Abstract
Kinase D-interacting substrate of 220 kDa/ankyrin repeat-rich membrane spanning (Kidins220/ARMS) is a conserved membrane protein mainly expressed in brain and neuroendocrine cells, which is a downstream target of the signaling cascades initiated by neurotrophins and ephrins. We identified kinesin light chain 1 (KLC1) as a binding partner for Kidins220/ARMS by a yeast two-hybrid screen. The interaction between Kidins220/ARMS and the kinesin-1 motor complex was confirmed by glutathione S-transferase-pull-down and coimmunoprecipitation experiments. In addition, Kidins220/ARMS and kinesin-1 were shown to colocalize in nerve growth factor (NGF)-differentiated PC12 cells. Using Kidins220/ARMS and KLC1 mutants, we mapped the regions responsible for the binding to a short sequence of Kidins220/ARMS, termed KLC-interacting motif (KIM), which is sufficient for the interaction with KLC1. Optimal binding of KIM requires a region of KLC1 spanning both the tetratricopeptide repeats and the heptad repeats, previously not involved in cargo recognition. Overexpression of KIM in differentiating PC12 cells impairs the formation and transport of EGFP-Kidins220/ARMS carriers to the tips of growing neurites, leaving other kinesin-1 dependent processes unaffected. Furthermore, KIM overexpression interferes with the activation of the mitogen-activated protein kinase signaling and neurite outgrowth in NGF-treated PC12 cells. Our results suggest that Kidins220/ARMS-positive carriers undergo a kinesin-1–dependent transport linked to neurotrophin action.
INTRODUCTION
The correct development of the nervous system relies on the ability of axons and dendrites to recognize their targets. During this highly regulated process, neurons migrate and differentiate in response to gradients of specific growth factors, collectively termed neurotrophins, which regulate multiple neuronal functions, including cell survival, differentiation, synaptic function, and plasticity (Bibel and Barde, 2000; Chao, 2003; Huang and Reichardt, 2003). The mammalian neurotrophin family includes four different members: nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), and neurotrophin 3 and 4/5 (NT3 and NT4/5). Their binding to the p75 neurotrophin receptor (p75NTR) and to members of the Trk family of tyrosine kinases initiates signaling by triggering receptor dimerization and autophosphorylation. Activated receptors are then able to recruit several adaptor proteins, which will in turn start different and often antagonistic signaling cascades. Despite the essential role of neurotrophins in the development and maintenance of the nervous system, several aspects of their trafficking and biological activity are still elusive.
Neurotrophin function undergoes multiple levels of regulation. The pathways activated by a specific neurotrophin are determined not only by the ligand itself but also by the temporal and spatial pattern of stimulation (Segal, 2003). In addition, the specificity of their cellular response relies on the balance between local and distal action of the receptor–neurotrophin complex, which may derive from the interaction with specific modulators and/or their recruitment to long-range axonal transport routes (Smith and Scott, 2002; Huang and Reichardt, 2003). The characterization of new factors involved in these processes thus represents a crucial step toward a better understanding of the regulation of neurotrophin signaling.
Among these players, kinase D-interacting substrate of 220 kDa (Kidins220) (Iglesias et al., 2000), also known as ankyrin repeat-rich membrane spanning (ARMS) (Kong et al., 2001), forms a ternary complex with p75NTR and TrkA, and recruits CrkL, an adaptor protein mediating the sustained activation of mitogen-activated protein kinase (MAPK) in response to neurotrophins (Arevalo et al., 2004; Chang et al., 2004). Kidins220/ARMS is a highly conserved integral membrane protein, which was initially isolated as a substrate for protein kinase D (PKD), a serine/threonine kinase involved in the modulation of several cellular processes, such as proliferation and Golgi trafficking (Ghanekar and Lowe, 2005). PKD has recently been shown to regulate Kidins220/ARMS transport from the trans-Golgi network (TGN) to the plasma membrane (Sanchez-Ruiloba et al., 2006).
Primary sequence analysis of Kidins220/ARMS predicts 11 ankyrin repeats within the N-terminal region, whereas the C-terminal segment contains a proline-rich stretch, a SAM-like domain, and a PDZ-binding motif (Figure 1A). Both the N and C terminus are exposed to the cytoplasm (Iglesias et al., 2000; Kong et al., 2001). Sequence homology and the presence of a Walker A and B domains determined the inclusion of Kidins220/ARMS in the new KAP family of P-loop NTPases, whose members are predicted to mediate the assembly and disassembly of protein complexes associated with the inner surface of cell membranes (Aravind et al., 2004). Kidins220/ARMS is mainly expressed in brain and neuroendocrine cells, such as PC12 cells, where it concentrates at the tip of neurites upon differentiation (Iglesias et al., 2000). Kidins220/ARMS has also been shown to be associated to lipid rafts in PC12 cells, cortical neurons, and synaptosomes (Cabrera-Poch et al., 2004). These observations, together with the fact that this protein is a downstream effector of neurotrophins and ephrins (Kong et al., 2001), have led to the suggestion that Kidins220/ARMS might play a role in the process of neuronal differentiation (Arevalo et al., 2004).
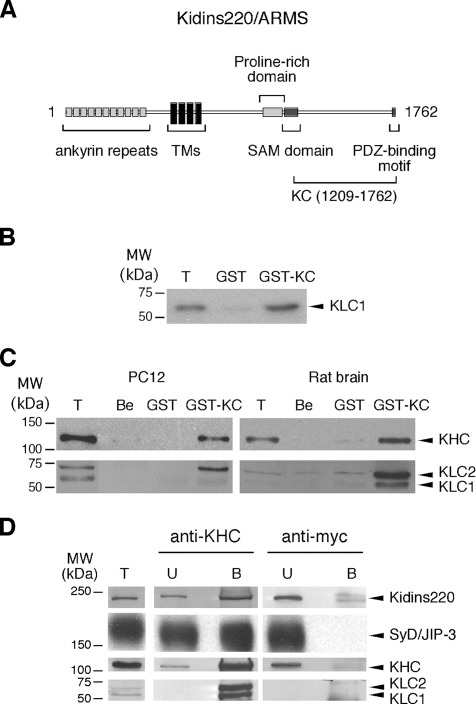
Figure 1.
