p150, p60, and p48 subunits, is highly conserved from yeast to humans and facilitates nucleosome assembly of newly replicated DNA in vitro. To investigate roles of CAF-1 in vertebrates, we generated two conditional DT40 mutants, respectively, devoid of CAF-1p150 and p60. Depletion of each of these CAF-1 subunits led to delayed S-phase progression concomitant with slow DNA synthesis, followed by accumulation in late S/G2 phase and aberrant mitosis associated with extra centrosomes, and then the final consequence was cell death. We demonstrated that CAF-1 is necessary for rapid nucleosome formation during DNA replication in vivo as well as in vitro. Loss of CAF-1 was not associated with the apparent induction of phosphorylations of S-checkpoint kinases Chk1 and Chk2. To elucidate the precise role of domain(s) in CAF-1p150, functional dissection analyses including rescue assays were preformed. Results showed that the binding abilities of CAF-1p150 with CAF-1p60 and DNA polymerase sliding clamp proliferating cell nuclear antigen (PCNA) but not with heterochromatin protein HP1-γ are required for cell viability. These observations highlighted the essential role of CAF-1–dependent nucleosome assembly in DNA replication and cell proliferation through its interaction with PCNA.
INTRODUCTION
During and after S phase of the cell cycle, newly synthesized DNA must be assembled into a copy of the parental chromatin structure to maintain proper genomic function (McNairn and Gilbert, 2003). Through the passage of the replication fork, parental nucleosomes are transiently disassembled to be transferred onto newly replicated DNA behind the fork (parental nucleosome segregation), and newly synthesized (de novo) histones H3 and H4 are also deposited as tetramers onto replicating DNA (de novo nucleosome assembly), followed by loading two sets of histone H2A/H2B dimers to form complete nucleosome (Gruss et al., 1993; Gasser et al., 1996).
CAF-1, a well-conserved protein complex composed of p150, p60, and p48 subunits, was originally purified from human cell nuclear extract as an active factor to promote nucleosome assembly of replicating DNA in the simian virus40 (SV40) replication system (Smith and Stillman, 1989). CAF-1p48, known as RbAp48, forms part of multiple complexes involved in different aspects of histone metabolism (Roth and Allis, 1996; Verreault et al., 1996). The other two subunits, p150 and p60, can directly interact with each other (Kaufman et al., 1995) and are recruited in replication foci together with the DNA polymerase sliding clamp proliferating cell nuclear antigen (PCNA) during S phase (Shibahara and Stillman, 1999). In addition, CAF-1 forms a complex with de novo histones H3 and H4 in proliferating cells (Krude, 1995; Verreault et al., 1996). These results support that CAF-1 is the prime candidate factor for the de novo nucleosome assembly (Krude and Keller, 2001; Mello and Almouzni, 2001; Verreault, 2003). Noticeably, CAF-1 also mediates nucleosome assembly during nucleotide excision repair after UV-induced DNA damages (Gaillard et al., 1996; Mello and Almouzni, 2001).
Despite the established role of CAF-1 in nucleosome assembly during DNA replication in vitro, deletion of each gene of the CAF-1 subunits (CAC1, -2, and -3) in Saccharomyces cerevisiae exhibits a minimal effect on growth (Kaufman et al., 1997). These observations are indicative of the possible existence of at least another nucleosome assembly pathway in yeast. Other histone binding proteins, ASF1 and HIRA, which interact with histones H3 and H4, may act in this pathway (Lorain et al., 1998; Tyler et al., 1999). In accord with this, recent studies indicated that there are two distinct pathways of nucleosome assembly in vertebrate cells: CAF-1 facilitates replication-dependent nucleosome assembly with the major histone H3 (H3.1), whereas HIRA is involved in replication-independent nucleosome assembly with the histone H3 variant (H3.3) (Ahmad and Henikoff, 2002; Ray-Gallet et al., 2002; Tagami et al., 2004).
In higher eukaryotes, evidences for the in vivo function of CAF-1 are being accumulated. For example, the expression of a dominant-negative p150 protein or knockdown of p150 protein by small interfering RNA (siRNA) in mammalian cells resulted in S phase block, inducing DNA damage linked checkpoint activation (Hoek and Stillman, 2003; Ye et al., 2003), and knockdown of p60 protein by siRNA led to a rapid cell death (Nabatiyan and Krude, 2004).
In mammalian cells, two protein kinases of the phosphatidylinositol-3–related protein family, ataxia telangiectasia mutated (ATM) and ATM and Rad3 related (ATR), are key components of the pathway for DNA damage and replication checkpoint (Nyberg et al., 2002; McGowan and Russell, 2004). ATM is primarily involved in response to double strand DNA breaks (DSBs) and activates downstream Chk2 kinase (Matsuoka et al., 2000). In contrast, ATR responds to both DSBs and other damages causing replication stress, and it activates downstream kinase Chk1 (Zhao and Piwnica-Worms, 2001). Although the molecular mechanisms of ATM- and ATR-mediated checkpoints have been elucidated for a decade, how eukaryotic cells monitor the proper formation of chromatin and how chromatin assembly-related factors are linked to checkpoints during DNA replication remain unsolved.
In this study, we generated two conditional knockout cells devoid of CAF-1p150 and p60, respectively, by using the gene-targeting technique in the chicken DT40 B-cell line. Depletion of either CAF-1 subunit led to delayed S-phase progression with retarded DNA synthesis and defects in a rapid nucleosome formation of newly replicated DNA, and then the final consequence was lethal. Lethal effect by p150 depletion depended on loss of p150 binding function as to p60 and PCNA, but not to heterochromatin protein HP1-γ. We also provide the evidence implying the possible functional link between CAF-1 and a Chk1-mediated replication checkpoint.
MATERIALS AND METHODS
Cell Culture and Transfection
DT40 cells and their mutants were cultured as described previously (Takami and Nakayama, 1997). To determine growth rate, the cells were counted at indicated times. Transfections and selections of drug-resistant cells were conducted as described previously (Takami and Nakayama, 1997).
Cloning of cDNA and Genomic DNA for Chicken CAF-1p150 and p60
We amplified a part of chicken p150 or p60 cDNA from total RNA prepared from DT40 cells by reverse transcription-polymerase chain reaction (PCR) by using degenerate primers based on human p150 or p60 amino acid (aa) sequences, and we used the resultant PCR products as probes for screening a DT40 λ ZAP II cDNA library as described previously (Takami et al., 1999). Both the 5′-truncated p150 cDNA and the full-length p60 cDNA could be isolated. To obtain full-length p150 cDNA, the 5′ portion of p150 cDNA was PCR amplified from total RNA of DT40 cells based on the sequence from the chicken Web site bursal expressed sequence tag database (http://www.chick.umist.ac.uk/index.html) and fused to the 5′-truncated p150 cDNA. Sequencing of each cDNA was performed using the dye terminator method (Applied Biosystems Division, PerkinElmer-Cetus, Wellesley, MA). Genomic DNA clones of p150 and p60 were isolated from the DT40 λFIX II genomic library (Takami et al., 1999) by using p150 and p60 cDNAs as probes.
Vector Constructions
To create p150-disruption constructs, 1.5-kilobase (kb) upstream and 2.5-kb downstream fragments were excised from the p150 genomic clone and transferred into a pBluescript II vector (Stratagene, La Jolla, CA). The neo or hisD drug resistance cassette was inserted between the upstream and downstream arms of pBluescript. Gene targeting by these constructs was expected to delete the coding sequence corresponding to p150 613-654 aa, which is located between exons 8 and 9 of the p150 gene. The p60-disruption constructs were made in a pBluescript II by subcloning 4-kb upstream and 2.5-kb downstream fragments of the p60 gene, followed by inserting the neo cassette flanked by a loxP site or the bsr/Cre-ER cassette (Fukagawa et al., 1999) between the upstream and downstream arms. Gene targeting with these constructs was expected to insert the drug resistance cassettes into as the position in exon 4 of the p60 gene, corresponding to 200 aa of p60 protein.
The p150 and p60 tet-responsive expression vectors were constructed by inserting the HA-tagged full-length p150 and p60 cDNAs into pUHD10-3 plasmids (Gossen and Bujard, 1992). To obtain the ptTA-bleo construct, a cassette of the bleomycin (bleo)-resistance gene driven by the β-actin promoter was inserted into the pUHD15-1 plasmid, which contains the tet-responsive transactivator gene controlled by the cytomegalovirus promoter.
FLAG-tagged wild-type and various mutant p150 cDNAs were generated by PCR amplification or by a QuikChange method, by using Pfu turbo DNA polymerase (Stratagene), followed by insertion into pApuro, which allows their expressions under the chicken β-actin promoter and carries a puro-resistance gene driven by the SV40 promoter (Takata et al., 1994). CENP-H-GFP targeting construct was described previously (Fukagawa et al., 2001). To construct the replacement targeting vector for HP1-γ-DsRed, we cloned a 5-kb fragment containing the last exon of HP1-γ gene into pBluescript and replaced the stop codon in the last exon with a DsRed sequence, followed by a hyg cassette driven by the β-actin promoter. To construct the replacement targeting vector for INCENP-FLAG, we cloned a 5-kb fragment containing the last exon of INCENP gene into pBluescript and replaced the stop codon in the last exon with 3x FLAG sequences, followed by a hyg cassette driven by the β-actin promoter.
