Abstract
Spindle assembly and accurate chromosome segregation require the proper regulation of microtubule dynamics. MCAK, a Kinesin-13, catalytically depolymerizes microtubules, regulates physiological microtubule dynamics, and is the major catastrophe factor in egg extracts. Purified GFP-tagged MCAK domain mutants were assayed to address how the different MCAK domains contribute to in vitro microtubule depolymerization activity and physiological spindle assembly activity in egg extracts. Our biochemical results demonstrate that both the neck and the C-terminal domain are necessary for robust in vitro microtubule depolymerization activity. In particular, the neck is essential for microtubule end binding, and the C-terminal domain is essential for tight microtubule binding in the presence of excess tubulin heterodimer. Our physiological results illustrate that the N-terminal domain is essential for regulating microtubule dynamics, stimulating spindle bipolarity, and kinetochore targeting; whereas the C-terminal domain is necessary for robust microtubule depolymerization activity, limiting spindle bipolarity, and enhancing kinetochore targeting. Unexpectedly, robust MCAK microtubule (MT) depolymerization activity is not needed for sperm-induced spindle assembly. However, high activity is necessary for proper physiological MT dynamics as assayed by Ran-induced aster assembly. We propose that MCAK activity is spatially controlled by an interplay between the N- and C-terminal domains during spindle assembly.
INTRODUCTION
Bipolar spindle assembly is essential for the faithful segregation of chromosomes into two daughter nuclei. Defects in this process are often associated with aneuploidy, which can lead to cancer or birth defects. The bipolar spindle is composed of a dynamic array of microtubules (MTs) and their associated proteins, including various MT-associated proteins and motor proteins, which are essential for proper spindle assembly (Gadde and Heald, 2004; Mogilner et al., 2006). Proper MT dynamics are highly regulated during the transition between interphase and mitosis, and many regulators of MT dynamics are critical for proper spindle assembly both in cells and in Xenopus laevis egg extracts (Kline-Smith and Walczak, 2004). In egg extracts, spindle assembly is dominated by chromatin-mediated MT nucleation and organization (Sawin and Mitchison, 1991). During chromatin-mediated spindle assembly, randomly nucleated MTs around chromatin become organized into an astral array that leads to the formation of a bipolar spindle due to the generation of anti-parallel overlapping MTs that are extended and focused into poles by molecular motors and MT-associated proteins (Walczak et al., 1997, 1998; Sharp et al., 2000; Gaetz and Kapoor, 2004; Mitchison et al., 2004, 2005; Goshima et al., 2005; Koffa et al., 2006). In addition to relying on molecular motors to organize and slide MTs, bipolarity also depends on the regulation of proper physiological MT dynamics by depolymerizing proteins that include depolymerizing kinesins and Op18/stathmin (Andersen et al., 1997; Ganem and Compton, 2004; Rogers et al., 2004; Sharp et al., 2005). One major signal chromatin uses to promote spindle assembly is the activation of the small GTPase Ran via its chromatin-linked exchange factor, RCC1 (Bilbao-Cortes et al., 2002; Moore et al., 2002; Li et al., 2003). Addition of Ran-GTP, the activated form of Ran, to egg extracts results in bipolar spindle formation from the proper regulation of MT nucleation, MT dynamics, and molecular motor activities (Wilde and Zheng, 1999; Carazo-Salas et al., 2001; Wilde et al., 2001). Although Ran-induced spindle activities are dependent on chromatin, it is unclear how many chromatin-induced activities are independent of Ran. One such activity is the stabilization of MTs around chromatin by the chromosome passenger complex through the regulation of the depolymerizing kinesin MCAK and Op18/stathmin (Sampath et al., 2004; Gadea and Ruderman, 2006).
Kinesins are molecular motors that convert the chemical energy of ATP into mechanical work specifically on MTs. The majority of kinesins are homodimeric proteins that walk along MTs carrying various cargos such as vesicles, proteins, and chromosomes (Hirokawa and Takemura, 2004; Wozniak et al., 2004; Miki et al., 2005). Many kinesins are important for mitosis and spindle assembly such that upon depletion by RNAi in cells or by immunodepletion in extracts, defects occur that either inhibit mitosis or impair spindle assembly (Walczak et al., 1998; Goshima and Vale, 2005; Zhu et al., 2005). Kinesins are categorized into 14 families (Lawrence et al., 2004; Miki et al., 2005) and are composed of four different domains: a conserved catalytic MT-stimulated ATPase domain; a class-specific neck domain that is essential for motility (Vale and Goldstein, 1990; Case et al., 1997; Endow and Waligora, 1998); a “stalk” domain that often functions to dimerize the protein; and the “tail” domain that binds to cargo and/or regulates activity (Hirokawa and Takemura, 2004). The location of these domains within the polypeptide is dependent on the specific family of kinesins, but the neck domain is always adjacent to the catalytic domain (Vale et al., 2000). Although most kinesins walk along MTs, members of the Kinesin-13 family catalytically induce MT depolymerization at both ends of a MT and are necessary to regulate MT dynamics (Desai et al., 1999a, 1999b; Hunter et al., 2003; Helenius et al., 2006).
Structurally, the Kinesin-13s have a central catalytic domain with the neck domain amino-terminal to the catalytic core. The neck and catalytic core are sufficient for MT depolymerization activity in vitro and in vivo (Maney et al., 2001; Newton et al., 2004; Ogawa et al., 2004; Hertzer et al., 2006), with the positively charged neck domain being particularly important for depolymerization activity (Ovechkina et al., 2002). The N-terminal domain of MCAK is necessary (Maney et al., 1998; Wordeman et al., 1999) and perhaps sufficient (Walczak et al., 2002) to target MCAK to kinetochores. The C-terminal domain is responsible for dimerization and plays a role in regulating the ATPase activity of MCAK (Maney et al., 2001; Moore and Wordeman, 2004; Hertzer et al., 2006). Biochemically, MCAK specifically binds to the ends of MTs where it either stabilizes an already bent conformation or induces a curvature at the MT end (Desai et al., 1999b; Moore and Wordeman, 2004; Ogawa et al., 2004; Helenius et al., 2006; Hertzer et al., 2006). In dynamic MTs, this curved protofilament within the MT is unstable and is thought to cause the MT polymer to depolymerize (Desai and Mitchison, 1997; Nogales, 2000). For in vitro–stabilized MTs, it is unclear whether MCAK depolymerizes MTs by removing individual tubulin dimers or tubulin oligomers (Desai et al., 1999b; Hunter et al., 2003; Moores et al., 2006; Tan et al., 2006), but it is thought that the ultimate end product from MCAK-induced MT depolymerization is tubulin heterodimer (Desai et al., 1999b). MCAK may processively depolymerize MTs by a mechanism that involves one-dimensional (1-D) diffusion of MCAK along the MT lattice or by the accumulation of tubulin-MCAK rings at the MT end (Hunter et al., 2003; Helenius et al., 2006; Tan et al., 2006), but it remains unclear how these mechanisms are utilized during depolymerization of dynamic MTs and how the different domains of MCAK contribute to this depolymerization activity.
MCAK predominantly localizes to three regions during mitosis: the spindle poles, the cytoplasm, and the centomeres/kinetochores (Wordeman and Mitchison, 1995; Walczak et al., 1996; Maney et al., 1998). It is unclear what function MCAK plays at the spindle poles, but more is known about the functions of MCAK in the cytoplasm and at the kinetochore (Kinoshita et al., 2006). MCAK was originally identified in cells as a kinesin that associates with mitotic centromeres (Wordeman and Mitchison, 1995) and is now known to regulate cytoplasmic MT dynamics in interphase and mitosis (Maney et al., 2001; Kline-Smith and Walczak, 2002). In extracts, MCAK is the major catastrophe regulator, and depletion perturbs MT dynamics, resulting in large asters with increased amounts of MT polymer that are unable to assemble into a bipolar spindle (Walczak et al., 1996; Tournebize et al., 2000). Antibody inhibition or RNAi of MCAK in cells causes excess polymer formation and/or monopolar spindles that interfere with normal bipolar spindle assembly (Walczak et al., 1996; Kline-Smith and Walczak, 2002; Cassimeris and Morabito, 2004; Ganem and Compton, 2004; Stout et al., 2006). The working model for MCAK inhibition is that the excess MT polymer formation and monopolar spindles are due to suppressed cytoplasmic MT dynamics, which prevent the appropriate capture of MTs by kinetochores and/or the appropriate sorting by molecular motors around chromatin.
MCAK localizes to the inner kinetochore via its N-terminal kinetochore targeting domain and regulates proper kinetochore-MT attachments by depolymerizing MTs that attach incorrectly (Maney et al., 1998; Wordeman et al., 1999; Walczak et al., 2002; Kline-Smith et al., 2004). Overexpression or injection of a dominant negative construct in cells or extracts that replaces endogenous MCAK at the centromere results in normal bipolar spindle assembly, but chromosome alignment and segregation are disrupted (Maney et al., 1998; Walczak et al., 2002; Kline-Smith et al., 2004). Other members of the Kinesin-13 family are also implicated in driving chromosome segregation in anaphase by depolymerizing MTs from both ends (Ganem and Compton, 2004; Rogers et al., 2004; Ganem et al., 2005; Sharp et al., 2005). MCAK and the related Kinesin-13, Klp10A, also associate with the +TIP-tracking proteins APC and EB1, and the interactions may be critical for their ability to +TIP-track on MTs (Banks and Heald, 2004; Mennella et al., 2005; Moore et al., 2005). It is still unknown how or if +TIP tracking is required for spindle assembly or for regulation of MT dynamics in extracts or in cells. MCAK kinetochore targeting, depolymerization activity, tip-tracking, corrections of mal-attached kinetochore MTs, and spindle assembly are all regulated by phosphorylation (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Holmfeldt et al., 2005; Moore et al., 2005; Knowlton et al., 2006), but the detailed molecular mechanisms of these complex regulated processes remain unknown. Despite the progress that has been made in identifying the mechanism of MCAK-induced MT depolymerization and understanding its physiological functions, many questions remain unanswered. Here we present the first comprehensive domain analysis of MCAK that correlates the roles of different domains both biochemically with purified proteins and physiologically in egg extracts. We show that there is an interplay between the N- and C-terminal domains that is necessary for physiological MT dynamics in extracts and that the N-terminal domain is necessary for efficient spindle assembly even though it is not required for in vitro MT depolymerization activity, MT and tubulin binding, or MT end binding. Additionally, we identify MCAK as an important regulator of spindle bipolarity, which is controlled by the N- and C-terminal domains of MCAK.