Direct interaction between Kidins220/ARMS and kinesin-1. (A) Schematic representation of Kidins220/ARMS. The KC domain of Kidins220/ARMS (residues 1209-1762), which was used as bait in the yeast two-hybrid screen, is indicated. TMs, transmembrane domains. (B) KLC1 binds GST-KC in an in vitro pull-down assay using 35S-labeled KLC1. The Coomassie staining of the SDS-PAGE gel corresponding to this experiment is available in Supplemental Figure S3Aa. (C) The kinesin-1 motor complex binds to GST-KC. PC12 cells and rat brain extracts were incubated with GST-KC, GST prebound to glutathione beads, or with empty beads (Be). The bound material was analyzed by Western blot by using anti-KHC and anti-KLC antibodies. The Ponceau staining of the nitrocellulose membranes corresponding to this experiment is available in Supplemental Figure S3Ab. (D) Native Kidins220/ARMS and kinesin-1 form a complex in PC12 cells. PC12 cell lysates were incubated with either anti-KHC or anti-myc monoclonal antibodies, and the immunoprecipitated material was subsequently analyzed by Western blot by using anti-Kidins220/ARMS, anti-SyD/JIP-3, anti-KHC, and anti-KLC antibodies. Lanes T show 1/10 (B) or 1/100 (C and D) of the starting material for comparison. In lanes U and B, ∼1 and 80% of the unbound (U) and bound (B) material are loaded, respectively. The amount of SyD/JIP-3 associated with kinesin-1 is about fivefold the amount of Kidins220/ARMS. The nonlinear detection of KLC versus KHC in C and D is likely to be dependent on the properties of the anti-KLC antibody, which does not allow a concentration-dependent recognition of KLC in the same range of KHC and Kidins220/ARMS. The limitations of the anti-KLC antibody for the quantitative visualization of KLC have been documented previously (Pfister et al., 1989; Stenoien and Brady, 1997).
Despite the recent studies elucidating the role of Kidins220/ARMS in neurotrophin response (Arevalo et al., 2004, 2006; Chang et al., 2004), many questions remain open regarding the relationship between the biological function of this protein and its targeting to growth cones and/or specific domains of the plasma membrane. To elucidate this issue, we undertook a yeast two-hybrid approach, which allowed us to identify kinesin light chain 1 (KLC1), a subunit of conventional kinesin, as Kidins220/ARMS-interacting partner and to investigate the role of this complex in NGF-dependent responses.
MATERIALS AND METHODS
Chemicals and Antibodies
Reagents were from Sigma-Aldrich (St. Louis, MO), unless otherwise specified. The monoclonal antibody against Kidins220/ARMS and the rabbit polyclonal antibody anti-synaptotagmin I (Syt) were raised against the glutathione S-transferase C-terminal region (1209–1762) of Kidins220/ARMS (GST-KC) fusion protein (see below) and the C2A domain of Syt, respectively, by the Cancer Research UK Antibody Service (London, United Kingdom). Mouse monoclonal antibodies anti-kinesin heavy chain (KHC) (H2) and kinesin light chain (KLC) (L1) are from Chemicon International (Temecula, CA). Rabbit polyclonal antibodies were as follows: Kidins220/ARMS (Iglesias et al., 2000), KLC (BAbCO, Richmond, CA), phospho-mitogen–activated protein kinase (P-MAPK) (9101S; Cell Signaling Technology, Beverly, MA), Sunday Driver/c-Jun NH2-terminal kinase (JNK) interacting protein-3 (SyD/JIP-3) (Cavalli et al., 2005), whereas the goat polyclonal antibody anti-kinesin-3 was from Santa Cruz Biotechnology (Santa Cruz, CA). AlexaFluor488- and Texas Red-conjugated secondary antibodies were from Invitrogen (Paisley, United Kingdom).
Plasmids and Yeast Two-Hybrid Screen
pcDNA3-HA-KLC-HR and pcDNA3-HA-KLC-TPR were a kind gift from Dr. K. J. Verhey (Harvard Medical School, Boston, MA) (Verhey et al., 1998, 2001). The construct encoding full-length SyD/JIP-3 was a gift from Dr. V. Cavalli (Washington University, St. Louis, MO). The construct encoding full-length protein phosphatase 5 (Pp5) was a gift from D. Barford and J. Yang (The Institute of Cancer Research, London, United Kingdom). KLC1 deletion mutants were amplified using the primers listed in Supplemental Table S1 and then subcloned into the pPCR-Script Amp SK(+) cloning vector by using the PCR-Script Amp Cloning kit (Stratagene, La Jolla, CA), according to the manufacturer's instructions. The C-terminal region (1209–1762) of Kidins220/ARMS (KC) was amplified by polymerase chain reaction (PCR) by using rat Kidins220/ARMS cDNA (Iglesias et al., 2000) as template and inserted in frame in the pGEX-KG vector (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). KC deletion mutants were amplified using the primers listed in Supplemental Table S2 and then subcloned into the EcoRI/NotI sites of pGEX-4T3-HA (Lalli et al., 1999). Constructs used in the yeast two-hybrid pairwise testing were amplified using the primers listed in Supplemental Table S3 and then subcloned into the EcoRI/NotI site of the pGBKT7 vector (baits) or in the EcoRI/XhoI sites of the pGADT7 vector (preys), both from Clontech (Mountain View, CA). KC(Y1379A) was derived from the pGBKT7 vector encoding KC, by using the QuikChange XL site-directed mutagenesis kit and the same primers used for mRFP-KIM(Y24A) (see below). Monomeric red fluorescent protein (mRFP)-KLC-interacting motif (KIM) was prepared using a vector derived from pEGFP-C2 (BD Biosciences, San Jose, CA), where the sequence encoding enhanced green fluorescent protein (EGFP) had been substituted with the one of mRFP (Campbell et al., 2002). The plasmids mRFP-KIM(Y24A) and EGFP-Kidins220/ARMSΔKIM were derived from vectors encoding mRFP-KIM and EGFP-Kidins220/ARMS, respectively, by using the QuikChange XL site-directed mutagenesis kit and the primers listed in Supplemental Table S2. The plasmid pE/L-KIM was obtained by inserting the coding sequence of KIM in a vector bearing the pE/L virus promoter (Frischknecht et al., 1999). KLC1 splice variant C (accession no. M75148) was amplified from a rat brain cDNA library (OriGene Technologies, Rockville, MD) and cloned in pGEM-T easy vector (Promega, Madison, WI). All the constructs were verified by direct sequencing.