Flow Cytometric Analysis of Cell Cycles
Flow cytometric analyses were carried out using an FACSCalibur (BD Biosciences, Mountain View, CA) as described previously (Takami and Nakayama, 2000). For synchronization of cells into the mitotic phase, cells were cultured for 17 h in the presence or absence of tet, treated with nocodazole (500 ng/ml) for 7 h, released from the block by washing with medium three times, and cultured further. At 2-h intervals, cells were collected, fixed in 70% ethanol, stained with propidium iodide (PI), and then analyzed. Synchronization of cells into G1/S phase was achieved by treating with 1 mM mimosine for last 8 h in the presence or absence of tet for 24 h, and cells were then processed as described above. For two-dimensional cell cycle analyses, cells were cultured in the presence of bromodeoxyuridine (BrdU) for 10 min, fixed in 70% ethanol, and stained with a fluorescein isothiocyanate (FITC)-labeled anti-BrdU antibody (BD PharMingen, San Diego, CA) and PI. To estimate the mitotic index, cells were fixed in 70% methanol and stained with anti-phospho Ser10 of histone H3 antibody (Upstate Biotechnology, Lake Placid, NY), followed by AlexaFluor 488-conjugated anti-rabbit IgG antibody (Invitrogen, Carlsbad, CA) and PI staining.
SV40 DNA Replication-coupled Nucleosome Assembly Reaction
The S100 extracts for SV40 DNA replication were prepared from human 293 cells as described previously (Stillman, 1986). DNA replication-coupled nucleosome assembly reaction was performed as described previously (Verreault et al., 1996), with indicated amounts of nuclear extracts prepared from chicken DT40 and mutant cells. DT40, p150−/− and p60−/− cell lines were grown in the presence or absence of 1 μg/ml tet for 24 h, and nuclear extracts were prepared as described previously (Smith and Stillman, 1989) with modifications as follows. The nuclei were homogenized with 20 mM HEPES, pH 8.0, 20% glycerol, 1.5 mM MgCl2, 420 mM KCl, 0.1 mM dithiothreitol (DTT), and protease inhibitors, and rotated for 30 min at 4°C. The residual nuclear material was removed by centrifugation at 12,000 × g for 15 min.
DNA Synthesis and Micrococcal Nuclease (MNase) Sensitivity
To monitor DNA synthesis rate during CAF-1 depletion, cells were cultured in the presence or absence of 1 μg/ml tet for indicated times and pulse-labeled by the addition of 2 μCi/ml [3H]thymidine (PerkinElmer Life and Analytical Sciences, Boston, MA) for 10 min. After cells were lysed by NaOH, DNA was precipitated with trichloroacetic acid (TCA) and trapped onto filter. The filter was washed with 5% TCA, 70% ethanol, and 100% ethanol, and then dried. Incorporated radioactivities onto filter were counted using a liquid scintillation counter.
To examine stability of nascent chromatin structure, cells were pulse labeled with 20 μCi/ml [3H]thymidine for 5 min and labeled nuclei were prepared as follows. Cells were washed with cold phosphate-buffered saline (PBS) and incubated in the presence of 0.1% NP-40 in NB (15 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 2 mM magnesium acetate, 2 mM CaCl2, 1 mM DTT, and protease inhibitor cocktail [Sigma-Aldrich, St. Louis, MO]). The resultant nuclei were washed with NB twice, suspended at 20 A260/ml (A260 was measured in 2 M NaCl and 5 M urea) in NB, and digested at 37°C for 8 min with 0.0025–0.2 U/ml MNase (Sigma-Aldrich). The reactions were stopped by adding EDTA and SDS to final concentrations of 10 mM and 0.5%, respectively, and then DNA was purified by incubation with 100 μg/ml proteinase K for 2 h at 37°C, followed by phenol-chloroform extraction and ethanol precipitation. DNA was electrophoresed in a 1.2% agarose gel, stained with ethidium bromide (EtBr), and transferred to a Hybond N+ membrane (GE Healthcare). To detect 3H-labeled DNA, blots were directly exposed on a BAS screen (TR2040) specific for tritium and visualized using a Mac BAS-1000 (Fuji Film, Tokyo, Japan).
Western Blotting and Immunoprecipitation
To generate polyclonal antibodies of chicken CAF-1p150, p60, and Chk2, a C-terminal fragment of p150, an N-terminal fragment of p60, and a C-terminal fragment of Chk2 were each expressed as glutathione S-transferase (GST)-fusion proteins in bacteria, and then the purified fusion proteins were injected into rabbits. Affinity purification of antibodies was performed as described previously (Takami et al., 1999).
Other antibodies used in Western blotting were anti-hemagglutinin (HA) (12CA5; Roche Diagnostics, Mannheim, Germany), anti-phospho H2AX (Upstate Biotechnology), anti-p48 (Fujisawa Pharmaceutical, Osaka, Japan), anti-PCNA (PC10; Sigma-Aldrich), anti-HP1-γ (Chemicon International, Temecula, CA), anti-phospho-Chk1 (P-Ser-317 and -345; Cell Signaling Technology, Beverly, MA), anti-Chk1 (G-4; Santa Cruz Biotechnology, Santa Cruz, CA), and horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit secondary antibodies (Dako, Glostrup, Denmark). Western blot signal was developed using a Super Signal-horseradish peroxidase system (Pierce Chemical, Rockford, IL) and visualized with an LAS-1000 imager (Fuji Film).
For immunoprecipitation analysis, cells (1 × 107) were lysed in 1 ml of radioimmunoprecipitation (RIPA) buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, and 1% Triton X-100) containing protease inhibitor cocktail (Sigma-Aldrich) for 30 min on ice, and then clarified lysates were immunoprecipitated with anti-FLAG M2 immobilized beads (Sigma-Aldrich) at 4°C for 2 h. After washing with RIPA buffer three times, beads were suspended in SDS sample buffer, followed by Western blotting.
Immunofluorescence Microscopy
DT40 subclones were fixed on slides in 3% paraformaldehyde (PFA) in HEPES buffer for 15 min, permeabilized with 0.5% NP-40 in PBS for 15 min, incubated in methanol at −20°C for 30 min, and blocked with 3% bovine serum albumin (BSA) in PBS for 20 min. For chromosome spreads, cells were swollen in 75 mM KCl for 10 min, fixed in ice-cold methanol, dropped onto slides, and washed with TEEN (1 mM triethanolamine, pH 8.5, 0.2 mM EDTA, and 0.25 mM NaCl) with 0.1% Triton X-100 and 0.1% BSA. After blocking, cells were incubated with primary antibodies for 60 min. After washing, cells were incubated with secondary antibody for 60 min and counterstained with 4′-6-diamidino-2-phenylindole (DAPI) at 0.2 μg/ml. Fluorescence images were obtained using a charge-coupled device camera (Orca-ER; Hamamatsu, Bridgewater, NJ) mounted on an Axiovert microscope (Carl Zeiss, Jena, Germany) equipped with a 40× objective or a Plan Apo 60×/numerical aperture 1.3 oil immersion objective. Primary antibodies used were as follows: FITC-labeled monoclonal anti-chicken α-tubulin (Sigma-Aldrich) at 1/50 dilution, rabbit anti-γ-tubulin (Sigma-Aldrich) at 1/2000 dilution, rabbit anti-CENP-C at 1/1000 dilution (Fukagawa et al., 2001), mouse anti-FLAG (Sigma-Aldrich) at 1/2000 dilution, rabbit anti-ScII at 1/2000 dilution, rabbit anti-phospho-Chk1 (P-Ser-317) at 1/100 dilution, and mouse anti-phospho H2AX at 1/500 dilution. To detect the primary antibodies, AlexaFluor 488- and 594-conjugated anti-rabbit and anti-mouse IgG antibodies (Invitrogen) (1/400 dilution) were used.
Complementation Assay
The p150-conditional knockout cells (1 × 107) were transfected with pApuro-p150-FLAG and its derivatives and split into two portions. One portion was incubated in medium containing 0.4 μg/ml puromycin (puro) in 96-well plates, and the other portion was incubated in medium containing 0.4 μg/ml puro plus 1 μg/ml tet in 96-well plates. After incubation for 10 d, surviving colonies were counted. The expression levels of transfected genes in several puro-resistant clones were checked by Western blotting. One of each of these clones, which exhibited similar expression levels as endogenous p150 protein, was chosen and used for cell growth and binding assays.