MATERIALS AND METHODS
Protein Expression and Purification
Green fluorescent protein (GFP) tagged expression constructs used in this work were created by subcloning the MCAK coding sequence from pEGFPC1 deletion constructs (Hertzer et al., 2006) into pFastBac1-GFP using SacI and KpnI restriction sites. Point mutations within the catalytic domain were created using the QuikChange Site-Directed Mutagenesis System (Stratagene, La Jolla, CA). All clones were verified by sequencing. The GFP-tagged proteins were then expressed using the Bac-to-Bac baculovirus expression system (Invitrogen, Carlsbad, CA) in HighFive insect cells. GFP-MCAK (GMCAK) and GMCAK mutants were purified to near homogeneity by conventional chromatography using ion exchange and gel filtration as previously described (Desai et al., 1999b). Modifications of GMCAK are designated as GM for GFP-MCAK followed by the amino acid numbers in parentheses. Protein concentrations are expressed in terms of monomer concentration and were quantified from Colloidal Coomassie Blue G-250–stained SDS polyacrylamide gels and densitometry using BSA as a standard. Densitometry was done using ImageJ software (NIH). At least two individual preparations of proteins were used in the described experiments. His-RanL43E was expressed, purified, and loaded with GTP as previously described (Ems-McClung et al., 2004). 6His-GFP was expressed in BL21(DE3) bacterial cells at 16°C for 24 h and was purified using NiNTA agarose as previously described (Ems-McClung et al., 2004).
Microtubule Preparation
Microtubules were polymerized using either guanylyl-(a,b)-methylene-diphosphonate (GMPCPP; Jena Scientific, Jena, Germany) alone or GMPCPP and paclitaxel to create stable tubulin polymer (Desai and Walczak, 2001; Hertzer et al., 2006). Briefly, recycled tubulin was clarified at 45,000 rpm for 5 min at 2°C in a TLA100 rotor (Beckman, Fullerton, CA). The tubulin concentration was determined by absorbance at 280 nm using 115,000 M−1 cm−1 as the extinction coefficient. Clarified tubulin was diluted to 10 μM for depolymerization and end binding assays or 40 μM for tubulin binding assays in BRB80 (80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT) and polymerized with 0.5 mM GMPCPP at 37°C for 30 min. For GMPCPP/paclitaxel stabilized MTs, 10 μM paclitaxel was added 20 min into the polymerization of the MTs, and the MTs were then allowed to polymerize an additional 10 min. All MT substrates were sedimented at 45,000 rpm at 35°C in a TLA100 (5 min) or TLA100.3 (11 min) rotor. For GMPCPP stabilized MTs, the MT pellet was either resuspend in BRB80 or BRB49 (49 mM PIPES, pH 6.8, 0.61 mM MgCl2, 0.61 mM EGTA, 1 mM DTT). For GMPCPP/paclitaxel stabilized MTs, the MT pellet was resuspended in BRB80 containing 10 μM paclitaxel.
MT EC50 Assays
MT EC50 assays were done essentially as previously described (Hertzer et al., 2006), except that the reactions were performed at a total ionic strength of 100 mM and included 0.2 μg/μl casein. GMPCPP-stabilized MTs were used at 1 μM, and enzyme concentrations ranged from 0 to 1 μM. MTs and enzyme were incubated at room temperature for 30 min and then sedimented at 45,000 rpm at 22°C in a TLA100 rotor for 5 min. The supernatant was removed and mixed with an equal volume of 2× sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 4% β-mercaptoethanol). The pellet was resuspended in one reaction volume of 2× sample buffer, added to an equal volume of BRB80, and then the samples were boiled. An equal volume of the supernatant and pellet fractions were electrophoresed on 10% SDS-PAGE gels according to the method of Laemmli (1970) and stained with Colloidal Coomassie Blue. Stained gels were scanned, and the percentage of soluble tubulin heterodimer was determined using ImageJ software. The percentage of soluble tubulin was determined for each reaction condition in Excel (Microsoft, Redmond, WA) and plotted against the log of the enzyme concentration using Prism 4 software (GraphPad, San Diego, CA). For each enzyme, the percentage of soluble tubulin was normalized to the lowest point for the no-enzyme condition and to 100% depolymerization for the maximum response. The best fit curve for the four parameter logistic equation or dose response curve was determined with Prism using the following equation:
where Response is the percentage of tubulin in the supernatant at a particular enzyme concentration; Amin is the percentage of tubulin in the supernatant in the absence of enzyme; Amax is the highest percentage of tubulin in the supernatant attained by the enzyme; logEC50 is the logarithm of the enzyme concentration that causes a response halfway between Amin and Amax; X is the logarithm of the enzyme concentration at a particular concentration; and H is the Hill slope of the curve. The best fit EC50 curve was plotted against the enzyme concentration on a log scale with the mean response for each enzyme concentration and its corresponding standard error of the mean (SEM). Statistical significance between the EC50 and maximum percent soluble tubulin concentrations was determined with an F-test. The F-test compared a model that constrained both curves to have identical EC50 or Amax concentrations to a model that did not have constrained concentrations. Significant differences were considered if p < 0.05. Four or more experiments were performed per enzyme.
Immunofluorescence MT End Binding
MT end binding assays were performed as described previously (Desai and Walczak, 2001) except that reactions were performed at 100 mM total ionic strength, with 1 μM GMPCPP-stabilized MTs, 0.2 μg/μl casein, 2 mM MgATP, and 22.5 nM enzyme. 6His-GFP was used as a control for background staining. Reactions were allowed to proceed for 3 min at room temperature, fixed in 1% gluteraldehyde for 2 min, and then sedimented onto poly-l-lysine–coated coverslips through a 10% glycerol, BRB80 cushion at 12,000 rpm for 45 min in a Beckman JS13.1 rotor at 20°C. The coverslips were then fixed in −20°C methanol for 5 min, rehydrated in TBSTx (20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100), washed, and then processed for immunofluorescence as follows. Coverslips were blocked in AbDil-Tx (2% bovine serum albumin, 20 mM Tris, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.1% sodium azide) for 30 min. To detect the GFP-tagged proteins, the coverslips were probed with 2 μg/ml rabbit anti-GFP antibody (Hertzer et al., 2006) in AbDil-Tx, washed with TBSTx, and then probed with 1:50 dilution donkey anti-rabbit Alexa488 secondary antibody (Invitrogen). The MTs were visualized by probing the coverslips with 1:250 dilution of mouse DM1α anti-tubulin antibody (Sigma-Aldrich, St. Louis, MO), washing with TBSTx, and probing with 1:50 dilution goat anti-mouse Alexa594 secondary antibody (Invitrogen). The coverslips were then mounted onto slides with anti-fade (0.5% p-phenylenediamine, 20 mM Tris-Cl, pH 8.8, 90% glycerol) and sealed with clear nail polish.
Analysis of the extent of MT binding by the different mutants was determined from five pictures taken blindly from each coverslip in four independent experiments. More than 300 microtubules were scored per condition per experiment. Images were taken with a Nikon 90i microscope equipped with a 100×/1.3 NA Plan Fluor oil objective (Nikon, Melville, NY). Digital images were collected at equivalent exposures with a Photometrics Coolsnap HQ cooled CCD camera (Roper Scientific, Trenton, NJ). The camera and shutter were controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). Per experiment, images were scaled equally to optimize GMCAK binding, and then the remaining images were scaled identically. Each image was quantified for the percentage of MTs that had either end binding events, lattice binding events, or no binding. The mean percentage of binding was plotted with the SEM, and the significant difference from GMCAK was determined using a Student's t test in Excel. All images were processed equivalently in Photoshop (Adobe, San Jose, CA) and assembled in Illustrator (Adobe).
Tubulin Competition Binding Assays
Tubulin competition binding assays were performed similarly to Hertzer et al. (2006). In this experiment, 200 nM of the GFP-tagged MCAK proteins were incubated in BRB27, 88 mM KCl (total ionic strength of 140 mM), with 1.2 μM doubly stabilized (GMPCPP and paclitaxel) MTs, in the presence or absence of 4.7 μM soluble GDP tubulin heterodimer for 15 min at room temperature. Reactions were sedimented at 45,000 rpm in a Beckman TLA100 rotor for 5 min at 22°C. After centrifugation, the supernatant and pellet of each reaction were adjusted to equal volumes and electrophoresed on 10% SDS polyacrylamide gels. Colloidal Coomassie–stained gels from three independent experiments representing multiple protein preparations were scanned, and then the percentage of MCAK partitioning to either the supernatant or the pellet was quantified using ImageJ software. The mean percentage of enzyme bound to MTs or tubulin are graphed with the SEM. Statistically significant differences from GMCAK were determined using a Student's t test in Excel.