The yeast two-hybrid screen was carried out using a Matchmaker System 2 (Clontech) following the manufacturer's instructions. The KC fragment of Kidins220/ARMS fused with GAL4-binding domain in pGBKT7 was used as bait to screen a rat brain cDNA library in the pACT2 vector by using the AH109 yeast reporter strain. Approximately 7 × 105 transformants were screened. Positive interacting clones were selected for growth on Ade−/His−/Trp−/Leu−/X-α-gal plates and analyzed by direct sequencing. β-Galactosidase activity was measured using the yeast β-galactosidase assay kit (Pierce Chemical, Rockford, IL), according to the manufacturer's instructions.
In Vitro Transcription-Translation, GST Pull-Down, and Coimmunoprecipitation Assays
Recombinant GST-KC and its deletion mutants were expressed at 30°C in the Escherichia coli TG1 strain (Lalli et al., 1999). 35S-labeled proteins were generated using TnT Quick-coupled transcription/translation system (Promega) and Redivue l-[35S]methionine, 15 mCi/ml (GE Healthcare). For pull-down assays, GST-fusion proteins were bound to glutathione-Sepharose beads in Hank's buffer (20 mM HEPES-NaOH, pH 7.4, 0.44 mM KH2PO4, 0.42 mM NaH2PO4, 5.36 mM KCl, 136 mM NaCl, 0.81 mM MgSO4, 1.26 mM CaCl2, and 6.1 mM glucose) containing 0.1% bovine serum albumin (BSA) (Hank's-BSA) for 1 h at room temperature. Beads were then blocked with 2% BSA in Hank's for 1 h at 4°C and washed with Hank's-BSA. In vitro-transcribed/translated proteins were precleared on glutathione-Sepharose beads for 1 h at 4°C and then incubated with either prebound GST-KC fragments or GST alone for 2 h at 4°C in Hank's-BSA. Beads were then washed six times with ice-cold Hank's-BSA in the presence of 250 mM NaCl, 0.1% Triton X-100, and resuspended in loading buffer. Eluted proteins were then analyzed by SDS-PAGE and autoradiography.
PC12 or rat brain extracts were used as a source of endogenous kinesins. PC12 cells were scraped and lysed in 10 mM Tris-HCl, pH 8.0, 150 mM KCl, 1% NP-40, 1% glycerol, 1 mM EDTA, 0.1 mM dithiothreitol, and protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) for 30 min 4°C under constant agitation. Rat brains were extracted in the same buffer by using a Teflon homogenizer (Wheaton Science Products, Milville, NJ) and then incubated as described above. After centrifugation at 16,000 × g for 60 min at 4°C, extracts were incubated with immobilized GST-KC fragments or GST overnight at 4°C. Alternatively, cell lysates were incubated with anti-KHC or anti-myc monoclonal antibodies, and immunocomplexes were isolated by the addition of protein G-Sepharose Fast Flow (GE Healthcare) for 1 h at 4°C. After six washes with lysis buffer, bound proteins were eluted in loading buffer and analyzed in SDS-PAGE followed by Western blot by using the appropriate primary antibody. After incubation with horseradish peroxidase-conjugated secondary antibodies (Dako UK, Ely, Cambridgeshire, United Kingdom), immunoreactive bands were revealed by enhanced chemiluminescence (GE Healthcare). Intensity of the bands in Coomassie-stained protein gels, Ponceau-stained nitrocellulose membranes, and immunoblots were quantified by using the NIH Image software (http://rsb.info.nih.gov/nih-image/). The amount of GST-fusion protein loaded in each lane was normalized to the amount of GST in the corresponding control, taking into consideration the different molecular weights.