RESULTS
Generation of p150- and p60-Conditional Knockout Cells
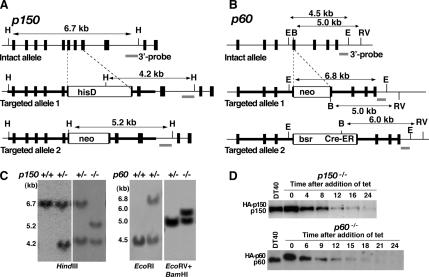
We isolated cDNAs and genomic DNAs of chicken CAF-1p150 and p60 (see Materials and Methods). Open reading frames of chicken p150 and p60 cDNAs encoded predicted proteins of 983 and 580 aa, respectively (DNA Data Bank of Japan accession nos. AB195692 and AB195693). The overall homology between chicken and human p150 proteins was calculated to be 57%. Similarly, the chicken p60 showed 74% overall identity to its human counterpart. Gene disruption constructs were designed based on the genomic DNAs isolated (Figure 1, A and B, top). We could not obtain homozygous knockout clones, p150−/− and p60−/−, even after several attempts. Therefore, we tried to obtain tet-responsive, conditional null mutants of p150 and p60, because this system has been applied successfully to generate conditional homozygous mutants for other essential genes (Takami and Nakayama, 2000). After obtaining p150−/+ and p60−/+ clones, where one allele of p150 and p60 had been disrupted by the hisD and neo disruption constructs, respectively (Figure 1, A and B, middle), we produced p150−/++tetHAp150 and p60−/++tetHAp60 cell lines, in which N-terminal HA-tagged p150 and p60 were expressed under the control of a tet-responsive promoter. After confirmation of the tet-repressive expression of HA-p150 or HA-p60, one of each cell line was transfected for the second round disruption of the remaining allele of p150 or p60 by neo or bsr/Cre-ER disruption construct (Figure 1, A and B, bottom). We obtained three independent p150−/−+tetHAp150 clones and four independent p60−/−+tetHAp60 clones, and then Southern blotting for one of these clones was shown in Figure 1C. Hereafter, we referred to these cell lines as p150- and p60-conditional knockout cells, respectively. In these cells, levels of HA-p150 and HA-p60 proteins were slightly higher than those of endogenous p150 and p60 proteins in DT40 cells, but upon the addition of tet, production of these proteins was gradually declined to below detection levels by 24 h (Figure 1D). Because all of the p150- or p60-conditional knockout clones exhibited essentially the same phenotypic properties after the treatment with tet, we analyzed one clone each for these two knockout clones in detail.
Figure 1.
Generation of p150- and p60-conditional knockout DT40 cells. (A and B) Schematic representations of the chicken p150 and p60 genome loci (top), and two targeted alleles changed by different targeting vectors (middle and bottom). Solid boxes and horizontal lines indicate positions of exons and introns. White boxes indicate drug resistance cassettes: hisD, histidinol dehydrogenase; neo, neomycin; bsr, blasticidin S; Cre-ER, cre-recombinase fused with part of estrogen receptor. The 3′ outer probes are indicated by gray lines. B, BamHI site; H, HindIII site; E, EcoRI site; ER, EcoRV site. (C) Southern blot analysis of wild-type (+/+), heterozygous mutant (+/−), and homozygous mutant (−/−) clones. Genomic DNA digested by indicated enzymes was hybridized with 3′ probes shown in A and B. The sizes of DNA fragments are shown on the left. (D). Western blot analysis for p150 and p60 expression after the addition of tet. Equal amounts of whole cell extracts prepared from DT40 (top and bottom), p150- (top) and p60 (bottom)-conditional knockout cells treated with tet for indicated times were immunoblotted with polyclonal antibodies against either chicken p150 or p60. HA-p150, endogenous p150, HA-p60, and endogenous p60 are indicated on the left.
CAF-1 Depletion Induces Delayed S Phase Progression Concomitant with Slower DNA Replication
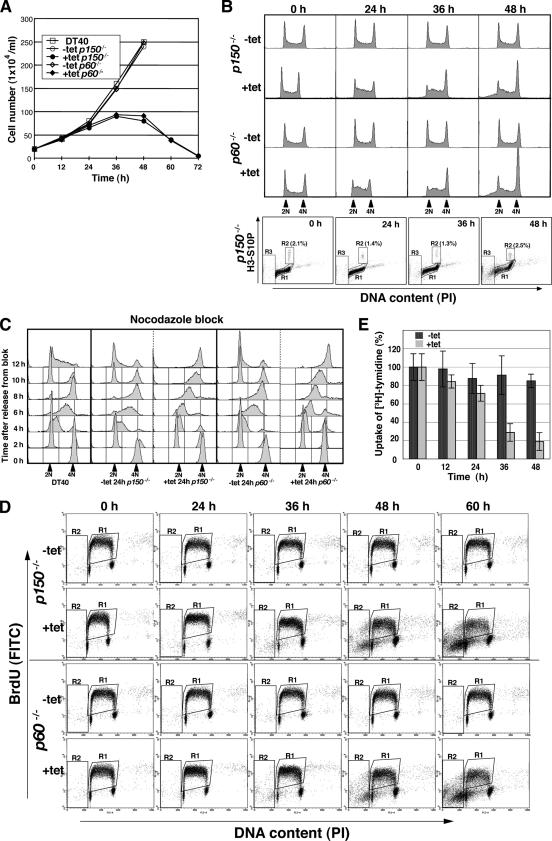
First, we examined proliferative properties of the p150- or p60-conditional knockout cells. In the absence of tet, the growth curves of these two mutant cells were similar to that of wild-type DT40 cells, with an approximate doubling time of 11–12 h (data not shown). However, after the addition of tet, p150- or p60-conditional knockout cells began to grow slowly at 24 h (Figure 2A), concomitant with the decrease in HA-p150 or -p60 protein production (Figure 1D). In addition, dead cells occurred at 48 h; and finally, almost all of the cells were dead by 60–72 h (Figure 2A; our unpublished data), indicating that both p150 and p60 are essential for viability of chicken DT40 cells.
Figure 2.
Growth curves and cell cycle analyses of p150- and p60-conditional knockout cells. (A) Growth curves for DT40, and p150- and p60-conditional knockout cells. The numbers of indicated genotype cells in the absence (−tet) or presence (+tet) of tet were counted. Each time point was examined in triplicate. (B) Flow cytometric analysis of asynchronous p150- and p60-conditional knockout cells. Asynchronous p150 (two top panels) and p60 (two middle panels) conditional cells were fixed and stained with PI to detect total DNA (x-axis; linear scale) at indicated time points in the absence (−tet) and presence (+tet) of tet. Bottom, cells were fixed, stained with anti-histone H3 phosphorylated at Ser10 antibody (H3-S10P) followed by a secondary AlexaFluor 488-labeled anti-rabbit antibody (y-axis; log scale), and finally counterstained with PI to detect total DNA (x-axis; linear scale). Boxed regions R1, R2, and R3 represent interphase, mitotic and apoptotic cells, respectively. (C) Flow cytometric analysis of synchronized p150- and p60-conditional knockout cells. For synchronization into mitotic phase, cells were cultured in the presence (+tet 24 h) or absence of tet (−tet 24 h) for 17 h, followed by culture with nocodazole (500 ng/ml) for 7 h. After release from the cell cycle block, cells were collected at 2 h intervals, fixed, and stained with PI to detect total DNA (x-axis; linear scale). (D) Two-dimensional flow cytometric analysis of p150- and p60-conditional knockout cells. At indicated times, p150 (two top panels) and p60 (two bottom panels) conditional cells were pulse labeled with BrdU for 10 min, fixed, stained with FITC-labeled anti-BrdU antibody (y-axis; log scale), and then stained with PI to detect total DNA (x-axis, linear scale). Boxed regions R1 and R2 indicate BrdU-incorporated S phase and apoptotic cells, respectively. (E) Relative DNA synthesis rate following p150 depletion. The [3H]thymidine incorporations at indicated times after cultivation with (+tet) or without (−tet) tet were normalized by dividing total cell numbers at each time point by those at 0 h, defined as 100%. Data are from two separate experiments.
Fluorescence-activated cell sorting (FACS) analysis, in terms of DNA content, indicated that either p150- or p60-depletion led to accumulation of early-to mid-S–phase cells by 24–36 h, followed by further increases in late S- or G2/M-phase cells up to 48 h (Figure 2B, middle and top). When we evaluated the mitotic index by two-dimensional FACS analysis of DNA content and phosphorylation at Ser-10 residue of histone H3, a marker of mitosis, insignificant accumulation of mitotic cells was observed after the addition of tet (Figure 2B, bottom). Thus, the relatively large population of cells with 4N DNA content at 36–48 h might represent an accumulation in late S or G2 phase rather than in M phase. The treatment with tet of wild-type DT40 cells did not induce aberrant cell cycle progression (data not shown). A cell population with sub-G1 content became apparent at 48 h (Figure 2B), consistent with the accumulation of apoptotic cells with characteristic microscopic morphology of condensed and fragmented chromatin (data not shown).