Cytostatic Factor Extract Preparation and MCAK Immunodepletion
Cytostatic factor (CSF)-arrested extracts were prepared as previously described (Desai et al., 1999a) from X. laevis laid eggs. Extracts were supplemented with rhodamine-labeled tubulin for visualization of spindle structures. Extracts were immunodepleted of endogenous MCAK by incubation with 4 μg anti-MCAK-NT or nonimmune IgG per 10 μl protein G Dynabeads (Invitrogen) per 40 μl extract for 90 min on ice. Beads were isolated on a magnet for 30 min, and the extract was removed. Depleted extracts were then cycled into interphase with the addition of CaCl2 and X. laevis sperm for 90 min and recombined on ice for 20 min. An equal volume of CSF-depleted extract was then added to the interphase extract to send it into mitosis. For each add-back reaction, 20 μl of extract was used with add-back of a final concentration of 100 nM 6His-GFP or GFP-tagged enzyme. The spindle assembly reactions were allowed to proceed for 90 min, fixed with 2% formaldehyde, 30% glycerol, BRB80, 0.5% TX-100, 2.5 mM MgCl2 for 10 min, and then sedimented onto coverslips through a 40% glycerol, BRB80 cushion at 6000 rpm for 20 min at 18°C in a Beckman JS7.5 rotor. Coverslips were postfixed in −20°C methanol for 5 min, rehydrated in TBSTx, stained with 2 μg/ml Hoechst No. 33258 (Sigma-Aldrich) for 5 min, mounted onto slides with anti-fade, and sealed with clear nail polish.
Sperm containing structures were counted on each coverslip as either being part of a spindle, large aster, or monopolar structure. A sperm containing structure was classified as a spindle if it contained two (bipolar spindle) or more (multipolar spindle) focused poles with the majority of the chromatin arranged between the poles. Asters were classified as large when the diameter of the aster exceeded the width of a normal spindle. These large asters typically contained qualitatively more tubulin polymer than small asters and were typical of MCAK depletion (Walczak et al., 1996). Because bipolar spindle formation progresses through intermediate structures, which include half-spindles and small asters, we grouped these in intermediate structures into one category and called it monopolar structures. Sperm containing structures were classified as a small aster when the diameter of the aster was equal to or less than the diameter of a normal bipolar spindle. Chromatin within the small asters were either located close to the aster pole or arranged around the periphery of the astral microtubules. Four separate experiments were analyzed from three different extract preparations with ∼100 structures counted per reaction. The mean percentages of each structure category are graphed with the SEM. Digital images were acquired with a 60 × 1.4 NA Plan Apo oil objective mounted on a Nikon 90i microscope. Images were processed in Photoshop and assembled in Illustrator (Adobe).
Kinetochore Targeting
Kinetochore targeting experiments were performed similarly to those previously described (Walczak et al., 2002). Briefly, 10 μg/ml nocodazole was added to MCAK-immunodepleted extracts that were then mixed with X. laevis sperm and 100 or 500 nM of the various GFP-tagged MCAK proteins on ice. Reactions were started by transferring the reaction to room temperature and were incubated for 45 min. Reactions were fixed and processed for immunofluorescence as described above except that they were immunostained with 1:430 dilution of rabbit anti-xCENP-A (a kind gift from Aaron Straight) and 1:20,000 donkey anti-rabbit Alexa594 (Invitrogen).
Z-stepped images were taken with a Nikon 90i microscope equipped with a 100×/1.3 NA Plan Fluor oil objective. Three-dimensional maximal projection images were scaled to best represent GMCAK localization, and then the remaining images were scaled identically. All images were processed in Photoshop and assembled in Illustrator (Adobe).
Ran Aster Analysis
Fresh CSF extract supplemented with X-Rhodamine-tubulin was immunodepleted of MCAK or nonimmune IgG as described above. His-RanL43E was added to the extract to a final concentration of 25 μM and GFP, and GFP-tagged MCAK proteins were added back to a final concentration of 100 or 200 nM. Protein addition did not exceed one-tenth the volume of the extract. Reactions were incubated at room temperature for 45 min, fixed, and processed as described above for sperm-assembled spindles. Approximately 300 MT structures were counted per condition per experiment from three separate experiments from two independent extracts and were classified as spindles, small asters, large asters, or MT aggregates. Similar results were seen in two additional independent extracts.
An aster was classified as a small aster if it did not exceed the diameter of the majority of asters observed in the control IgG depletion reaction, which were typical of Ran-induced asters (Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999; Carazo-Salas et al., 2001; Gruss et al., 2001). When an aster exceeded the size of a typical Ran-induced aster (small aster), which was typical of MCAK-depleted extract, the aster was categorized as a large aster. MT aggregates formed in MCAK-depleted extracts and were typified by a size that grossly exceeded the size of large asters in the MCAK-depleted extract. These MT aggregates did not have a focused pole, and the MTs were often arranged parallel to one another. In Figure 4B, spindles and small asters were grouped into the category called “Small Asters and Spindles” to represent structures that were rescued in comparison to large asters, which were nonrescued structures typical of the MCAK-depleted phenotype. In Figure 4C, small asters and large asters were grouped into an “Aster” category to exemplify the increased spindle formation that was observed with GM(2-592). The data are graphed as either the mean percentage of small asters and spindles or large asters from the total number of asters and spindles for each construct (Figure 4B) or as the mean percentage of asters, spindles, or MT aggregates from the total number of structures for each construct (Figure 4C). The mean percentages were determined from three independent experiments and graphed with the SEM using Excel (Microsoft). Statistical significance was determined with a Student's t test performed in Excel and was considered significant if p < 0.05. Images were taken with a 40× 1.0 NA Plan Fluor oil objective on a Nikon 90i microscope and processed equivalently in Photoshop and assembled in Illustrator (Adobe).
Figure 4.
The N-terminal domain regulates physiological MT dynamics and the C-terminal domain regulates spindle bipolarity. Asters and spindles were induced by the addition of 25 μM RanL43E to control or MCAK-depleted extracts to which 200 nM GFP, GMCAK, GM(187-731), GM(2-592), GM(187-582), GM(R522A), GM(E529A), and GM(296-308Δ) were added. Reactions were incubated for 45 min at room temperature, fixed, sedimented onto coverslips, and mounted with anti-fade. (A) Representative images are shown and were scaled identically. Scale bar, 10 μm. (B) Quantification of the structure type is graphed as the mean percentage of the total structures plus the SEM from three independent experiments. (C) Quantification of the aster types is graphed as the mean percentage from the aster and spindle total plus the SEM from three independent experiments. Small asters and spindles, which represent rescued structures, were combined and are compared with large asters. An asterisk indicates significant difference relative to GMCAK add-back.
RESULTS
The C-terminal Domain and Neck Are Required for Robust MT Depolymerization Activity
To study MCAK domain function biochemically with purified proteins and physiologically in egg extracts, five deletion constructs with N-terminally tagged GFP were expressed in insect cells and purified to near homogeneity using conventional chromatography (Desai et al., 1999b; Supplementary Figure 1). The modifications removed the N-terminal domain, the neck, and/or the C-terminal domain (Figure 1A). All constructs contained the catalytic domain. For simplicity, GFP-MCAK is referred to as GMCAK, and the GMCAK modified proteins are referenced as GM followed by the amino acids included in the protein. From hydrodynamic analysis, GMCAK has a native molecular weight of 262 kDa, which is slightly larger than its predicted dimeric molecular weight of 220 kDa, suggesting an extended conformation (Hertzer et al., 2006). In contrast, GM(187-592) has a native molecular weight of 74 kDa, which is almost identical to its predicted monomeric molecular weight of 75 kDa (Hertzer et al., 2006). Consistent with the C-terminal domain being responsible for dimerization and being extended, GM(187-731) and GM(263-731) also migrated on gel-filtration with molecular weights that were larger than that predicted for dimers (our unpublished data). GM(263-592), which lacked both the N- and C-terminal domains, eluted as a monomeric protein by gel filtration chromatography (our unpublished data). GM(2-592) migrated on gel filtration and sucrose gradients with a molecular weight that was larger than a monomer but smaller than a dimer. The N-terminal domain alone was not sufficient for dimerization in either two-hybrid or hydrodynamic assays (our unpublished data); therefore we conclude that GM(2-592) is most likely a monomeric protein, although we cannot rule out the possibility that it forms a weak dimer (Maney et al., 2001). Together, these data suggest that the N- and C-terminal domains may exist in an extended state, whereas the neck and catalytic domain in combination or the catalytic domain alone may be more globular.
Figure 1.
The C-terminal domain and neck of MCAK are necessary for efficient MT depolymerization activity. (A) Schematic diagram of expressed and purified MCAK proteins. The GFP tag is located on the N-terminus but was omitted from the diagrams for simplicity. (B and C) GMPCPP stabilized MTs (1 μM) were incubated with increasing concentrations of enzyme (0–1 μM) for 30 min in saturating MgATP at room temperature. MTs were sedimented, and the soluble tubulin heterodimer removed. Equal volumes of soluble tubulin and MT pellets were electrophoresed on 10% SDS polyacrylamide gels, and the gels were stained with Colloidal Coomassie Blue. The percentage of soluble tubulin was quantified from the stained gels and plotted against the log of the enzyme concentration. The four-parameter logistic equation was used to determine the EC50 concentrations of each enzyme from four or more individual experiments. The curves represent the best fit curve to the data with each point represented by the mean ± SEM.
MCAK is a potent MT depolymerase (Desai et al., 1999b; Hunter et al., 2003; Hertzer et al., 2006), and to study subtle differences in MT depolymerization activity, we used the dose response assay described previously (Hertzer et al., 2006), but changed the salt conditions to physiological ionic strength (100 mM) because we wanted to compare these in vitro activities to physiological activities in Xenopus extracts that are described below. The dose response or EC50 assay provided us an overview of the catalytic efficiency of each protein as a function of the enzyme concentration needed to depolymerize a given concentration of MTs at a fixed time point. In addition, to elucidate the mechanisms for any differences in MT depolymerization activities between the mutants, MT end binding and tubulin competition assays were used to give snap shots of two steps in the MT depolymerization cycle (Moore and Wordeman, 2004; Moore et al., 2005; Hertzer et al., 2006). The visual MT end binding assay illustrated the specificity by which the various mutants bound MTs and MT ends. Thus, we expected the proficient mutant MT depolymerases to bind MTs and MT ends well and the poor MT depolymerases to bind poorly to MTs and MT ends. The gel-based tubulin competition assay demonstrated how well the mutant proteins bound MTs in the presence of excess tubulin as an indication of how well the enzymes bound tubulin heterodimer. We predict that the robust MT depolymerases will bind MTs preferentially and only minimally associate with tubulin heterodimer as seen previously with wild-type MCAK (Moore et al., 2005; Hertzer et al., 2006). Together, these analyses provided a means to quantify the biochemical competency of each enzyme as described below.