Immunofluorescence
PC12 cells were grown on poly-l-lysine–coated coverslips and differentiated with 100 ng/ml NGF (Alomone Labs, Jerusalem, Israel) in DMEM for 72 h (Herreros et al., 2001). Rat spinal cord motor neurons (MNs) were purified from E14 Sprague-Dawley embryos and maintained in culture as described previously (Bohnert and Schiavo, 2005). For immunocytochemistry experiments, cells were fixed with 3.7% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature, washed with PBS, incubated with 50 mM NH4Cl for 10 min, and rinsed and permeabilized with blocking buffer (2% BSA, 0.25% porcine skin gelatin, 0.2% glycine, 15% fetal calf serum, and 0.1% Triton X-100, in PBS) for 1 h. Primary antibodies were diluted in PBS containing 1% BSA, 0.25% porcine skin gelatin, and 3% fetal calf serum (antibody dilution buffer) and incubated for 1 h at room temperature. After rinsing with PBS, secondary antibodies diluted in antibody dilution buffer were applied for 30 min at room temperature. Cells were then washed and mounted with Mowiol 4-88 (Harco, Lynchburg, VA). Images were acquired by confocal microscopy (LSM510; Carl Zeiss, Jena, Germany) by using a 63× Plan-Apochromat oil immersion objective. Colocalization was quantified using the LSM510 software. For each couple of proteins analyzed, the percentage of Kidins220/ARMS, SyD/JIP-3, or Syt-positive pixels that overlapped with KHC- or KIF1A-positive pixels was calculated.
Transfection and Microinjection
PC12 cells were transfected with 0.5 μg of DNA and 1–2 μl of Lipofectamine (Invitrogen) in Opti-MEM (Invitrogen). Five hours after transfection, the medium was replaced with DMEM containing 100 ng/ml NGF, and 72 h later, the cells were fixed and analyzed by confocal microscopy. In selected experiments, HeLa cells were infected with the A36R-YdF recombinant vaccinia virus (Rietdorf et al., 2001). To induce high levels of protein expression in infected cells, all transfected genes were under the control of the viral pE/L promoter. At 4 h postinfection, cells were transfected with pE/L mRFP, pE/L mRFP-KIM, or pE/L mRFP-TPR (Rietdorf et al., 2001) by using Lipofectin (Invitrogen) according to manufacturer's instructions and processed for immunofluorescence after 4 h. Virus particles were visualized by labeling infected cells without permeabilization with an anti-B5R antibody (Schmelz et al., 1994), followed by incubation with AlexaFluor488-conjugated anti-rat antibodies.
For microinjection, PC12 cells were plated on MatTek dishes (MatTek, Ashland, MA) and differentiated for 2–4 d in DMEM containing 100 ng/ml NGF, 0.1% fetal calf serum, and 0.1% horse serum. Cells were microinjected with 50 μg/ml EGFP-Kidins220/ARMS plasmid alone or in combination with 20 μg/ml mRFP-KIM(Y24A) or 20 μg/ml mRFP-KIM. After overnight recovery, cells were washed with nonfluorescent DMEM without phenol red, riboflavin, folic acid, penicillin/streptomycin, and supplemented with 30 mM HEPES-NaOH, pH 7.3 (DMEM−), and imaged by low-light time-lapse microscopy. Images were taken every 2 s with a Nikon Diaphot 200 inverted microscope equipped with a Nikon 100×, 1.25 numerical aperture Plan differential interference contrast oil immersion objective by using an XF-100 Filter set (Omega Filters; Omega Optical, Brattleboro, VT) to detect EGFP-tagged proteins and an XF-102 Filter set (Omega Filters) to detect mRFP-tagged proteins. Images were acquired with a Hamamatsu C4742-95 Orcal cooled charge-coupled device camera (Hamamatsu Photonic Systems, Bridgewater, NJ) controlled by the Kinetic Acquisition Manager 2000 software (Kinetic Imaging, Nottingham, United Kingdom). Exposure times varied between 55 and 333 ms. Moving carriers were tracked using the MetaMorph software (Molecular Devices, Sunnyvale, CA), and the point tracking data were imported into a custom-written Mathematica 5.2 (Wolfram Research, Long Hanborough, Oxfordshire, United Kingdom) notebook, which identifies the extremes of the tracked particles and the linear displacement from their origin. The notebook also calculates the total distance traveled (obtained by summation of the distances between tracked points), and the average speed (obtained from “total distance traveled”/time). The length of the movies analyzed was constant, and all the visible particles were tracked. For the speed analysis, we considered only tracking data from carriers that were moving more than 4 μm from their origin.
For the quantitative analysis of neurite outgrowth, PC12 cells were transfected with mRFP, mRFP-KIM(Y24A), or mRFP-KIM and differentiated with 100 ng/ml NGF for 3 d. Images were acquired by confocal microscopy (LSM510; Carl Zeiss) by using a 40× Plan-Apochromat oil immersion objective. We considered differentiated cells as those with at least one neurite longer than twice the diameter of the cell body.
P-MAPK Signaling
PC12 cells were transfected with the indicated constructs and treated with NGF as described above. After 62 h, the medium was replaced by complete growth medium without NGF for 5 h. To stimulate sustained P-MAPK signaling, cells were treated with 100 ng/ml NGF in DMEM, 0.1% fetal calf serum, 0.1% horse serum for 40 min, and then fixed and immunostained for P-MAPK. For quantitation, the P-MAPK mean fluorescence intensity of individual cells was measured using ImageJ 1.34S (http://rsb.info.nih.gov/ij/). The average value for untransfected cells was set to 100% and compared with the mean fluorescence intensity of transfected cells. To determine the basal fluorescence intensity of P-MAPK (corresponding to 0%), cells were fixed and stained for P-MAPK after the 5-h incubation in complete growth medium. The specificity of the P-MAPK signal was verified by preincubating cells for 30 min with mitogen-activated protein kinase kinase inhibitor PD98059 (5 μg/ml; Calbiochem, San Diego, CA) and performing the NGF treatment in the presence of the inhibitor, followed by fixing and staining for P-MAPK. The P-MAPK mean fluorescence intensity of these cells was similar to the basal fluorescent intensity described above (data not shown).