To verify effects of p150 or p60 depletion on S-phase progression, p150- or p60-conditional knockout cells pretreated with or without tet were synchronized in mitosis using nocodazole, ∼90% of the cells being arrested at M phase (Figure 2C). After the release from the nocodazole block, DNA content was monitored by FACS at 2 h intervals. In the absence of tet, both p150- and p60-conditional knockout cells, like DT40 cells, had proceeded to G1/S and G2/M phases by 2 and 8 h, respectively (Figure 2C). By contrast, in the presence of tet, these two mutant cells exhibited the prolonged S-phase progression (Figure 2C, 4, 6, and 8 h), although the progression from M to G1/S phases was not affected (Figure 2C, 2 h). We also monitored cell cycle events after synchronization with mimosine, wherein >60% of the cell population being arrested at the G1/S boundary (Supplemental Figure 1). Delayed S-phase progression, especially from the mid-to-late S phase, was observed, when each of the p150- and p60-conditional knockout cells had been pretreated with tet for 24 h (Supplemental Figure 1, right, 4, 6, and 8 h).
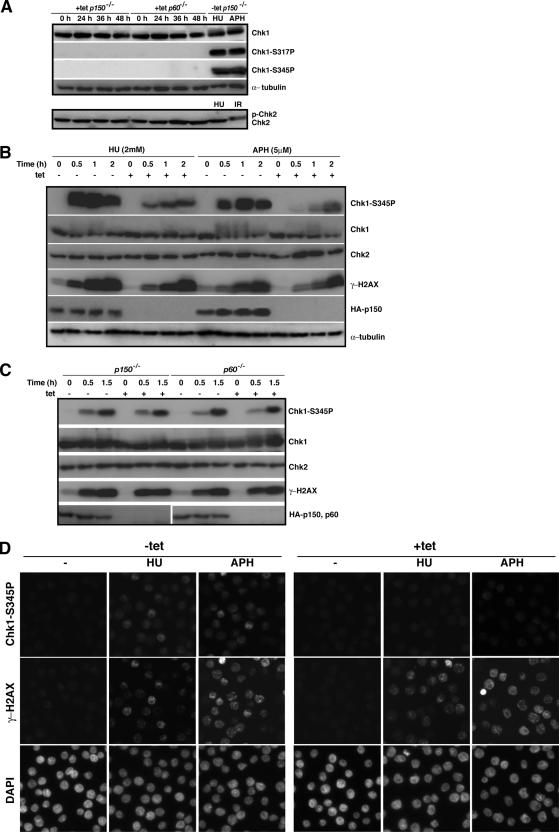
Figure 4.
Checkpoint activation in p150- and p60-deficient cells. (A) The p150- and p60-conditional knockout cells were cultured in the presence (+tet) of tet for indicated times, and whole cell extracts were prepared for Western blotting, by using antibodies for Chk1, phosphorylated Chk1 at Ser317 (Chk1-S317P), phosphorylated Chk1 at Ser345 (Chk1-S345P), Chk2, and α-tubulin as a loading control. Extracts from the p150-conditional knockout cells exposed to 2 mM HU and 5 μM APH for 1 h, or x-ray irradiation (IR; 5 Gy) in the absence of tet (−tet) were also subjected to Western blotting to examine whether Chk1 and Chk2 activations are intact in the p150-conditional knockout cells. (B) The p150-conditional knockout cells were cultured in the presence (+) or absence (−) of tet for 24 h, and treated with 2 mM HU or 5 μM APH for indicated times. The same amounts of whole cell extracts were analyzed by Western blotting, by using antibodies for phosphorylated Chk1 at Ser345 (Chk1-S345P), Chk1, Chk2, γ-H2AX, HA, and α-tubulin as a loading control. Antibody for HA was used to confirm the p150 depletion. (C) The p150- and p60-conditional knockout cells were cultured in the presence (+) or absence (−) of tet for 24 h and irradiated with UV light (UV-C; 3 J/mm2) for indicated times. Western blotting was performed as described in A. Antibody for HA was used to confirm the depletion of p150 and p60. (D) The p150-conditional knockout cells were cultured in the presence (+tet) or absence (−tet) of tet for 24 h, treated with 1 mM HU or 5 μM APH 1.5 h and examined by immunofluorescence microscopy by using antibodies for phosphorylated-Chk1 at Ser345 (Chk1-S345P) (top) and γ-H2AX (middle). Nuclei were counterstained with DAPI (bottom). More than 300 cells were examined at each time to determine the percentage of cells displaying signals of phosphorylated Chk1 and γ-H2AX.
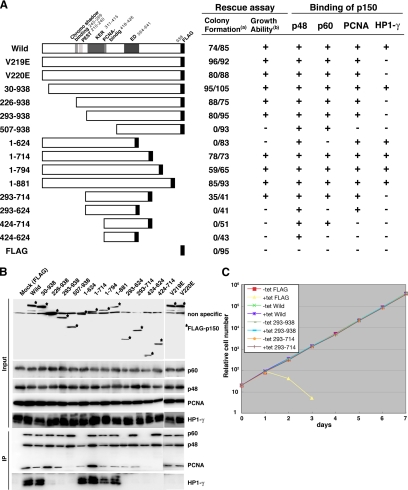
Figure 6.
Regions of p150 required for protein binding and cell viability. (A) N-terminal or C-terminal deletion and missense mutant p150 proteins used are schematically represented on the left, and results are summarized on the right. For rescue assay, the p150-conditional knockout cells (1 × 107) were transfected with pApuro-p150-FLAG and its derivatives and cultured in the presence of 0.4 μg/ml puro with or without tet. (a) Numbers of surviving colonies were counted at 10 d after transfection and shown in numerator (+tet) and denominator (−tet), respectively. (b) From several puro-resistant clones in each transfection, one clone expressing almost similar levels of a series of p150-FLAG proteins was chosen and examined for growth capacity with or without tet. In the presence of tet, clones that were dead within three days are indicated with −, and clones that continuously grew for >2 wk are indicated with + in the table. (B) Interaction of p150 and its derivatives with p60, p48, PCNA, and HP1-γ. Stable p150-conditional knockout cell lines, expressing p150-FLAG protein and its derivatives, were used. Cell extracts were prepared from indicated clones (top lanes) at 24 h after the addition of tet and immunoprecipitated with anti-FLAG antibody-coupled beads (M2; Sigma-Aldrich). Input samples before the immunoprecipitation (top; corresponding to 8% of the materials used) and immunoprecipitated samples (bottom) were resolved in 8 or 12.5% SDS-PAGE and then analyzed by Western blotting, by using antibodies for FLAG, p60, p48, PCNA, and HP1-γ. The results obtained are schematically indicated with + and − in the table in A. (C) Growth curves of p150-conditional knockout cell lines expressing FLAG (as a control), wild-type p150-FLAG protein, and its derivatives. Indicated cells were cultured in the presence (+tet) or absence (−tet) of tet. Cells were inoculated in 24-well dish with 1 ml of medium at 2 × 105 cells/ml at day 0. At indicated times, cells were counted and passaged by diluting culture to be 2 × 105 cells/ml.
With defects in S-phase progression, DNA replication was also impaired in p150- and p60-depleted cells. Two-dimensional FACS analysis for DNA content and BrdU uptake showed that proportion of S-phase cells incorporating BrdU was increased upon the depletion of either subunit of CAF-1, but the amount of incorporated BrdU was slightly reduced at 24 h and greatly reduced by 36–60 h (compare levels of y-axis and shapes of BrdU-arc), indicating an impaired DNA synthesis during S phase (Figure 2D). Reduction of DNA synthesis rate after p150-depletion was also confirmed by [3H]thymidine uptake assay (Figure 2E). Together, we concluded that both p150 and p60 subunits of CAF-1 are essential for efficient DNA replication that links to proper S-phase progression.
Depletion of CAF-1 Causes Failure of Rapid Nucleosome Formation during DNA Replication
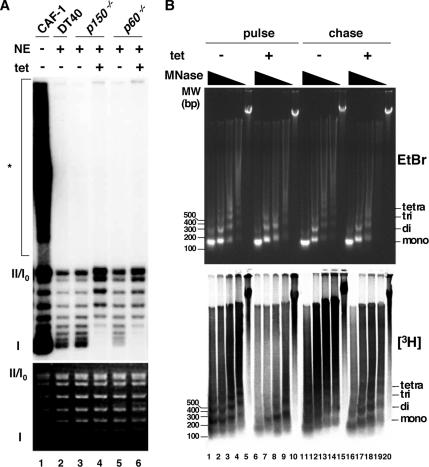
Next, we examined whether there is any impairment in replication-coupled nucleosome assembly in CAF-1–depleted cells. It has been shown that yeast CAF-1 prepared from its nuclear extracts can promote preferentially nucleosome assembly for in vitro replicated SV40 DNA in the presence of the human cell cytosolic extracts, S100, that supported SV40 DNA replication (Smith and Stillman, 1989). Using this system, we assessed whether chicken nuclear extracts could promote replication-coupled nucleosome assembly, and if so, whether this activity is dependent on chicken CAF-1. All nuclear extracts from DT40 p150- and p60-conditional knockout cells (−tet) could induce increased supercoiling of the replicated template (Figure 3A, lanes 2, 3, and 5). However, nuclear extracts from either of p150- and p60-depleted mutant cells (+tet) could not form supercoiled DNA (Figure 3A, lanes 4 and 6). Thus, both CAF-1p150 and p60 are factors essentially required for DNA replication-coupled nucleosome assembly in chicken nuclear extracts.