Under physiological buffer conditions, 1.45 nM GMCAK depolymerized 50% of 1 μM MTs in 30 min (Figure 1B; Supplementary Table 1). Similar to previous work, MT end binding assays illustrated that full-length GMCAK bound MTs well and bound MT ends with high specificity (Supplementary Figure 2; Moore and Wordeman, 2004; Hertzer et al., 2006). Similarly, tubulin competition assays demonstrated that GMCAK only marginally released from MTs in the presence of excess tubulin heterodimer (Supplementary Figure 3), which was consistent with previous results (Moore et al., 2005; Hertzer et al., 2006). Deletion of the N-terminal domain in GM(187-731) resulted in an EC50 of 1.44 nM, which was not significantly different from GMCAK. This indicated that GM(187-731) is also a potent MT depolymerase. MT end binding and tubulin competition assays confirmed that GM(187-731) is a potent MT depolymerase because it bound MTs and released from MTs in the presence of excess tubulin in a manner essentially indistinguishable from GMCAK (Supplementary Figures 2 and 3). In sharp contrast, further truncation of the N-terminus, which deleted the neck in GM(263-731), resulted in a ∼150-fold reduction in the EC50 for MT depolymerization (213 nM; p < 0.0001; Figure 1B; Supplementary Table 1). MT end binding experiments indicated that GM(263-731) did not depolymerize MTs well because it bound MTs poorly and with reduced MT end specificity (Supplementary Figure 2). Interestingly, in the tubulin competition assays only twofold more GM(263-731) released from MTs in the presence of excess tubulin over that which did not bind MTs, suggesting that GM(263-731) has a low tubulin affinity similar to GMCAK (Supplementary Figure 3). These results are in agreement with previous results showing that the neck domain is very important for robust MT depolymerization activity (Maney et al., 2001; Ovechkina et al., 2002; Hertzer et al., 2006).
Removal of the C-terminal domain in GM(2-592) resulted in a modest decrease (∼2-fold) in the EC50 for MT depolymerization (2.63 nM) that was statistically different in comparison to GMCAK (p < 0.001; Figure 1C; Supplementary Table 1). The MT end binding results suggest that in comparison to GMCAK, GM(2-592) had a very small but statistically significant reduction in MT end binding (p < 0.01; Supplementary Figure 2). In the tubulin competition assays, there was a slightly reduced ability to bind MTs (p < 0.01), but a nearly fourfold increase in the amount of GM(2-592) that released from MTs in the presence of excess tubulin (p < 0.0001; Supplementary Figure 3). Given the magnitude of the differences between GM(2-592) and GMCAK tubulin binding, we favor the idea that GM(2-592) is likely defective in its ability to release tubulin dimer at the end of its catalytic cycle, similar to what has been observed previously for another monomeric MCAK construct (Hertzer et al., 2006). However, we cannot exclude the possibility that this result is due to a decreased ability of GM(2-592) to diffuse along the MT lattice. Together our in vitro results so far support the ideas that the C-terminal domain is necessary and that the N-terminal domain is dispensable for efficient MT depolymerization (summarized in Figure 5).
Figure 5.
Summary of the in vitro and extract activities of GMCAK and its various mutants. The data from each experiment are summarized with plus signs to delineate the extent of activity. Four consecutive plus signs indicate the highest activity (+ + + +) and a minus sign indicates no activity (−). ND indicates activity was not determined for that assay. Each GMCAK mutant is represented schematically based on the domain(s) deleted or mutated. The domains are as depicted in Figure 1. The catalytic domain mutants are full-length GMCAK proteins with either a point mutation (R522A or E529A) or a 13-amino acid deletion (296-308Δ) within the catalytic domain.
Similar to our previous report (Hertzer et al., 2006), GM(187-592) had a fourfold reduction in the EC50 for MT depolymerization (5.46 nM) in relation to GMCAK (p < 0.0001; Figure 1C; Supplementary Table 1). Overall, GM(187-592) bound MT ends with slightly lower specificity (p < 0.001), bound MTs with a slightly lower affinity (p < 0.05), and bound tubulin dimer fivefold more robustly (p < 0.01) than GMCAK (Supplementary Figures 2 and 3). This indicates that perhaps GM(187-592) is less adept at targeting specifically to MT ends or diffusing along the MT lattice as well as releasing from tubulin dimer in the final step of MT depolymerization, which is consistent with our previous results (Hertzer et al., 2006). Because removal of the N-terminal domain severely affected the depolymerization activity of GM(187-592) relative to GM(2-592), these results suggest that the N-terminal domain of MCAK may play a small role in regulating the in vitro MT depolymerization activity, especially when the C-terminal domain is absent. From our previous work with minimal domain MCAK, it is likely that GM(2-592) and GM(187-592) both get stuck at tubulin release, whereas GM(187-592) also gets stuck on the MT lattice, thereby further reducing its MT depolymerization potency (Hertzer et al., 2006). These results indicate that GM(2-592) and GM(187-592) likely have similar biochemical mechanisms, which in part is due to them being monomeric.
Interestingly, removal of the neck domain from GM(187-592) to create GM(263-592) resulted in a protein with a 13-fold higher MT depolymerization activity (16 nM) than GM(263-731; p < 0.0001) and an 11-fold lower MT depolymerization activity than GMCAK (p < 0.0001; Figure 1C; Supplementary Table 1). MT end binding and tubulin competition assays indicate that GM(263-592) was able to bind MTs better than GM(263-731) but bound to MT ends less well than GMCAK, consistent with their differences in MT depolymerization activities (Supplementary Figures 2 and 3). In addition, GM(263-592) released from MTs in the presence of excess tubulin more robustly than GMCAK and GM(263-731; Supplementary Figure 3). These results support the idea that the neck is minimally required for 1-D diffusion (Ovechkina et al., 2002; Helenius et al., 2006), because all proteins except the neckless mutants targeted to MT ends effectively.
Overall, our data support previous work showing that the neck domain is critical and that dimerization is important to MCAK-induced in vitro MT depolymerization activity (Maney et al., 2001; Ovechkina et al., 2002; Hertzer et al., 2006). However, our results extend previous analyses to indicate that the C-terminal domain positively influences MCAK MT depolymerization activity when the neck domain is present, but becomes inhibitory when the neck domain is absent, suggesting a mechanistic interplay between the C-terminal domain and neck that is necessary for efficient MT depolymerization activity.
The N-Terminal Domain Is Required for Efficient Spindle Assembly in MCAK-depleted Extracts
Although our in vitro work gave us new insights into the biochemical mechanism of MCAK, we were interested in understanding how it related to the physiological function of MCAK. Thus, Xenopus CSF extracts were depleted of endogenous MCAK, reconstituted with GMCAK or the various domain mutants (Supplementary Figure 4), and assayed for spindle assembly. The extracts were cycled into interphase in the absence of MCAK, and GMCAK add-back was done at the time of the second CSF addition to ensure that the rescue would show only the effects of the mutants on spindle assembly and not any defects in the intervening interphase. MCAK immunodepletion resulted in 80–90% depletion of endogenous MCAK as determined by Western blot (Supplementary Figure 4A). Cycled spindle assembly of control IgG depleted extract with the add-back of recombinant GFP (IgGΔ + GFP) resulted in a large proportion of spindles (56 ± 10%), a small percentage of large asters (20 ± 5%), and a small percentage of intermediary monopolar structures (23 ± 8%) that consisted of small asters and half spindles (Figure 2; Supplementary Table 2). The spindles in the control IgG depleted extract were of normal morphology, with the majority having normal MT polymer levels and aligned chromosomes, indicating normal physiological MT dynamics (our unpublished observations). We considered this control IgG depletion phenotype as an example of efficient spindle assembly. In contrast, MCAK depletion and add-back of recombinant GFP (MΔ + GFP) resulted in the complete failure to undergo efficient spindle assembly due to suppressed MT dynamics. This was exemplified by very few spindles (4.0 ± 2%, p < 0.05) and the formation of mainly large asters (61 ± 9%, p < 0.01), in comparison to control IgG depleted extract (Figure 2; Supplementary Table 2). The few spindles that formed had excess MT polymer and unaligned chromosomes, indicating suppressed physiological MT dynamics (our unpublished results). This MCAK depletion phenotype is similar to previously reported results (Walczak et al., 1996). In comparison to the MCAK depleted extract, add-back of GMCAK (MΔ + GMCAK) completely rescued spindle assembly as the percentage of spindles, large asters, and monopolar structures were statistically identical to control IgG depletion (Figure 2; Supplementary Table 2). The majority of the spindles had normal MT polymer levels and aligned chromosomes, indicating normal physiological MT dynamics.
Figure 2.
The N-terminal domain of MCAK is required for efficient spindle assembly. CSF extracts were depleted of endogenous MCAK (MΔ) or mock-depleted with IgG (IgGΔ), cycled into interphase with CaCl2, and cycled back into mitosis with additional depleted CSF extract. GFP-tagged proteins were added back to 100 nM final concentration with the second addition of CSF extract. Spindles were assembled for 90 min, fixed, and sedimented onto coverslips, and the chromatin was stained with Hoechst and mounted with anti-fade. (A) Images of representative structures from the add-back experiments are shown with MTs in magenta and chromatin in green. Scale bar, 20 μm. (B) Quantification of the spindles, large asters, and monopolar structures observed in the extracts that are represented in A. The data are graphed as the mean percentage plus the SEM from four independent extracts. An asterisk indicates a significant difference relative to GMCAK add-back.