RESULTS
Kidins220/ARMS Directly Interacts with KLC
To gain insight on the function of Kidins220/ARMS, we sought to identify its interacting partners by performing a yeast two-hybrid screen of a rat brain cDNA library by using the C-terminal part of Kidins220/ARMS (KC) as bait (Figure 1A). Among other hits (see Supplemental Table S4), we identified KLC1 splice variant C, a subunit of kinesin-1 (also known as conventional kinesin or KIF5A), as an interacting protein. Kinesin-1 is a heterotetramer of two heavy chains (KHC) and two light chains. KHCs contain the ATPase motor domain responsible for the movement along microtubules, whereas KLCs are involved in the modulation of KHC activity and cargo binding. Four genes encoding KLCs have been identified in mice: KLC1 is enriched in the nervous system, whereas KLC2 and KLC4 are ubiquitously expressed, and KLC3 shows a restricted expression to spermatids (Rahman et al., 1998; Junco et al., 2001; Carninci et al., 2005). In addition, three KLC1 splice variants (KLC1 A–C) have been identified in rat brain, which are characterized by different C termini (Cyr et al., 1991). The observation that Kidins220/ARMS concentrates at the neurite tips in differentiated PC12 cells (Iglesias et al., 2000) prompted us to consider the possibility that Kidins220/ARMS trafficking is mediated by kinesin-1 via a direct interaction with KLC.
The binding of KC to KLC1 was first verified by an independent pairwise yeast two-hybrid analysis on high-stringency medium (see Supplemental Figure S1A). The number of colonies obtained by cotransformation of yeast with KC- and KLC1-encoding plasmids was in the range of that obtained with KLC1 and SyD/JIP-3, a well known KLC1-binding protein (Verhey et al., 2001). Furthermore, we confirmed this interaction by GST pull-down, by using a GST-KC fusion protein and in vitro-translated rat KLC1. The binding of KLC1 to GST-KC was specific, because no interaction of KLC1 with GST alone was observed (Figure 1B). We also performed GST pull-down experiments on PC12 cell lysates and rat brain extracts. In both cases, we were able to detect a specific interaction between GST-KC and the native kinesin-1 holocomplex, because both the heavy and light chains of the motor complex were found associated with GST-KC beads (Figure 1C). Neither control glutathione beads nor immobilized GST bound endogenous kinesin-1 under these conditions.
The ability of the endogenous Kidins220/ARMS to interact with the kinesin-1 motor complex was then assessed in PC12 cell lysates by coimmunoprecipitation with an antibody directed against KHC, and immunoblotting with a Kidins220/ARMS antiserum as well as with anti-KHC and anti-KLC antibodies. Confirming the results obtained with GST pull-downs, we detected the association of endogenous Kidins220/ARMS with KHC, KLC1, and KLC2. This binding was specific because a control antibody did not lead to the recovery of this complex with the beads. Under the same experimental conditions, kinesin-1 displays a robust interaction with other proteins, such as SyD/JIP-3 (Figure 1D). Quantification of the amount of Kidins220/ARMS interacting with kinesin-1 in PC12 cell lysates suggests that only a fraction of Kidins220/ARMS (∼1%) is bound to this molecular motor under these conditions, whereas the recovery of SyD/JIP-3, which we have chosen as a control for the coimmunoprecipitation efficiency, is 5% of the total. Unfortunately, we were unable to coimmunoprecipitate kinesin-1 with anti-Kidins220/ARMS antibodies. This negative result may be due to steric hindrance caused by the binding of the anti-Kidins220/ARMS antibody to the KC domain, which could block KLC binding. The binding of Kidins220/ARMS to kinesin-1 was observed in lysates from cells either treated or untreated with NGF, indicating that the formation of this complex is independent of neurotrophin stimulation (data not shown). These data confirm the interaction detected by the yeast two-hybrid screen and suggest that the endogenous Kidins220/ARMS and kinesin-1 form a complex in PC12 cells and rat brain.
Kidins220/ARMS and Kinesin-1 Colocalize in Differentiated PC12 Cells
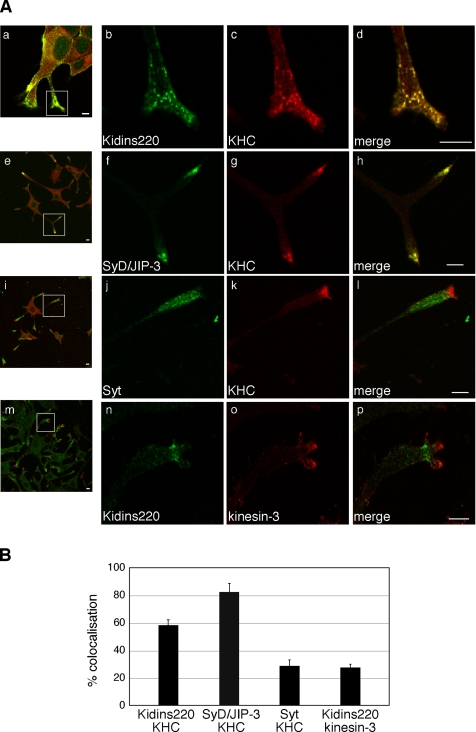
We then asked whether Kidins220/ARMS colocalizes with kinesin-1 in NGF-differentiated PC12 cells. Kidins220/ARMS and KHC displayed a punctate distribution in cell bodies as well as neurites, characterized by extensive clustering at the neurite tips (Figure 2A, a–d). To provide a quantitative analysis of the level of codistribution of Kidins220/ARMS and the kinesin-1 complex, we determined the degree of colocalization of Kidins220/ARMS as well as that of a range of control proteins, with different members of the kinesin family (Figure 2B). SyD/JIP-3, used here as a positive control, exhibited an 82% overlap with KHC (Figure 2A, e–h), whereas Syt, which is known to be transported by kinesin-3/KIF1A and not by kinesin-1 (Okada et al., 1995), showed only 28% colocalization with KHC (Figure 2A, i–l). We found that 58% of Kidins220/ARMS colocalized with kinesin-1 (Figure 2A, a–d), whereas only 28% overlapped with kinesin-3 (Figure 2A, m–p). A partial overlap between the distribution of Kidins220/ARMS and both KHC and KLC was also observed in MNs, mainly in dendrites and growth cones (see Supplemental Figure S2). These results indicate that Kidins220/ARMS and kinesin-1 specifically colocalize to an internal compartment targeted to neuronal processes in PC12 cells and primary neurons.