Figure 3.
Effect of p150 or p60 depletion on nucleosome assembly during DNA replication. (A) SV40 DNA replication was performed in the presence of [32P]dATP with (+) or without (−) nuclear extracts (NE) from wild-type DT40 (DT40), and p150- and p60-conditional knockout cells (p150−/− and p60−/−) (lanes 2–6). As a control, recombinant CAF-1 was used (lane 1). To examine supercoil formation, the reaction products were deproteinized and subjected to 1.0% agarose gel electrophoresis. The extent of DNA synthesis monitored by dAMP incorporations was as follows: 32.9, 3.5, 3.9, 4.3, 3.3, and 3.0 pmol (lanes 1, 2, 3, 4, 5, and 6). The top and bottom panels show the radiolabeled DNA in autoradiograph, and the bulk DNA stained with EtBr, respectively. The positions of form I, form II, and replication intermediates or unresolved catenates (*) are indicated to left. Same results were obtained when reactions were titrated differently. (B) MNase digestion assay of nascent chromatin in p150-conditional knockout cells. After cultivation in the presence (+) or absence (−) of tet for 24 h, cells were pulse labeled with [3H]thymidine for 5 min, washed off, and incubated in [3H]thymidine free medium for 60 min. Nuclei were prepared soon after pulse labeling (pulse) or after 60-min chase period (chase). Same numbers of isolated nuclei were treated with MNase at 0.2, 0.068, 0.022, 0.0074, and 0 U/ml. Purified DNAs were resolved in 1.2% agarose gels, stained with EtBr (top), transferred onto Hybond N+ membrane, and autoradiographed (bottom).
CAF-1 is predicted to facilitate rapid nucleosome formation behind the DNA replication fork in coordination with PCNA (Shibahara and Stillman, 1999; Krude and Keller, 2001; Mello and Almouzni, 2001). Therefore, to examine effects of CAF-1 depletion on the nucleosome formation onto replicating DNA in vivo, we analyzed stability of nascent chromatin structure by labeling DNA with [3H]thymidine, followed by MNase digestion assay. The p150-conditional knockout cells were cultured in the presence or absence of tet for 24 h, pulse labeled with [3H]thymidine for 5 min, and divided into two portions, one portion of which was chased for 60 min in [3H]thymidine-free medium. Equal numbers of nuclei were subjected to MNase digestion assay soon after radiolabeling (pulse) or after 60-min chase (chase) (Figure 3B). In the presence of p150, pulse-labeled DNA was resistant to MNase digestion, and characteristic ladders of mono-, di-, and trinucleosomes were clearly observed, showing that nucleosomes were formed rapidly after DNA replication in vivo (Figure 3B, bottom, lanes 1–5). However, in the absence of p150, pulse-labeled DNA became highly sensitive to MNase digestion, indicating that rapid nucleosome formation of newly replicated DNA was retarded (Figure 3B, bottom, lanes 6–10), although the bulk DNA was comparatively resistant to MNase digestion even in the absence of p150 (Figure 3B, top, compare lanes 1–5 with 6–10). Importantly, after chased for 60 min, labeled DNA from the p150-depleted cells restored resistance to MNase digestion, although it was still more sensitive to the digestion (Figure 3B, bottom, compare lanes 11–15 with 16–20), suggesting that some nucleosome assembly during replication occurred in the absence of CAF-1. Similar results were also observed for p60-depleted cells (data not shown). Of note, the slightly reduced DNA synthesis rate (Figure 2E) could not be accounted for this observed MNase hypersensitivity of the nascent chromatin in CAF-1 depleted cells, because the aphidicoline (APH)-treated DT40 cells showed a significantly reduced DNA synthesis rate but did not cause an increased MNase sensitivity of newly synthesized DNA (data not shown). In addition, no apoptosis-related nuclease activities were detected (Figure 3B, lanes 5, 10, 15, and 20). These results suggest that rapid nucleosome assembly soon after DNA synthesis is specifically compromised in CAF-1–depleted cells or that lack of CAF-1 somehow impedes or slows down the rate of conversion of mature nucleosomes into stable higher order chromatin fibers that are more resistant to MNase digestion.
CAF-1 Is Associated with Full Activation of Chk1 in Response to Replication Stalling
Rapid nucleosome assembly during DNA replication was impaired, but slowly and gradually progressive pathway of nucleosome assembly could continue even in the absence of CAF-1 (Figure 3B). Nevertheless, unexpectedly, the defects in DNA replication and S-phase progression were much more severe in CAF-1–depleted cells (Figure 2). Therefore, we examined whether any intra-S-phase checkpoints are activated by CAF-1 depletion. The ATM signal responds primarily to damages causing DSBs and phosphorylates a downstream kinase Chk2 (Matsuoka et al., 2000). The ATR signal is prominent in inducing checkpoint responses to damages causing replication blocks and activates a downstream kinase Chk1 (Zhao and Piwnica-Worms, 2001). Activation of Chk2 is manifested by gel mobility shift due to its phosphorylation (Zachos et al., 2003), and activation of Chk1 is hallmarked by its phosphorylation at Ser-317 and Ser-345 residues (Zhao and Piwnica-Worms, 2001). Hydroxyurea (HU) causes replication blocks by depleting the available dNTP pool, and APH is an inhibitor of DNA polymerases α and δ. In the presence of CAF-1, treatment with either HU or APH induced massive phosphorylations of Chk1 at Ser-317 and Ser-345 residues (Figure 4A, top, lanes 9 and 10), and x-ray-irradiation that creates DSBs, but not HU-treatment, activated Chk2 (Figure 4A, bottom, lanes 9 and 10), indicating that the pathways for ATR-Chk1 and ATM-Chk2 are intact in chicken DT40 cells. When these activations were examined during CAF-1 depletion, unexpectedly, neither induction of Chk1 phosphorylation nor phosphorylated form of Chk2 could be detected at any time after the tet treatment both in p150- and p60-conditional knockout cells (Figure 4A, lanes 1–4 and 5–8). These results suggest that, despite the apparent delayed S phase in CAF-1–depleted cells, certain levels of intra-S-phase checkpoints are compromised.
To assess whether CAF-1 is involved in activations of some S-phase checkpoint pathways, we compared efficiencies of phosphorylations of Chk1, Chk2, and H2AX, a marker for DNA damage, after the treatment with HU or APH of CAF-1–depleted and –nondepleted cells. To minimize the effects of DNA replication defect in CAF-1–depleted cells, we treated p150-conditional knockout cells with tet only for 24 h, at which time point the p150 protein was barely detectable, but ability of DNA synthesis was not so compromised (Figure 2, D and E). HU or APH treatment led to rapid and remarkable phosphorylation of Chk1 at Ser-345 in the presence of p150, but in the absence of p150 the induction was delayed and the level of the Chk1 phosphorylation was lowered significantly (Figure 4B, first and second panels). HU or APH treatment also induced a time-dependent increase in H2AX phosphorylation either in the presence or absence of p150, with slightly lesser extent in p150-depleted cells (Figure 4B, fourth panel, 0.5 and 1.0 h). Treatment of the mutant cells with HU or APH did not result in the band shift representing phosphorylation of Chk2 (Figure 4B, third panel). Similar results also were observed in p60-depleted cells (data not shown). These results imply that CAF-1 is involved in full activation of Chk1 in response to replication stalling. Contrary to the cases of APH and HU, UV-induced Chk1 phosphorylation level was not compromised in p150- and p60-depleted cells (Figure 4C, first and second panels). Thus, the requirement of CAF-1 for efficient Chk1 activation is likely to be specific in response to APH- or HU-induced replication stalling but not to UV-induced DNA lesions.
Phosphorylations of both Chk1 and H2AX also were examined by immunofluorescence analysis. Exposures to HU and APH produced cells containing intense fluorescent signals for phosphorylated Chk1 (41.9% for HU; 52.1% for APH) and phosphorylated H2AX (52.3% for HU; 64.9% for APH) in the presence of p150 (Figure 4D, left). By contrast, there were no cells containing intense signals, if any, with weaker signals, for phosphorylated Chk1 (<1%) in p150-depleted cells (Figure 4D, right). In contrast, intense signals for γ-H2AX were observed in the p150-depleted cells after treatments with HU and APH, but the percentages of cells containing intense signals were slightly reduced to 48.7 and 50.0, respectively (Figure 4D, right). These results give additional support for the involvement of CAF-1 in the Chk1 activation in response to replication stress.