Surprisingly, add-back of GM(187-731) [MΔ + GM(187-731)], which lacked the N-terminal domain and had full in vitro MT depolymerization activity, only partially rescued the MCAK depletion phenotype in comparison to GMCAK. This was exemplified by a twofold decrease in spindle formation (27 ± 8%, p < 0.05; Figure 2; Supplementary Table 2). Of the spindles in the GM(187-731) add-back, many of them often had excess MT polymer and poor chromosome alignment in comparison to control IgG or GMCAK add-back, indicating suppressed MT dynamics (our unpublished results). This result was very surprising based on the MT depolymerization and MT binding abilities of GM(187-731). Further truncation of the N-terminus to remove the neck in GM(263-731) and add-back to MCAK depleted extracts resulted in GM(263-731) being unable to rescue spindle assembly in comparison to GMCAK (Figure 2). Specifically, GM(263-731) add-back resulted in an approximate threefold decrease in spindle formation (20 ± 8%, p < 0.05) and a threefold increase in large asters (54 ± 10%, p < 0.05; Figure 2; Supplementary Table 2). The few spindles that formed in the GM(263-731) add-back were characterized by excess MT polymer and unaligned chromosomes. These results strongly imply that the N-terminal domain is important and that the neck is essential for spindle assembly and normal physiological MT dynamics. Surprisingly, these results indicate that competent in vitro depolymerization activity of GM(187-731) did not predict its efficient spindle assembly in extracts. In addition, these results imply that the N-terminal domain of MCAK is critical for efficient spindle assembly.
Because the N-terminal domain appeared to be important for spindle assembly in extracts, we next tested the importance of the N-terminal domain in spindle assembly by adding back GM(2-592). Interestingly, GM(2-592) rescued spindle assembly to levels statistically identical to the IgG control depletion and the GMCAK add-back reactions, indicating that GM(2-592) was efficient at inducing spindle assembly (Figure 2; Supplementary Table 2). In comparison to GMCAK, GM(2-592) add-back resulted in 57% spindle formation, 20% large asters, and 23% monopolar structures (Supplementary Table 2). Morphologically the majority of the spindles that formed in the GM(2-592) add-back extract had more MT polymer and had more poorly aligned chromosomes than GMCAK spindles, suggesting that perhaps MT dynamics were not completely normal (our unpublished results). Overall, these results imply that the N-terminal domain of MCAK is critical and the C-terminal domain is dispensable for spindle assembly. This is completely opposite our in vitro MT depolymerization results in which the N-terminal domain is dispensable but the C-terminal domain is more important for activity. These results suggest that GM(2-592), which has a twofold reduction in MT depolymerization activity, is able to regulate MT dynamics sufficiently for efficient spindle assembly.
To further test the theory that the N-terminal domain is necessary for efficient spindle assembly, we added back GM(187-592), which is missing both the N-terminal and C-terminal domains, to MCAK depleted extracts. Similar to GM(187-731), add-back of GM(187-592) only partially rescued spindle assembly. In comparison to GMCAK add-back, add-back of GM(187-592) resulted an approximate twofold decrease in spindle formation (30 ± 7%; p < 0.05), which was identical to what we found for GM(187-731) (Figure 2; Supplementary Table 2). This result is interesting because GM(187-731) and GM(187-592) differed in their in vitro MT depolymerization, MT end binding, and tubulin binding assays. These results suggest that the C-terminal domain of MCAK may not play a major role in spindle assembly, despite its importance in regulating MT depolymerization activity in vitro or that a twofold reduction in depolymerization activity is sufficient for efficient spindle assembly. Finally, add-back of GM(263-592) to MCAK-depleted extracts rescued spindle assembly poorly and was essentially identical to GM(263-731) phenotypically and morphologically (Figure 2). In comparison to GMCAK add-back, GM(263-592) add-back resulted in a threefold decrease in spindle formation (17 ± 5%; p < 0.01) and a threefold increase in large asters (59 ± 8; p < 0.01; Figure 2; Supplementary Table 2). Overall, these add-back experiments show that for efficient spindle assembly in egg extracts, the N-terminal domain is required and the C-terminal domain is not essential (summarized in Figure 5).
High MCAK MT Depolymerization Activity Is Not Necessary for Efficient Sperm-induced Spindle Assembly
We were surprised that GM(187-731), which had full in vitro MT depolymerization activity, did not effectively rescue spindle assembly and that GM(2-592) fully rescued spindle assembly despite its decreased in vitro MT depolymerization activity. These results suggest that high MT depolymerization activity is not critical for spindle assembly. Because this finding was unexpected, we next asked if three different catalytic domain mutations in full-length MCAK that interfere with catalytic activity to varying degrees would promote spindle assembly in MCAK-depleted extracts. The first mutation was an arginine-to-alanine modification at residue 522 (R522A) within helix 4 that is analogous to conventional kinesin K256, which reduces MT binding without affecting catalysis or MT gliding (Woehlke et al., 1997). In our in vitro depolymerization assay, GM(R522A) MT depolymerization activity was reduced by about twofold (3.15 nM; p < 0.001) in comparison to GMCAK (1.45 nM), which was similar to the decrease caused by removal of the C-terminal domain in GM(2-592) (Figure 3A; Supplementary Table 1). In comparison to GMCAK add-back, add-back of GM(R522A) to MCAK depleted extracts resulted in similar spindle assembly efficiency (Figure 3B). Specifically, add-back of GM(R522A) to MCAK depleted extracts resulted in only a 1.4-fold lower percentage of spindles (40 ± 4%) and a twofold higher percentage of large asters (36 ± 11%) in comparison to GMCAK add-back (Figure 3B; Supplementary Table 2). Despite rescuing spindle assembly, the spindles in the GM(R522A) add-back displayed increased MT polymer and unaligned chromosomes, suggesting reduced physiological MT dynamics (our unpublished data).
Figure 3.
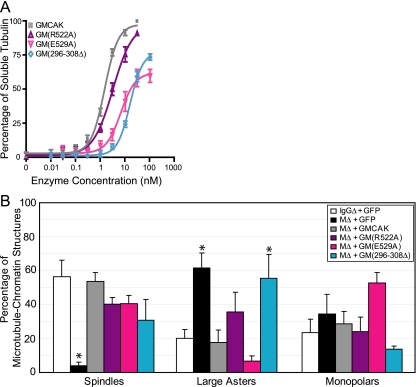
High MT depolymerization activity is not necessary for efficient sperm-induced spindle assembly. (A) The three catalytic domain mutants, GM(R522A), GM(E529A), and GM(296-308Δ), were assayed for MT depolymerization activity, quantified, and graphed as described in Figure 1, B and C. (B) The catalytic domain mutants were added back to cycled MCAK-depleted extracts (MΔ), and assayed for their ability to rescue spindle assembly as described for Figure 2. Quantification of the observed structures is graphed as the mean percentage plus the SEM from three independent experiments. An asterisk denotes a significant difference relative to add-back of GMCAK.
The second mutation we tested was a glutamic acid to alanine mutation at residue 529 (E529A) in helix 4, which is within the conserved KECIRAL motif of Kinesin-13s and is predicted to be critical for MT depolymerization (Niederstasser et al., 2002; Hertzer et al., 2003; Ogawa et al., 2004). GM(E529A) had a 4.5-fold reduced MT depolymerization activity (6.53 nM; p < 0.0001) in comparison to GMCAK and did not completely depolymerize the MTs like GMCAK (Figure 3A; Supplementary Table 1). Add-back of GM(E529A) to MCAK depleted extracts showed that this mutant rescued spindle formation similar to GM(R522A) (Figure 3B). In comparison to GMCAK add-back, add-back of GM(E529A) resulted in a 1.3-fold decrease in spindle formation (41 ± 5%), an approximate threefold decrease in large asters (6.7 ± 3%), and an approximate twofold increase in monopolar structures (53 ± 6%; Figure 3B; Supplementary Table 2). In addition, the bipolar spindles in the GM(E529A) did have more MT polymer and unaligned chromosomes similar to GM(R522A) (our unpublished data). Thus, up to an approximate fivefold reduction in MCAK depolymerization activity did not significantly affect spindle assembly, suggesting robust MT depolymerization activity is not necessary for efficient sperm-induced spindle assembly.
The third mutation we analyzed was the deletion of the KVD finger (amino acids 296-308) that is critical for the ability of MCAK to depolymerize MTs in cells (Ogawa et al., 2004; Shipley et al., 2004). The MT depolymerization activity of GM(296-308Δ) was 11-fold decreased (15.7 nM; p < 0.0001) in comparison to GMCAK (1.45 nM) and was similar to the depolymerization activity of GM(263-592), a very poor depolymerase (Figure 3A; Supplementary Table 1). GM(296-308Δ) add-back partially rescued spindle assembly in MCAK-depleted extracts, suggesting that GM(296-308Δ) was less efficient at promoting spindle assembly (Figure 3B). In comparison to GMCAK, GM(296-308Δ) add-back resulted in spindle formation that was reduced by only 1.7-fold (31 ± 12%), large asters were increased threefold (56 ± 14%; p < 0.05), and monopolar structures were twofold reduced (14 ± 2%; Figure 3B; Supplementary Table 2). Essentially all the spindles that formed in the GM(296-308Δ) reactions had excess MT polymer and unaligned chromosomes, indicating suppressed MT dynamics (our unpublished results). Overall, this data suggests that robust in vitro MT depolymerization activity is not necessary for efficient sperm-induced spindle assembly such that up to a 4.5-fold reduction in depolymerization activity is sufficient (summarized in Figure 5).