Figure 2.
Kidins220/ARMS and kinesin-1 colocalize in PC12 cells. (A) PC12 cells treated for 3 d with NGF were stained with polyclonal (b) or monoclonal (n) anti-Kidins220/ARMS, polyclonal anti-SyD/JIP-3 (f), polyclonal anti-Syt (j), monoclonal anti-KHC (c, g, and k), or polyclonal anti-kinesin-3 (o) antibodies and analyzed by confocal microscopy. Bars, 5 μm. (B) The percentage of colocalization of the indicated proteins in neurites is shown. Kidins220/ARMS:KHC, n = 16 neurites; SyD/JIP-3:KHC, n = 11 neurites; Syt:KHC, n = 16 neurites; for Kidins220/ARMS:kinesin-3, n = 22 neurites. Error bars represent SEM.
The Region 83-296 of KLC1 Mediates the Binding to Kidins220/ARMS
KLCs are characterized by two main regions: a series of heptad repeats (HR) at the N terminus, and six tetratricopeptide repeats (TPRs) at the C terminus (Figure 3A; Gindhart and Goldstein, 1996). The HR-containing region is known to mediate the interaction with KHC, whereas the TPR motifs have been shown to dock kinesin-1 to specific cargoes (Karcher et al., 2002; Hirokawa and Takemura, 2005). We tested the ability of KC to interact with the TPR motifs of KLC by pairwise yeast two-hybrid analysis. Surprisingly, yeast growth was severely reduced in cotransformation with KC- and KLC-TPR–expressing plasmids and was abolished with KC and Pp5-TPR, an unrelated TPR-containing protein (Das et al., 1998; see Supplemental Figure S1). To identify the region of KLC involved in the binding to Kidins220/ARMS, we generated a series of truncated versions of rat KLC1 and tested them in pull-down experiments using GST-KC (Figure 3). In agreement with the data obtained from the yeast two-hybrid analysis, we found that the KLC region containing the TPR motifs (KLC1-C and -F; Figure 3) binds KC with very low efficiency and that a portion of the HR domain and the linker region (aa 83-199) together with the first two TPR motifs (aa 199-296) are required for the interaction (KLC1-H; Figure 3). The need of a portion of the HR region for the binding to KC has been independently confirmed by pairwise yeast two-hybrid analysis (see Supplemental Figure S1). Interestingly, the removal of the region 1–63 from KLC1 seems to enhance the binding to KC above the levels observed with full-length KLC1 in GST pull-down, raising the possibility that it might act as a regulatory region (Figure 3). Altogether, these results suggest that Kidins220/ARMS binds to kinesin-1 by using a novel docking site on KLC1, which includes both HR and TPR motifs.
Figure 3.
The region 83–296 of KLC1 mediates the binding to Kidins220/ARMS. (A) Scheme of the KLC1 mutants used in this study. The empty box corresponds to the heptad repeats region, whereas the gray ovals represent the TPR motifs. The putative inhibitory region for the Kidins220/ARMS–KLC interaction (aa 1-63) is marked by a dashed line. The ability of each mutant to bind Kidins220/ARMS is reported on the right: +, ≅100%; ++, ≅200%; +/−, ≅ <25%; and −, no binding. (B) The ability of in vitro 35S-labeled KLC1 mutants to bind GST-KC is shown. Intensity of the bands was quantified as described in Materials and Methods. 1/25 of the starting in vitro transcription/translation mixes is loaded in T for comparison. The Coomassie staining of the SDS-PAGE gels corresponding to this experiment is available in Supplemental Figure S3B.
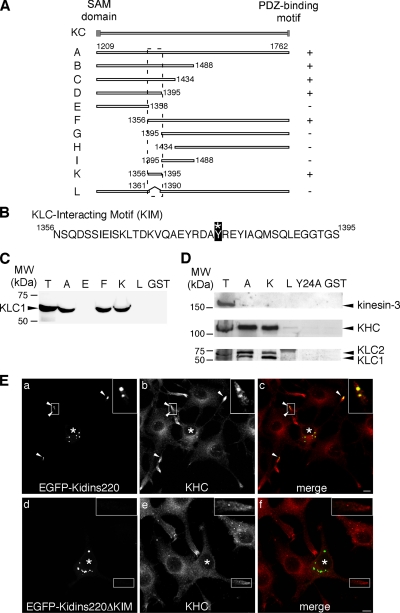
The KIM Domain of Kidins220/ARMS Is Sufficient for KLC1 Binding
Ligands usually interact with KLC via a short sequence close to their C termini (Terlecky et al., 1995; Gatto et al., 2000; Scheufler et al., 2000). The absence of sequence homology between Kidins220/ARMS and other known KLC-binding proteins, together with the new binding determinants recognized by Kidins220/ARMS on KLC1, suggest that the interaction between these two proteins is mediated by a novel motif. To map this domain, we generated a series of KC deletion mutants (Figure 4A), which we expressed as GST-fusion proteins and tested in pull-down assays using in vitro transcribed-translated KLC1 (Figure 4C) or PC12 cell lysates (Figure 4D). Using this approach, we were able to identify the region between residues 1356 and 1395 as sufficient, and between 1361 and 1390 as necessary, for the binding to KLC1. We named the region 1356–1395, which is predicted to fold as a helix (http://robetta.bakerlab.org/), KIM (Figure 4B). KIM represents a novel kinesin-1–interacting signature, because it does not bear any similarities to other proteins reported to interact with KLC. Furthermore, it specifically binds kinesin-1, as shown by its inability to interact with other kinesins, such as kinesin-3 (Figure 4D). The interaction between KIM and KLC is highly specific, because single point mutations of several of the amino acid residues of KIM to alanine, such as Tyr24 in KIM(Y24A) (corresponding to residue 1379 of the full-length protein), abolished the binding to kinesin-1 in GST pull-down (Figure 4D). The inhibitory effect of the Y1379A mutation on the binding of KC to KLC1 was confirmed by pairwise yeast two-hybrid analysis, as shown by both number of colonies and β-galactosidase activity (Supplemental Figure S1).