Loss of CAF-1 Induces the Formation of Extra Centrosomes Leading to Mitotic Aberration
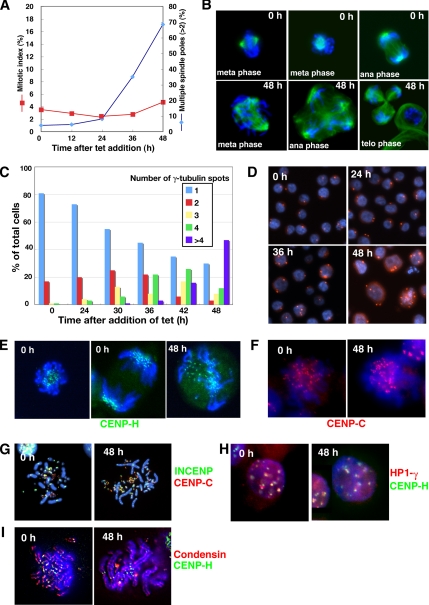
Our FACS analyses revealed that CAF-1 depletion resulted in slight decrease in mitotic index at 24 and 36 h after the addition of tet and increase in cell population of late S/G2 phase up to 48 h (Figure 2B), suggesting a delay or arrest in the cell cycle before mitosis. To define more precisely the effect of CAF-1-deficiency on individual cells, we examined the status of nuclei and microtubulin by using immunofluorescence microscopy. p150-conditional knockout cells were stained by DAPI and anti α-tubulin or γ-tubulin antibodies at various times after the addition of tet. As shown in Figure 5A, mitotic index was slightly decreased during 24–36 h, indicating a delay in G2 phase. However, finally, many of p150-depleted cells seemed to escape the G2 checkpoint block so as to enter into mitotic phase. Although most of these cells were accumulated in prometaphase, some cell population continued to progress thorough mitosis. Interestingly, many mitotic cells exhibited aberrant phenotypes, such as unaligned chromosomes in a metaphase plate accompanied with multipolar (>2) spindles (Figure 5B). Consistent with this, cells at interphase showing multiple centrosomes (>2) began to accumulate significantly at 30 h, and by 48 h, >70% of the cells had extra number of centrosomes (>2) (Figure 5, C and D). Increase in proportion of mitotic cells with multiple spindle poles started at 36 h, and then 70% of mitotic cells had spindle abnormalities by 48 h (Figure 5A). As a consequence, these led to a highly aberrant mitotic exit, such as unequally segregating anaphases (Figure 5B) or cytokinesis failure, followed by dying before or early in the next cell cycle.
Figure 5.
p150-deficiency induces multiple spindle poles and aberrant chromosome alignment in mitosis. (A) Mitotic index and frequency of mitotic cells with multiple spindle poles (>2) during p150 depletion. The p150-conditional knockout cells were cultured in the presence of tet. At indicated times, cells were fixed and stained with FITC-labeled α-tubulin antibody. Nuclei were counterstained by DAPI. Numbers of mitotic cells (red) and cells having multiple spindle poles (>2) per one mitotic cell (blue) were counted under a fluorescence microscope. At least 1000 cells were counted at each time for mitotic index, and at least 200 mitotic cells were examined for mitotic aberration at each time. (B) Mitotic aberration in p150-depleted cells. Cells were cultured in the absence (0 h) or presence of tet for 48 h (48 h), spotted onto slide glasses, fixed by 3% PFA, and stained with FITC-conjugated anti-α-tubulin antibody (green). DNA was counterstained with DAPI (blue). Top, well-ordered chromosomes with bipolar spindle at the metaphase and anaphase in the absence of tet. Bottom, misaligned chromosomes with multipolar spindles at metaphase, anaphase, and telophase at 48 h in the presence of tet. (C) Centrosome amplification in p150-deficient cells. The numbers of γ-tubulin spots were counted at each time point after the addition of tet. Total of 200 cells were counted at each time point. (D) Immunofluorescence microscopy of γ-tubulin spots in p150-deficient cells. The p150-conditional knockout cells were cultured for indicated times in the presence of tet, spotted onto slide glasses, fixed by 3% PFA, and stained with anti-γ-tubulin antibody (red). DNA was counterstained with DAPI (blue). (E) Localization of CENP-H during p150 depletion. p150-conditional knockout cells, in which enhanced green fluorescent protein (EGFP) had been knocked in last exon of CENP-H allele, were cultured in the absence (0 h) or presence of tet for 48 h (48 h). At indicated times, CENP-H-GFP signal and Hoechst 33258 stained DNA were directly observed by fluorescence microscopy in live mitotic cells. (F) Localization of CENP-C in metaphase spread of p150-conditional knockout cells. Cells were cultured in the absence (0 h) or presence of tet for 48 h (48 h), treated with colcemid for last 1.5 h, fixed by methanol, and spotted onto slide glasses to make chromosome spread. Chromosomes were stained with anti CENP-C antibody (red). DNA was counterstained with DAPI (blue). (G) Colocalization of CENP-C and INCENP in metaphase spread. p150-conditional knockout cells, in which FLAG had been knocked in last exon of INCENP allele, were cultured in the absence (0 h) or presence of tet for 48 h (48 h), and then chromosome spread was made and doubly stained with anti-FLAG (green) and CENP-C (red) antibodies. DNA was counterstained with DAPI (blue). (H) Colocalization of HP1-γ and CENP-H in interphase cells. p150-conditional knockout cells, in which DsRed and EGFP had been tagged into respective last exons of HP1-γ and CENP-H alleles, were cultured in the absence (0 h) or presence of tet for 48 h (48 h). Cells were cytospun onto slide glasses and fixed by 3% PFA. DNA was counterstained with DAPI (blue). Signals for HP1-γ-DsRed (red) and CENP-H-GFP (green) were directly observed. (I). Localization of condensin and CENP-H in metaphase spread. p150-conditional knockout cells, in which EGFP had been tagged into the CAEP-H last exon, were cultured in the absence (0 h) or presence of tet for 48 h (48 h), treated with colcemid for last 1.5 h, fixed by methanol, and spotted onto slide glasses to make chromosome spread. Chromosomes were stained with anti-ScII antibody (red). CENP-H-GFP signals (green) were directly observed. DNA was counterstained with DAPI (blue).
Because CAF-1 has been implicated to function in kinetochore structure through pericentromeric heterochromatin organization in yeast, we determined whether the aberrant mitotic phenotypes accompany with impaired kinetochore structure or function. Localizations of several centromere or pericentromeric heterochromatin-associated proteins were examined by immunofluorescence or monitored by these endogenous proteins tagged with green fluorescent protein (GFP), DsRed, or FLAG (Fukagawa et al., 2001). Kinetochore localizations of both inner kinetochore proteins, CENP-H and CENP-C, were likely to be normal in p150-deficient cells (Figure 5, E and F). In addition, localization of chromosome passenger protein INCENP that was recruited to inner centromeres at metaphase did not seem to be disturbed in p150-depleted cells (Figure 5G). Likewise, association of the heterochromatin protein HP1-γ to pericentromeric heterochromatin regions, which was indicated by its colocalization with CENP-H, was not disturbed in p150-depleted cells (Figure 5H). These results suggested that inner kinetochore structure, including pericentromeric heterochromatin structure, was not severely compromised in CAF-1–deficient cells. However, we cannot exclude the possibility that CAF-1 is involved in formation or maintenance of pericentromeric heterochromatin, which is critical for proper formation of kinetochore and chromosome segregation. In relation to this, human Asf1a, another histone deposition protein, has been shown to promote the formation of specialized heterochromatin, known as senescence-associated heterochromatin foci (Zhang et al., 2005). The most reasonable explanation for such an aberrant mitosis could be due to resultant extra centrosome duplication, which presumably occurred during prolonged S and arrested G2 phases caused by slower DNA replication during CAF-1 depletion.