Because the N-terminus is necessary for MCAK kinetochore-targeting (Maney et al., 1998; Walczak et al., 2002), one possible explanation for the differences seen in spindle assembly assays relative to the in vitro depolymerization assays could be that the GMCAK mutants targeted differently to kinetochores. To test this idea, we analyzed the localization of each GMCAK mutant to kinetochores using MCAK-depleted extracts in which the microtubule polymerization was inhibited with nocodazole to enrich for kinetochore proteins. GMCAK localized prominently to kinetochores in these targeting assays as seen previously (Supplementary Figure 5B; Walczak et al., 2002). In contrast, GM(187-731) did not target to kinetochores (Supplementary Figure 5C). Unexpectedly, GM(2-592) also did not target to kinetochores when added back to endogenous levels (100 nM; Supplementary Figure 5D). However, when GM(2-592) was added back to five times the endogenous level (500 nM), it targeted to kinetochores nicely (Supplementary Figure 5D′), whereas add-back of GM(187-731) did not target to kinetochores at this level (Supplementary Figure 5C′). These results indicate that GM(2-592) targeted to kinetochores 5- to 10-fold less efficiently than GMCAK and that the C-terminal domain positively regulates the degree of kinetochore targeting, similar to that demonstrated in mammalian cells (Maney et al., 1998). In addition, the three catalytic domain mutant proteins targeted robustly to kinetochores in the targeting assay when added back to 100 nM (Supplementary Figure 5, E–G). Together, these results support the idea that robust kinetochore targeting of MCAK is not necessary for sperm-induced spindle assembly, but is likely important for proper chromosome alignment as reported previously (Walczak et al., 2002; Kline-Smith et al., 2004).
The N-Terminal Domain Regulates Physiological MT Dynamics, and the C-Terminal Domain Regulates Spindle Bipolarity
One complication of our analysis of spindle assembly reactions is that MTs are nucleated from both centrosomes and chromatin in the extracts, and therefore it was impossible to dissect whether these two sources of MTs were differentially affected by MCAK loss. We wanted to identify a physiological assay by which we could evaluate MT dynamics without any contribution from chromatin. Because mostly small asters and a small percentage of bipolar spindles form with the addition of activated Ran (Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999; Carazo-Salas et al., 2001; Gruss et al., 2001), this system may provide a means to directly test the ability of MCAK to regulate physiological MT dynamics independent of its role in chromatin-mediated spindle formation. We depleted MCAK and added back GMCAK to Ran-induced aster assembly reactions (Figure 4). Depletion of MCAK and induction of aster assembly by addition of His-RanL43E resulted in the formation of a very small percentage of spindles and small asters (15 ± 1%) with the majority of the structures being large asters (85 ± 1%) and large MT aggregates (28 ± 3%; Figure 4; Supplementary Table 3). This phenotype indicates that the physiological MT dynamics in the extract are severely suppressed in response to MCAK depletion, and is similar to the sperm-induced phenotype of MCAK depletion. Add-back of physiological levels of GMCAK (100 nM), as we did in the chromatin-induced spindle assembly assay, resulted in mostly small asters and a small percentage of spindles with a substantial percentage of large MT aggregates (our unpublished data). Instead, add-back of GMCAK to twice the physiological level (200 nM) was needed to reduce the large MT aggregates to near control IgG levels (6 ± 2%) while maintaining high percentages of small asters and spindles (86 ± 5%; Figure 4, B and C; Supplementary Table 3). Thus, the Ran system allowed us to study the contributions of the GMCAK mutants to the regulation of physiological MT dynamics.
Add-back of GM(187-731) poorly rescued the MCAK-depleted phenotype of mainly large asters, suggesting that GM(187-731) is unable to properly regulate physiological MT dynamics in the extract (Figure 4). Specifically, add-back of GM(187-731) resulted in mostly large asters (68 ± 4%; p < 0.001) and a small percentage of small asters and spindles (32 ± 4%; p < 0.001) with a high percentage of MT aggregates (23 ± 7%; p < 0.05) in comparison to add-back of GMCAK (Figure 4, B and C; Supplementary Table 3). This result strongly implies that GM(187-731) did not rescue sperm assembled spindles well because it was much less active in the extracts than in assays with purified MTs. Because GM(187-731) is a potent in vitro MT depolymerase, the extract results indicate that GM(187-731) activity was mis-regulated in the extract. Therefore, in addition to kinetochore targeting, the N-terminal domain of MCAK is likely necessary for the regulation of proper physiological MT dynamics.
If the hypothesis is true that the proper regulation of MCAK depolymerization activity via the N-terminal domain is necessary for spindle assembly, then we would expect GM(2-592) to rescue the MCAK-depletion phenotype in the Ran system. Indeed, add-back of GM(2-592) fully rescued the MCAK depletion phenotype (Figure 4), indicating that the N-terminal domain is necessary for regulating physiological MT dynamics. Specifically, add-back of GM(2-592) resulted in 85 ± 3% small asters and spindles and only 15 ± 3% large asters, which were statistically equivalent to add-back of GMCAK (Figure 4B; Supplementary Table 3). Surprisingly, GM(2-592) add-back to MCAK-depleted extracts dramatically increased the number of spindles that formed (52 ± 8%) to levels higher than control IgG depleted extract (10 ± 1%; p < 0.05; Figure 4, A and C; Supplementary Table 3). This increase in spindle assembly over control IgG depletion was also seen with add-back of 100 nM GM(2-592), but was not observed when GM(2-592) was added to non-MCAK depleted extracts (our unpublished results). Increased spindle assembly with GM(2-592) add-back was completely unexpected and suggests that MCAK may have an undefined role in promoting spindle bipolarity that is likely promoted by the N-terminal domain and inhibited by the C-terminal domain. To fully assess the importance of the N-terminal domain in MT depolymerization activity in the absence of the C-terminal domain, GM(187-592) was added back to the extract. Similar to the sperm induced add-back assay, GM(187-592) did not fully rescue the MCAK depletion phenotype in the Ran-induced assembly assay (Figure 4, A and B; Supplementary Table 3). Specifically, add-back of GM(187-592) resulted in a twofold decrease in small aster and spindle formation (41 ± 6%, p < 0.01), a fourfold increase in large aster formation (59 ± 6%, p < 0.01), and a threefold increase in MT aggregates (18 ± 3%, p < 0.05) in comparison to GMCAK add-back (Figure 4; Supplementary Table 3). These results support the idea that the N-terminal domain of MCAK is essential for the proper regulation of physiological MT dynamics in extracts.
Our sperm-induced spindle assembly assays with the catalytic domain mutants suggested that robust MT depolymerization activity is not critical for spindle assembly, but it was unclear how their decreased activity would affect physiological MT dynamics in the absence of chromatin. GM(R522A) add-back rescued the MCAK depletion phenotype similar to GMCAK, indicating that GM(R522A) has wild-type physiological MT depolymerization activity (Figure 4). In particular, GM(R522A) add-back resulted in a high percentage of small asters and spindles (73 ± 10%), a relatively low percentage of large asters (27 ± 10%), and a small percentage of MT aggregates (4 ± 1%), which were statistically identical to GMCAK add-back (Figure 4, B and C; Supplementary Table 3). In contrast, add-back of GM (E529A) poorly rescued the MCAK-depleted Ran aster phenotype and was similar to GM(187-731) and GM(187-592) add-back (Figure 4, A and B). Add-back of GM(E529A) resulted in a 2.5-fold decrease in small asters and spindles (35 ± 9%, p < 0.01) and an approximate fivefold increase in large asters (65 ± 9%, p < 0.01) in comparison to GMCAK add-back, suggesting that GM(E529A) had a reduced ability to regulate physiological MT dynamics (Figure 4B; Supplementary Table 3). Add-back of GM(296-308Δ) failed to rescue the MCAK-depleted phenotype entirely and had an approximate sixfold decrease in small asters and spindles (15 ± 5%; p < 0.001), a sixfold increase in large asters (85 ± 5%; p < 0.001), and a fivefold increase in MT aggregates (30 ± 8%; p < 0.05) in comparison to GMCAK add-back (Figure 4; Supplementary Table 3). The GM(296-308Δ) phenotype was essentially identical to the MCAK-depleted phenotype in which physiological MT dynamics are drastically suppressed (Figure 4; Supplementary Table 3). These results are different from the sperm-induced spindle assembly assay in which GM(E592A) and GM(296-308Δ) were able to rescue the MCAK depletion phenotype to some degree.
Together the results from the sperm-induced and Ran-induced spindle assembly assays indicate that in the presence of chromatin, robust MCAK MT depolymerization activity is not important for spindle assembly, but becomes critical in the absence of chromatin. We believe that it is likely that chromatin exerts an additional level of control over the regulation of MT dynamics, which may aid in spindle assembly when MCAK activity is less robust. Overall, our results indicate that high MCAK MT depolymerization activity in vitro (EC50 1.45–3.15 nM) is sufficient for the proper regulation of physiological MT dynamics, that physiological MCAK MT depolymerization activity is regulated through the N-terminal domain, and that MCAK promotes bipolar spindle assembly through an interplay between the N- and C-terminal domains (summarized in Figure 5).
DISCUSSION
The Interplay of the Neck and C-Terminal Domain Regulates MCAK In Vitro Activity
Using purified proteins, we demonstrate that robust in vitro MT depolymerization by MCAK relies on the neck for effective MT end binding and the C-terminal domain for efficient tubulin release, indicating an important interplay between the neck and C-terminal domain. Our current and previous data are consistent with the model that the C-terminal domain is necessary to keep the MCAK catalytic domain in an inactive state by inhibiting lattice-stimulated ATPase activity until the MT end is contacted where the C-terminal domain “primes” MCAK for MT depolymerization by activating the catalytic domain (Moore and Wordeman, 2004). We would like to extend this model to suggest that the neck and the C-terminal domain are flexible allowing them to bind and release one another directly to promote activity, that the neck is necessary to release the inhibitory hold of the C-terminal domain, that the neck is required for MT end targeting, and that the C-terminal domain is essential for quick tubulin dimer release. Such interplay between the domains of kinesin is not unheard of. The conventional kinesin tail domain folds back onto the stalk and hinge region, inactivating the enzyme, and upon binding substrate the stalk-tail unfolds, activating the enzyme (Coy et al., 1999; Friedman and Vale, 1999; Bathe et al., 2005; Adio et al., 2006). In addition, it is proposed that most kinesin necks provide the power stroke for motility by docking/undocking with the catalytic domain (Vale et al., 2000). With regards to MCAK, the crystal structure shows that the neck is flexible (Ogawa et al., 2004) and therefore may directly contact the catalytic domain and C-terminal domain. Additionally, our gel-filtration data and previous electron microscopy analysis of Kif2 (Noda et al., 1995) suggest that MCAK can exist in an extended or a compact state, supporting the idea that the N- and C-terminal domains are flexible. Our model also explains the unusual finding that GM(263-592) depolymerized MTs better than GM(263-731). The model predicts that GM(263-731) stays in a folded inactivated state, because it is missing the neck. In contrast, GM(263-592) is not as inhibited, due to the missing C-terminal domain. But, because GM(263-592) lacks both the neck and the C-terminal domain, it is a poor depolymerase. Thus, for robust MT depolymerization activity, we propose that MCAK requires an interaction between the neck and C-terminal domain to target effectively to MT ends, to activate MT end-stimulated ATPase activity, and to efficiently release from tubulin heterodimer in the final steps of catalysis.