Figure 4.
KIM is sufficient for the binding of Kidins220/ARMS to KLC. (A) Scheme of the Kidins220/ARMS mutants used in this study. The ability of each mutant to bind KLC1 is reported on the right (+, binding, −, no binding). The position of the KIM motif is highlighted by the dashed line. (B) Sequence of the KIM motif (KC-K). The asterisk indicates the tyrosine (residue 1379 of the full-length protein) that is mutated to alanine in KIM(Y24A). (C) The ability of some of the KC mutants expressed as GST-fusion proteins to bind in vitro 35S-labeled KLC1 is shown. We loaded 1/25 of the starting in vitro transcription/translation mix in T for comparison. The Coomassie staining of the SDS-PAGE gel corresponding to this experiment is available in Supplemental Figure S3Ca. (D) The KIM motif is sufficient to bind the native kinesin-1 motor complex. PC12 cells lysates were incubated with either GST or GST-fusion proteins prebound to glutathione-beads, as indicated. The bound material was analyzed by Western blot by using anti-kinesin-3, anti-KHC, and anti-KLC antibodies. T corresponds to 10 μg of total lysate. The Ponceau staining of the nitrocellulose membrane corresponding to this experiment is available in Supplemental Figure S3Cb. (E) PC12 cells were transfected with EGFP-Kidins220/ARMS (a–c) or EGFP-Kidins220/ARMS-ΔKIM (d–f), and stained with anti-KHC antibody (b and e). Full-length Kidins220/ARMS-positive puncta are positive for KHC (c; arrowheads and inset), whereas there is no significant overlapping between KHC and the mutant lacking the KIM domain (f; inset). Asterisks indicate transfected cells. Bars, 5 μm.
To confirm these findings for the full-length Kidins220/ARMS in a cellular context, we transfected PC12 cells with constructs encoding either the full-length EGFP-Kidins220/ARMS, or a mutant lacking the KIM domain (EGFP- Kidins220/ARMS-ΔKIM). We then stained the cells with an antibody against KHC and analyzed the distribution of the proteins by confocal microscopy (Figure 4E). Both the full-length Kidins220/ARMS (Figure 4E, a) and the ΔKIM mutant (Figure 4E, d) are expressed at the same level in NGF-differentiated PC12 cells, and accumulate in discrete structures upon transfection. However, although Kidins220/ARMS-positive structures were found both in the cell bodies and in the neurites (Figure 4E, a and c), the majority of Kidins220/ARMS-ΔKIM was confined to cell bodies (Figure 4E, d and f). A marked overlap of kinesin-1 with EGFP-Kidins220/ARMS was observed (Figure 4E, a–c, arrowheads, and insets), whereas no colocalization was found between KHC and the mutant lacking the KIM domain (Figure 4E, d–f, and insets). These findings indicate that the KIM domain of Kidins220/ARMS is responsible for the recruitment of the kinesin-1 complex both in vitro and in neuronal cells.
Kidins220/ARMS Is a Kinesin-1–specific Cargo
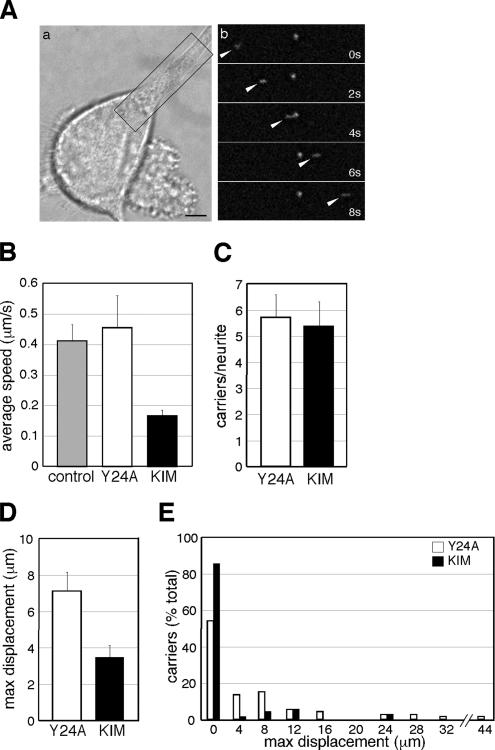
The accumulation of Kidins220/ARMS at neurite tips implies the existence of an active targeting mechanism responsible for its peripheral localization in differentiating PC12 cells. To visualize this process in real time, we imaged cells microinjected with EGFP-Kidins220/ARMS by time-lapse fluorescent microscopy. EGFP-Kidins220/ARMS is localized to discrete vesicular structures, which show a bidirectional movement along the neurites. On treatment with NGF, these Kidins220/ARMS-positive carriers accumulate at the tips of growing neuronal processes (see Supplemental Movie S1), in agreement with the immunofluorescence data on fixed cells. Figure 5A shows a time series of an EGFP-Kidins220/ARMS carrier (arrowhead) moving along a neurite toward the growth cone. The average speed of these carriers is 0.41 ± 0.05 μm/s (Figure 5B), which is compatible with the rate observed for kinesin-mediated transport (Woehlke and Schliwa, 2000; Hirokawa and Takemura, 2005).