We also examined localization of the condensin subunit ScII/SMC2, which participates in chromosome condensation (Hudson et al., 2003). ScII/SMC2 localized all along chromatids in metaphase spreads, and such staining patterns were not compromised in p150-depleted cells (Figure 5I), suggesting that formation of stable chromatin structure by CAF-1 was not linked with the processing of ScII-mediated chromosome condensation.
p150 Binding with p60 and PCNA, but Not with HP-1-γ, Is Required for Cell Viability
CAF-1p150 is well known to be associated with CAF-1p60 and p48, PCNA, and HP1 (Kaufman et al., 1995; Murzina et al., 1999; Shibahara and Stillman, 1999). To examine functional relationship between the binding of p150 with these four proteins and cell growth, we first produced a series of p150-conditional knockout cell-derived cell lines, in which FLAG-tagged p150 and its mutants were stably expressed (Figure 6A). Binding was assessed by coimmunoprecipitation assays with the same amount of whole cell extract from each clone, by using an anti-FLAG antibody. To eliminate the effect of HA-p150 expressed under the tet-responsible promoter, cell extracts were prepared from cells treated with tet for 24 h. Each immunoprecipitant was analyzed by Western blotting but using anti-p60, p48, PCNA, and HP1-γ antibodies. We confirmed that all four proteins could bind to full-length p150 (Figure 6B). N- and C-terminal deletion analyses revealed that 507-938, 1-714, and 424-714 aa of p150 could be precipitated with both p60 and p48 proteins, whereas 424-624 aa with p48 protein only, suggesting not only that its binding region with p60 lies within 507–714 aa, containing complete ED region, but also that the binding region with p48 lies within 507-624 aa (Figure 6, A and B). These results agreed overall with the in vitro results (Kaufman et al., 1995; Krawitz et al., 2002). In contrast, the binding region of p150 with PCNA was mapped within 293-624 aa, and the characterized PCNA-binding motif Qxx(I/L/V)xx(F/Y)(F/Y) was located at 418-426 aa (Figure 6B). This motif has been shown to be crucial for interaction with PCNA in yeast and human p150 (Krawitz et al., 2002; Ye et al., 2003). In addition, the N-terminal 1-226 aa of p150 was required for the binding with HP1-γ, and the binding ability was diminished by mutations at Val 219 or Val 220, which lied within the putative motif for binding with the HP1 chromo shadow domain (Figure 6B), indicating that the interaction with HP1-γ was mediated by this site, consistent with the previous report (Murzina et al., 1999).
We then performed a complementation assay to identify specific region(s) of p150 required for cell viability. The complementation abilities of p150 mutant proteins were examined based on the appearance of surviving colonies in the presence of both tet and puro after transfection with expression vectors encoding FLAG-tagged p150 and its mutants (Figure 6A). Survival under the conditions would indicate that the mutant proteins tested could rescue cells from the tet-induced lethality. As expected, the wild-type p150 protein could efficiently form the surviving colonies, whereas introduction of empty construct could not. As summarized in Figure 6A, the minimum region required for cell viability and cell growth was confined to 293-714 aa of p150. Of note, point mutations (V219E and V220E), which abolished the binding ability as to HP1-γ, retained its rescue ability. All the established cell lines expressing mutant p150 proteins, which retained rescue ability, grew continuously for more than two weeks with similar growth rates in the absence or presence of tet (Figure 6, A and C; our unpublished data). These results suggest that the region of p150 required for binding with p60 and PCNA, but not with HP1-γ, is essential for cell viability.
DISCUSSION
Recently, essential roles for p150 and p60 subunits of CAF-1 were clarified by studies with siRNA mediated p150 or p60 knockdown approach (Hoek and Stillman, 2003; Nabatiyan and Krude, 2004) and with overexpression of dominant-negative p150 (Ye et al., 2003). To better understand physiological functions of CAF-1, we genetically created conditional p150 and p60 knockout chicken DT40 cell lines and clearly demonstrated that both p150 and p60 are essential for cell viability. Depletion of either p150 or p60 caused defects in DNA replication and S-phase progression accompanied with defects in de novo chromatin assembly. We also revealed that the region of CAF-1p150 linked to cell proliferation is associated with its interacting capabilities as to p60 and PCNA in vivo. These observations highlighted the fact that CAF-1–dependent nucleosome assembly plays a crucial role in DNA replication and cell proliferation through its interaction with PCNA.
The involvement of CAF-1 in de novo nucleosome assembly during DNA replication has been suggested by several lines of evidence, including its localization at replication foci (Krude, 1999) and its association with de novo histones H3/H4 (Verreault et al., 1996) as well as PCNA (Shibahara and Stillman, 1999). We revealed that CAF-1–depleted nuclear extracts lack the ability of nucleosome assembly coupled with the DNA replication in vitro. In addition, newly synthesized DNA in CAF-1–depleted cells was highly sensitive to MNase digestion soon after DNA replication, although the MNase resistance was partially restored after continued DNA replication, indicating that rapid nucleosome formation of replicated DNA was significantly impaired in the absence of CAF-1 in living cells. Importantly, these results provided the first direct evidence for a slow and gradual nucleosome assembly pathway remaining intact even without CAF-1, suggesting that a distinct pathway independent of CAF-1 for nucleosome assembly during DNA replication is present. However, such additional assembly pathway might be insufficient for a rapid and complete assembly of nascent DNA into functional chromatin, probably leading to regional and stochastic disturbances of chromatin formation triggering various cellular defects, such as accumulation of damaged DNA in human cells (Ye et al., 2003; Nabatiyan and Krude, 2004) and aberrant mitotic process (Figure 5).
The reason for the defects in DNA replication and S-phase progression in CAF-1–depleted DT40 cells is an interesting question that remains to be addressed. Intimate linkage between nucleosome assembly and checkpoint cascade has become evident in yeast. Asf1 directly interacts with Rad53 and releases from it in response to DNA damage, although Asf1 is not required for S-phase checkpoint activation (Emili et al., 2001; Hu et al., 2001). Recently, it has been shown that when fork progression is disturbed by replication block, Asf1 is required for maintenance of a subset of replication machinery proteins at the stalled site (Franco et al., 2005). In mammalian cells, CAF-1 has also been proposed to contribute in fork progression during DNA replication through destabilization of parental nucleosomes ahead of fork or by maintaining topological state of genome DNA (Hoek and Stillman, 2003). Alternatively, the defects in rapid nucleosome assembly by CAF-1 depletion may trigger replication checkpoint to monitor the fidelity of chromatin formation behind the replication fork, leading to the arrest of fork progression in S phase. Indeed, previous studies have shown that CAF-1 inactivation in human cells leads to activations of ATR/ATM (Ye et al., 2003) or Chk1 and p53 (Hoek and Stillman, 2003), but these activations seem to occur after the onset of the replication defect (Hoek and Stillman, 2003). In DT40 cells, we could not detect an increase in phosphorylation of either Chk1 or Chk2 at any stage of CAF-1 depletion (Figure 4A), although the possible low level of phosphorylation cannot be ruled out. These suggest that the slower replication seen in CAF-1–depleted cells might be caused by a primary defect in DNA replication, not solely by the consequence of the activation of checkpoint machinery, implying that CAF-1 may function closely to DNA replication machinery during replication fork progression.
The exact reason for discrepancy between our result and others in Chk1 activation (Hoek and Stillman, 2003) is unclear, but it could be explained by several factors, such as cell types or CAF-1 depletion systems used. It is possible that DT40 cells may cause distinct checkpoint responses due to lack of p53 gene (Takao et al., 1999). Additionally, the depletion system used may cause different levels of decreasing rate and/or amount of CAF-1 proteins, which would definitely influence on triggering different levels of checkpoint responses. These possibilities need to be clarified in future. We also found that HU- and APH- but not UV-induced Chk1 phosphorylations were severely weakened in CAF-1–depleted DT40 cells (Figure 4B). This implies that CAF-1 is necessary for full activation of Chk1 in response to stalled replication. Alternatively, CAF-1 depletion enforced cells to accumulate in late S/G2 phase, suggesting that some sort of checkpoint activation occurs. One plausible explanation for such a contradictory finding is that CAF-1–depleted cells have already arrested at a point in cell cycle before that at which Chk1 phosphorylation occurs. Activation of the ATR/Chk1 pathway is initiated by the formation of collapsed DNA structure derived from replication stalling (McGowan and Russell, 2004). Because CAF-1 is required for DNA replication and would contribute to the efficient fork progression, its depletion could negatively affect on the formation of fork collapse after replication stalling, which may lead to a reduced amount of collapsed DNA, and, therefore, reduced checkpoint signaling. However, another possibility cannot be excluded out that CAF-1 may act as a part of the replication checkpoint signal transducer. For example, CAF-1 or CAF-1–mediated chromatin structure might suitably serve to amplify the signal by providing a platform for the retention of checkpoint proteins, including Rad9–Rad1–Hus1 complex, ATR or claspin, which are required for Chk1 activation (Volkmer and Karnitz, 1999; Nyberg et al., 2002; Lee et al., 2003; McGowan and Russell, 2004). Further studies will be required to resolve these considerations.
In yeast, loss of CAF-1 genes causes defects in gene silencing at heterochromatin, and their combined loss with Hir gene causes defects in kinetochore organization, indicating the important role of CAF-1 for heterochromatin and pericentromeric heterochromatin structures. Localization of p150 at heterochromatin sites in late S phase through direct interaction with heterochromatin protein HP1 (Murzina et al., 1999), and derepression of artificial silenced gene by a dominant-negative mutant of p150 (Tchenio et al., 2001) suggest a certain role of CAF-1 in heterochromatin organization in vertebrates. However, we could not observe any severe defects in kinetochore or pericentromeric heterochromatin structure in p150-deficient cells (Figure 5). Furthermore, complementation assay showed that p150 mutant protein lacking HP1-γ binding ability could continue cell growth with undetectable defect in heterochromatin structure (Figure 6; our unpublished data). However, these results do not exclude a possibility for involving of CAF-1 in heterochromatin organization, such as HP1 deposition (Murzina et al., 1999; Quivy et al., 2004), and rather suggest that redundant pathways for HP1-γ deposition exist to complement loss of interaction between CAF-1 and HP1-γ. Regardless, the established cell lines expressing exclusively particular mutant p150 proteins will be useful to dissect participation of CAF-1 in heterochromatin organization. Despite insignificant defect in kinetochore or pericentromeric heterochromatin structure, p150-deficient cells exhibited severe mitotic impairments, such as multipolar spindle formations and disturbed mitotic chromosome alignments at late stage of p150 depletion. Presumably, slowed DNA replication caused by CAF-1 deficiency might trigger such aberrant mitosis by interfering centrosome duplication, which is in line with the previous result that HU or APH treatment induced centrosome amplification in DT40 cells (Dodson et al., 2004).