Robust MT Depolymerization Activity Is Not Necessary for Sperm-induced Spindle Assembly
We were surprised by our finding that full MCAK depolymerization activity was not necessary to get efficient sperm-induced spindle assembly. One explanation could be that the maximum depolymerization activity at physiological ratios of MCAK to tubulin is most important. In our in vitro MT depolymerization assays, the EC50 of GMCAK is ∼1.5 nM using 1 μM MTs (or a ratio of ∼1 MCAK to 1000 tubulin dimers). In contrast, the physiological ratio of MCAK to tubulin is 1–100. Perhaps, if the EC50 value is <10 nM, any differences are insignificant. Alternatively, it may be more appropriate to ask how much of the MTs can be fully depolymerized in the in vitro assays when the concentration of enzyme is 10 nM. Using this analysis, GMCAK and GM(2-592) maximally depolymerize MTs at 10 nM enzyme, which suggests that GM(2-592) activity is sufficient. In contrast, GM(E529A), which rescued spindle assembly effectively, only depolymerizes ∼40% of the MTs at 10 nM enzyme. Is this sufficient? We think not, because the EC50 results of GM(2-592) and the catalytic domain mutants correlate with the extent of rescue in the Ran aster assay, implying that ineffective in vitro depolymerases are ineffective physiological depolymerases. Perhaps kinetochore and/or chromatin localization was a factor for the ability of GM(E529A) to rescue sperm-induced spindle assembly, and not solely depolymerization activity. Indeed, all the GMCAK mutants that did not bind kinetochores at all were poor rescuers of sperm-induced spindle assembly. However, GM(2-592) is a poor kinetochore binder, but was very effective at rescuing spindle assembly. This suggests that spindle assembly does not require robust MCAK kinetochore binding. Perhaps chromatin exerts an additional level of control over the regulation of MT dynamics independent of MCAK, which may aid in spindle assembly when MCAK activity is less robust (Heald et al., 1996; Karsenti and Vernos, 2001).
The Interplay of the N- and C-Terminal Domains Regulates Physiological MT Dynamics and Spindle Bipolarity
Surprisingly, physiological analysis of the MCAK domains demonstrated that the N-terminal domain is critical for regulating physiological MT dynamics, for promoting spindle bipolarity, and for controlling kinetochore targeting. Of the noncatalytic domain mutants, only GM(2-592) rescued spindle assembly in MCAK-depleted extracts and targeted to kinetochores, suggesting that physiologically the N-terminal domain regulates the activation of MT depolymerization by controlling the interplay of the neck with C-terminal domain. Despite missing the C-terminal domain, the N-terminal domain in GM(2-592) is sufficient to adequately regulate the activation of MT depolymerization activity to control physiological MT dynamics. In addition, GM(2-592) targets to kinetochores, albeit less efficiently, which may play into its ability to rescue spindle bipolarity. But interestingly, GM(2-592) dramatically enhanced spindle bipolarity in the Ran aster assay, suggesting that perhaps GM(2-592) fully rescued spindle assembly because GM(2-592) enhanced bipolarity and not simply because of its ability to regulate MT dynamics. This finding raises the question, “Is the presence of GM(2-592) sufficient to activate bipolarity by gain of function?” The answer appears to be “No,” because adding GM(2-592) to nondepleted extracts does not cause enhanced spindle bipolarity. Thus, MCAK might naturally promote spindle bipolarity by an unknown mechanism, and the C-terminal domain then limits the extent of bipolarity.
We are left with the question, “Why is the C-terminal domain needed?” Undoubtedly, the C-terminal domain is required for robust in vitro MT depolymerization activity via the interplay with the neck and efficient enzymatic recycling. We believe that robust MT depolymerization activity is physiologically important, but at spatially distinct places within the spindle. For example, an efficient MT depolymerase is critical at kinetochores to correct mal-attachments (Walczak et al., 2002; Kline-Smith et al., 2004; Knowlton et al., 2006). In support of this idea, GM(2-592) and GM(R522A) have reduced in vitro depolymerization activities and cause excess MT polymer and unaligned chromosomes within spindles. In addition, the C-terminal domain, in conjunction with the N-terminal domain, is needed for +TIP tracking (Moore et al., 2005). We also find that both the N- and C-terminal domains are needed for +TIP tracking, as neither GM(2-592) or GM(187-731) track MT ends (our unpublished results). These results imply that +TIP tracking is not required for spindle assembly, because GM(2-592) promotes spindle bipolarity, but does not +TIP track. Perhaps +TIP tracking is more important in interphase than in mitosis.
Our findings then leave open the question of how MCAK activity might be spatially and temporally controlled. Physiologically, this interplay will likely be regulated by proteins interacting with the N- and/or C-terminal domains. Perhaps MCAK phosphorylation regulates more than just MCAK depolymerization activity and localization to centromeres (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Sampath et al., 2004). Consistent with this idea, an Aurora B phospho-mutant of MCAK (MCAK-4A) targets effectively to centromeres, maintains wild-type MT depolymerization activity, but is unable to promote spindle bipolarity in MCAK-depleted extracts (Ohi et al., 2004). It is interesting to note that all of the MCAK Aurora B phosphorylation sites map to the N-terminal and neck domains, so one can readily envision how phosphorylation of these domains might alter interaction with the C-terminus and thus regulate the interplay between the domains. Given that MCAK is a substrate of multiple kinases, it is likely that phosphorylation will be a key mechanism to regulate MCAK activity during spindle assembly.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jane Stout and Chantal LeBlanc for critical review of the manuscript. We also thank Xiang Zhou for making the MCAK(296-308Δ) construct. Anti-xCENP-A was a kind gift from Aaron Straight (Stanford University). We thank H. Sosa for kindly sharing results before publication. K.M.H. and X.Z. are each supported by a predoctoral award from the American Heart Association. K.M.H. was also supported by National Institutes of Health (NIH) training Grant GM-007757. M.W.M. is supported by NIH award R15GM073688-01, and funds were made available by the Ohio Board of Regents. This work was supported by American Cancer Society Scholar Award RSG CSM-106128 to C.E.W. and in part by NIH Grant R01GM059618 to C.E.W. C.E.W. is a scholar of the Leukemia and Lymphoma Society.
Abbreviations used:
- MT
microtubule
- GMPCPP
guanylyl-(a,b)-methylene-diphosphonate
- GFP
green fluorescent protein
- GM
GFP-MCAK
- SEM
standard error of the mean.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0724)) on November 8, 2006.
REFERENCES
- Adio S., Reth J., Bathe F., Woehlke G. Review: regulation mechanisms of Kinesin-1. J. Muscle Res. Cell Motil. 2006;27:153–160. doi: 10.1007/s10974-005-9054-1. [DOI] [PubMed] [Google Scholar]
- Andersen S. S., Ashford A. J., Tournebize R., Gavet O., Sobel A., Hyman A. A., Karsenti E. Mitotic chromatin regulates phosphorylation of Stathmin/Op18. Nature. 1997;389:640–643. doi: 10.1038/39382. [DOI] [PubMed] [Google Scholar]
- Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., Swedlow J. R. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Banks J. D., Heald R. Adenomatous polyposis coli associates with the microtubule-destabilizing protein XMCAK. Curr. Biol. 2004;14:2033–2038. doi: 10.1016/j.cub.2004.10.049. [DOI] [PubMed] [Google Scholar]
- Bathe F., Hahlen K., Dombi R., Driller L., Schliwa M., Woehlke G. The complex interplay between the neck and hinge domains in kinesin-1 dimerization and motor activity. Mol. Biol. Cell. 2005;16:3529–3537. doi: 10.1091/mbc.E04-11-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbao-Cortes D., Hetzer M., Langst G., Becker P. B., Mattaj I. W. Ran binds to chromatin by two distinct mechanisms. Curr. Biol. 2002;12:1151–1156. doi: 10.1016/s0960-9822(02)00927-2. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas R. E., Gruss O. J., Mattaj I. W., Karsenti E. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- Case R. B., Pierce D. W., Hom-Booher N., Hart C. L., Vale R. D. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 1997;90:959–966. doi: 10.1016/s0092-8674(00)80360-8. [DOI] [PubMed] [Google Scholar]
- Cassimeris L., Morabito J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 2004;15:1580–1590. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy D. L., Hancock W. O., Wagenbach M., Howard J. Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat. Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- Desai A., Mitchison T. J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T. J., Walczak C. E. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999a;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T. J., Walczak C. E. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999b;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Desai A., Walczak C. E. Assays for microtubule destabilizing kinesins. In: Vernos I., editor. Kinesin Protocols. Totowa: Humana Press; 2001. pp. 109–121. [DOI] [PubMed] [Google Scholar]
- Ems-McClung S. C., Zheng Y., Walczak C. E. Importin α/β and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell. 2004;15:46–57. doi: 10.1091/mbc.E03-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S. A., Waligora K. W. Determinants of kinesin motor polarity. Science. 1998;281:1200–1202. doi: 10.1126/science.281.5380.1200. [DOI] [PubMed] [Google Scholar]
- Friedman D. S., Vale R. D. Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat. Cell Biol. 1999;1:293–297. doi: 10.1038/13008. [DOI] [PubMed] [Google Scholar]
- Gadde S., Heald R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004;14:797–805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Gadea B. B., Ruderman J. V. Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proc. Natl. Acad. Sci. USA. 2006;103:4493–4498. doi: 10.1073/pnas.0600702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaetz J., Kapoor T. M. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 2004;166:465–471. doi: 10.1083/jcb.200404015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Compton D. A. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N. J., Upton K., Compton D. A. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Goshima G., Nedelec F., Vale R. D. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R. D. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 2005;16:3896–3907. doi: 10.1091/mbc.E05-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O. J., Carazo-Salas R. E., Schatz C. A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., Mattaj I. W. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti W. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Helenius J., Brouhard G., Kalaidzidis Y., Diez S., Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- Hertzer K. M., Ems-McClung S. C., Kline-Smith S. L., Lipkin T. G., Gilbert S. P., Walczak C. E. Full-length dimeric MCAK is a more efficient microtubule depolymerase than minimal domain monomeric MCAK. Mol. Biol. Cell. 2006;17:700–710. doi: 10.1091/mbc.E05-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzer K. M., Ems-McClung S. C., Walczak C. E. Kin I kinesins: insights into the mechanism of depolymerization. Crit. Rev. Biochem. Mol. Biol. 2003;38:453–469. doi: 10.1080/10409230390267419. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Exp. Cell Res. 2004;301:50–59. doi: 10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Holmfeldt P., Zhang X., Stenmark S., Walczak C. E., Gullberg M. CaMKIIgamma-mediated inactivation of the Kin I kinesin MCAK is essential for bipolar spindle formation. EMBO J. 2005;24:1256–1266. doi: 10.1038/sj.emboj.7600601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A. W., Caplow M., Coy D. L., Hancock W. O., Diez S., Wordeman L., Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pu R. T., Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Karsenti E., Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Noetzel T. L., Arnal I., Drechsel D. N., Hyman A. A. Global and local control of microtubule destabilization promoted by a catastrophe kinesin MCAK/XKCM1. J. Muscle Res. Cell Motil. 2006:1–8. doi: 10.1007/s10974-005-9045-2. [DOI] [PubMed] [Google Scholar]
- Kline-Smith S. L., Khodjakov A., Hergert P., Walczak C. E. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol. Biol. Cell. 2002;13:2718–2731. doi: 10.1091/mbc.E01-12-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S. L., Walczak C. E. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Knowlton A. L., Lan W., Stukenberg P. T. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- Koffa M. D., Casanova C. M., Santarella R., Kocher T., Wilm M., Mattaj I. W. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S. L., Rosasco S. E., Barrett-Wilt G. A., Shabanowitz J., Hunt D. F., Walczak C. E., Stukenberg P. T. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., et al. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Y., Wirtz D., Zheng Y. A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 2003;160:635–644. doi: 10.1083/jcb.200211004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Hunter A. W., Wagenbach M., Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Wagenbach M., Wordeman L. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J. Biol. Chem. 2001;276:34753–34758. doi: 10.1074/jbc.M106626200. [DOI] [PubMed] [Google Scholar]
- Mennella V., Rogers G. C., Rogers S. L., Buster D. W., Vale R. D., Sharp D. J. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Miki H., Okada Y., Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Maddox P., Gaetz J., Groen A., Shirasu M., Desai A., Salmon E. D., Kapoor T. M. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. J., Maddox P., Groen A., Cameron L., Perlman Z., Ohi R., Desai A., Salmon E. D., Kapoor T. M. Bipolarization and poleward flux correlate during Xenopus extract spindle assembly. Mol. Biol. Cell. 2004;15:5603–5615. doi: 10.1091/mbc.E04-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A., Wollman R., Civelekoglu-Scholey G., Scholey J. Modeling mitosis. Trends Cell Biol. 2006;16:88–96. doi: 10.1016/j.tcb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Moore A., Wordeman L. C-terminus of mitotic centromere-associated kinesin (MCAK) inhibits its lattice-stimulated ATPase activity. Biochem. J. 2004;383:227–235. doi: 10.1042/BJ20040736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A. T., Rankin K. E., von Dassow G., Peris L., Wagenbach M., Ovechkina Y., Andrieux A., Job D., Wordeman L. MCAK associates with the tips of polymerizing microtubules. J. Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W. J., Zhang C., Clarke P. R. Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr. Biol. 2002;12:1442–1447. doi: 10.1016/s0960-9822(02)01076-x. [DOI] [PubMed] [Google Scholar]
- Moores C. A., Cooper J., Wagenbach M., Ovechkina Y., Wordeman L., Milligan R. A. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5:1812–1815. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- Newton C. N., Wagenbach M., Ovechkina Y., Wordeman L., Wilson L. MCAK, a Kin I kinesin, increases the catastrophe frequency of steady-state HeLa cell microtubules in an ATP-dependent manner in vitro. FEBS Lett. 2004;572:80–84. doi: 10.1016/j.febslet.2004.06.093. [DOI] [PubMed] [Google Scholar]
- Niederstasser H., Salehi-Had H., Gan E. C., Walczak C., Nogales E. XKCM1 acts on a single protofilament and requires the C terminus of tubulin. J. Mol. Biol. 2002;316:817–828. doi: 10.1006/jmbi.2001.5360. [DOI] [PubMed] [Google Scholar]
- Noda Y., Sato-Yoshitake R., Kondo S., Nangaku M., Hirokawa N. KIF2 is a new microtubule-based anterograde motor that transports membranous organelles distinct from those carried by kinesin heavy chain or KIF3A/B. J. Cell Biol. 1995;129:157–167. doi: 10.1083/jcb.129.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. Structural insights into microtubule function. Annu. Rev. Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Nitta R., Okada Y., Hirokawa N. A common mechanism for microtubule destabilizers—M-type kinesins curl the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- Ohba T., Nakamura M., Nishitani H., Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Ohi R., Sapra T., Howard J., Mitchison T. J. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina Y., Wagenbach M., Wordeman L. K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J. Cell Biol. 2002;159:557–562. doi: 10.1083/jcb.200205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. C., Rogers S. L., Schwimmer T. A., Ems-McClung S. C., Walczak C. E., Vale R. D., Scholey J. M., Sharp D. J. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Sampath S. C., Ohi R., Leismann O., Salic A., Pozniakovski A., Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Sawin K. E., Mitchison T. J. Mitotic spindle assembly by two different pathways in vitro. J. Cell Biol. 1991;112:925–940. doi: 10.1083/jcb.112.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Brown H. M., Kwon M., Rogers G. C., Holland G., Scholey J. M. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D. J., Mennella V., Buster D. W. KLP10A and KLP59C: the dynamic duo of microtubule depolymerization. Cell Cycle. 2005;4:1482–1485. doi: 10.4161/cc.4.11.2116. [DOI] [PubMed] [Google Scholar]
- Shipley K., Hekmat-Nejad M., Turner J., Moores C., Anderson R., Milligan R., Sakowicz R., Fletterick R. Structure of a kinesin microtubule depolymerization machine. EMBO J. 2004;23:1422–1432. doi: 10.1038/sj.emboj.7600165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout J. R., Rizk R. S., Kline S. L., Walczak C. E. Deciphering protein function during mitosis in PtK cells using RNAi. BMC Cell Biol. 2006;7:26. doi: 10.1186/1471-2121-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D., Asenjo A. B., Mennella V., Sharp D. J., Sosa H. Kinesin-13s form rings around microtubules. J. Cell Biol. 2006;175:25–31. doi: 10.1083/jcb.200605194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R., Popov A., Kinoshita K., Ashford A. J., Rybina S., Pozniakovsky A., Mayer T. U., Walczak C. E., Karsenti E., Hyman A. A. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- Vale R. D., Case R., Sablin E., Hart C., Fletterick R. Searching for kinesin's mechanical amplifier. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:449–457. doi: 10.1098/rstb.2000.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R. D., Goldstein L.S.B. One motor, many tails: an expanding repertoire of force-generating enzymes. Cell. 1990;60:883–885. doi: 10.1016/0092-8674(90)90334-b. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Gan E. C., Desai A., Mitchison T. J., Kline-Smith S. L. The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. Curr. Biol. 2002;12:1885–1889. doi: 10.1016/s0960-9822(02)01227-7. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Mitchison T. J., Desai A. XKCM1, a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Walczak C. E., Verma S., Mitchison T. J. XCTK2, a kinesin-related protein that promotes mitotic spindle assembly in Xenopus laevis egg extracts. J. Cell Biol. 1997;136:859–870. doi: 10.1083/jcb.136.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C. E., Vernos I., Mitchison T. J., Karsenti E., Heald R. A model for the proposed roles of different microtubule based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Wilde A., Lizarraga S. B., Zhang L., Wiese C., Gliksman N. R., Walczak C. E., Zheng Y. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- Wilde A., Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Woehlke G., Ruby A. K., Hart C. L., Ly B., Hom-Booher N., Vale R. D. Microtubule interaction site of the kinesin motor. Cell. 1997;90:207–216. doi: 10.1016/s0092-8674(00)80329-3. [DOI] [PubMed] [Google Scholar]
- Wordeman L., Mitchison T. J. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L., Wagenbach M., Maney T. Mutations in the ATP-binding domain affect the subcellular distribution of mitotic centromere-associated kinesin (MCAK) Cell Biol. Int. 1999;23:275–286. doi: 10.1006/cbir.1999.0359. [DOI] [PubMed] [Google Scholar]
- Wozniak M. J., Milner R., Allan V. N-terminal kinesins: many and various. Traffic. 2004;5:400–410. doi: 10.1111/j.1600-0854.2004.00191.x. [DOI] [PubMed] [Google Scholar]
- Zhu C., Zhao J., Bibikova M., Leverson J. D., Bossy-Wetzel E., Fan J. B., Abraham R. T., Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.