Our study, implying the essential role of CAF-1 in DNA replication machinery and its possible association with Chk1 activation, may help to understand the potential link among replication-coupled nucleosome assembly, DNA replication, and S-phase checkpoint machinery in vertebrate cells. We have now generated two HIRA-deficient and ASF1-conditional mutant DT40 cell lines (Ahmad et al., 2005; Sanematsu et al., 2006). Analyses of these mutant cells, together with the CAF-1–deficient conditional cells, will be of powerful tools for understanding the roles of CAF-1–dependent and CAF-1–independent nucleosome assembly, in relation to DNA replication, checkpoint response, DNA repair, and maintenance of genome integrity in vertebrate cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. H. Araki (National Institute of Genetics), S. Enomoto (Department of Genetics, Cell Biology, and Development, University of Minnesota, Minneapolis, MN), and Y. Murakami (Kyoto University, Kyoto, Japan) for critical comments and advice. We also thank N. Nagamatsu-Yamamoto for technical support, H. K. Barman for a critical reading of the manuscript, and Dr. B. Stillman (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for providing the human p150 cDNA clone. This work was supported in part by a grant-in-aid and The 21st Century COE Program (Life Science) from Ministry of Education, Culture, Sports, Science, and Technology; and Core Research for Evolutional Science and Technology, Precursory Research for Embryonic Science and Technology (PRESTO), and Human Frontier Science Program from the Science and Technology Agency of Japan.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0426) on October 25, 2006.
REFERENCES
- Ahmad A., Kikuchi H., Takami Y., Nakayama T. Different roles of N-terminal and C-terminal halves of HIRA in transcription regulation of cell cycle-related genes that contribute to control of vertebrate cell growth. J. Biol. Chem. 2005;280:32090–32100. doi: 10.1074/jbc.M501426200. [DOI] [PubMed] [Google Scholar]
- Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Dodson H., Bourke E., Jeffers L. J., Vagnarelli P., Sonoda E., Takeda S., Earnshaw W. C., Merdes A., Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A., Schieltz D. M., Yates J. R., 3rd, Hartwell L. H. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- Franco A. A., Lam W. M., Burgers P. M., Kaufman P. D. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Hayward N., Yang J., Azzalin C., Griffin D., Stewart A. F., Brown W. The chicken HPRT gene: a counter selectable marker for the DT40 cell line. Nucleic Acids Res. 1999;27:1966–1969. doi: 10.1093/nar/27.9.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard P. H., Martini E. M., Kaufman P. D., Stillman B., Moustacchi E., Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- Gasser R., Koller T., Sogo J. M. The stability of nucleosomes at the replication fork. J. Mol. Biol. 1996;258:224–239. doi: 10.1006/jmbi.1996.0245. [DOI] [PubMed] [Google Scholar]
- Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss C., Wu J., Koller T., Sogo J. M. Disruption of the nucleosomes at the replication fork. EMBO J. 1993;12:4533–4545. doi: 10.1002/j.1460-2075.1993.tb06142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek M., Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Alcasabas A. A., Elledge S. J. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D. F., Vagnarelli P., Gassmann R., Earnshaw W. C. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell. 2003;5:323–336. doi: 10.1016/s1534-5807(03)00199-0. [DOI] [PubMed] [Google Scholar]
- Kaufman P. D., Kobayashi R., Kessler N., Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Kaufman P. D., Kobayashi R., Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Krawitz D. C., Kama T., Kaufman P. D. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Chromatin. Nucleosome assembly during DNA replication. Curr. Biol. 1995;5:1232–1234. doi: 10.1016/s0960-9822(95)00245-4. [DOI] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly during DNA replication in somatic cells. Eur. J. Biochem. 1999;263:1–5. doi: 10.1046/j.1432-1327.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Krude T., Keller C. Chromatin assembly during S phase: contributions from histone deposition, DNA replication and the cell division cycle. Cell Mol. Life Sci. 2001;58:665–672. doi: 10.1007/PL00000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W. G. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Lorain S., Quivy J. P., Monier-Gavelle F., Scamps C., Lecluse Y., Almouzni G., Lipinski M. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol. Cell Biol. 1998;18:5546–5556. doi: 10.1128/mcb.18.9.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S. J. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan C. H., Russell P. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 2004;16:629–633. doi: 10.1016/j.ceb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- McNairn A. J., Gilbert D. M. Epigenomic replication: linking epigenetics to DNA replication. Bioessays. 2003;25:647–656. doi: 10.1002/bies.10305. [DOI] [PubMed] [Google Scholar]
- Mello J. A., Almouzni G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 2001;11:136–141. doi: 10.1016/s0959-437x(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Murzina N., Verreault A., Laue E., Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- Nabatiyan A., Krude T. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol. Cell Biol. 2004;24:2853–2862. doi: 10.1128/MCB.24.7.2853-2862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- Quivy J. P., Roche D., Kirschner D., Tagami H., Nakatani Y., Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D., Quivy J. P., Scamps C., Martini E. M., Lipinski M., Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Allis C. D. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- Sanematsu F., Takami Y., Barman H. K., Fukagawa T., Ono T., Shibahara K. I., Nakayama T. Asf1 is required for viability and chromatin assembly during DNA replication in vertebrate cells. J. Biol. Chem. 2006;281:13817–13827. doi: 10.1074/jbc.M511590200. [DOI] [PubMed] [Google Scholar]
- Shibahara K., Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Smith S., Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Takami Y., Kikuchi H., Nakayama T. Chicken histone deacetylase-2 controls the amount of the IgM H-chain at the steps of both transcription of its gene and alternative processing of its pre-mRNA in the DT40 cell line. J. Biol. Chem. 1999;274:23977–23990. doi: 10.1074/jbc.274.34.23977. [DOI] [PubMed] [Google Scholar]
- Takami Y., Nakayama T. A single copy of linker H1 genes is enough for proliferation of the DT40 chicken B cell line, and linker H1 variants participate in regulation of gene expression. Genes Cells. 1997;2:711–723. doi: 10.1046/j.1365-2443.1997.1550353.x. [DOI] [PubMed] [Google Scholar]
- Takami Y., Nakayama T. N-terminal region, C-terminal region, nuclear export signal, and deacetylation activity of histone deacetylase-3 are essential for the viability of the DT40 chicken B cell line. J. Biol. Chem. 2000;275:16191–16201. doi: 10.1074/jbc.M908066199. [DOI] [PubMed] [Google Scholar]
- Takao N., Kato H., Mori R., Morrison C., Sonada E., Sun X., Shimizu H., Yoshioka K., Takeda S., Yamamoto K. Disruption of ATM in p53-null cells causes multiple functional abnormalities in cellular response to ionizing radiation. Oncogene. 1999;18:7002–7009. doi: 10.1038/sj.onc.1203172. [DOI] [PubMed] [Google Scholar]
- Takata M., Sabe H., Hata A., Inazu T., Homma Y., Nukada T., Yamamura H., Kurosaki T. Tyrosine kinases Lyn and Syk regulate B cell receptor-coupled Ca2+ mobilization through distinct pathways. EMBO J. 1994;13:1341–1349. doi: 10.1002/j.1460-2075.1994.tb06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchenio T., Casella J. F., Heidmann T. A truncated form of the human CAF-1 p150 subunit impairs the maintenance of transcriptional gene silencing in mammalian cells. Mol. Cell Biol. 2001;21:1953–1961. doi: 10.1128/MCB.21.6.1953-1961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J. K., Adams C. R., Chen S. R., Kobayashi R., Kamakaka R. T., Kadonaga J. T. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Verreault A. Histone deposition at the replication fork: a matter of urgency. Mol. Cell. 2003;11:283–284. doi: 10.1016/s1097-2765(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman P. D., Kobayashi R., Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Volkmer E., Karnitz L. M. Human homologs of Schizosaccharomyces pombe rad1, hus1, and rad9 form a DNA damage-responsive protein complex. J. Biol. Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- Ye X., Franco A. A., Santos H., Nelson D. M., Kaufman P. D., Adams P. D. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Zachos G., Rainey M. D., Gillespie D. A. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 2003;22:713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Zhao H., Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.