Figure 5.
KIM overexpression impairs Kidins220/ARMS trafficking in PC12 cells. (A) EGFP-Kidins220/ARMS is transported along neurites in PC12 cells. (a) Phase image of a microinjected cell. (b) Time series of the boxed area. Arrowheads indicate an EGFP-Kidins220/ARMS–positive moving carrier. Bar, 5 μm. (B–E) NGF-treated PC12 cells were coinjected with plasmids driving the expression of EGFP-Kidins220/ARMS alone or in combination with either mRFP-KIM, or mRFP-KIM(Y24A). (B) The average speed of EGFP-Kidins220/ARMS–positive carriers is decreased by mRFP-KIM expression. (C) The average number of EGFP-Kidins220/ARMS–positive carriers per neurite is not affected by the overexpression of mRFP-KIM. n = 13 neurites were analyzed for both conditions. (D) The average maximum displacement of EGFP-Kidins220/ARMS–positive carriers is reduced in mRFP-KIM–expressing cells. The analysis of the movement of EGFP-Kidins220/ARMS positive carriers is shown in E (see text and Materials and Methods for details). A 4-μm binning starting from 0 was applied. n = 24 carriers for EGFP-Kidins220/ARMS–expressing cells, n = 70 carriers for EGFP-Kidins220/ARMS– and mRFP-KIM–expressing cells; n = 74 for EGFP-Kidins220/ARMS– and mRFP-KIM(Y24A)–expressing cells. Error bars represent SEM.
Given that Kidins220/ARMS is actively transported in PC12 cells and binds KLC via the KIM motif, kinesin-1 might be responsible for the intracellular trafficking of Kidins220/ARMS. If this is the case, the KIM peptide is predicted to have a dominant-negative effect, because its binding to KLC should block the interaction with full-length Kidins220/ARMS and impair its transport. To test this hypothesis, we coinjected EGFP-Kidins220/ARMS with either mRFP-tagged KIM or mRFP-KIM(Y24A) and analyzed their effect on the formation and transport of Kidins220/ARMS-positive structures by time-lapse microscopy. First, we monitored the transport of EGFP-Kidins220/ARMS–positive structures in the presence of KIM(Y24A), and we compared it to the case where only EGFP-Kidins220/ARMS was expressed. We found that the coexpression of KIM(Y24A) did not have any effect on the speed of the moving carriers, whose behavior was indistinguishable from the control samples (0.45 ± 0.10 μm/s; Figure 5B). In contrast, the average speed of the carriers in cells overexpressing KIM was reduced to 0.16 ± 0.02 μm/s (Figure 5B). However, the number of structures per neurite was unaffected by KIM overexpression (5.69 ± 0.87 structures for cells expressing KIM(Y24A), 5.38 ± 0.93 for cells expressing KIM; Figure 5C), suggesting that the presence of this peptide does not interfere with the formation of the EGFP-Kidins220/ARMS–positive structures but rather that it selectively impairs their trafficking. In agreement with this hypothesis, the average maximum displacement of the EGFP-Kidins220/ARMS–positive carriers was significantly reduced in KIM-expressing cells in comparison with KIM(Y24A)-microinjected samples (3.5 ± 0.63 μm versus 7.1 ± 1.03 μm) (Figure 5D and Supplemental Movies S2 and S3). Quantitative kinetic analysis revealed that in KIM-expressing cells, the majority (86%) of the Kidins220/ARMS-positive structures are stationary (maximum displacement between 0 and 3.9 μm), and only 14% move farther away (Figure 5E, black bars). In contrast, in cells microinjected with the inactive KIM(Y24A) mutant, 54% of the particles have a maximum displacement between 0 and 3.9 μm, and the remaining cover longer distances, up to 45 μm (Figure 5E, white bars). Overexpression of KIM, therefore, results in a net increase in the number of structures that are either stationary or undergo only short-range movements. Altogether, these findings suggest that KIM overexpression displays a dominant-negative effect on the transport of Kidins220/ARMS-positive carriers, causing a reduction in both their maximum displacement and average speed, and support the hypothesis that the trafficking of Kidins220/ARMS is mediated by the kinesin-1 complex.
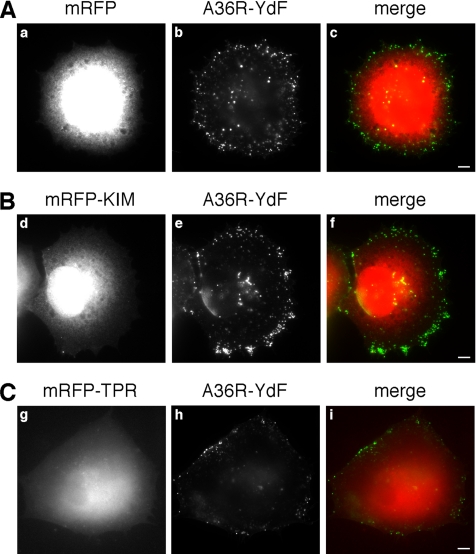
Does KIM selectively prevent Kidins220/ARMS transport or does it act as a general inhibitor for kinesin-1–dependent trafficking? To discriminate between these two possibilities, we tested whether different forms of kinesin-mediated transport are affected by KIM overexpression. Vaccinia virus reaches the surface of infected cells by exploiting a kinesin-1–dependent mechanism (Rietdorf et al., 2001). Once at the plasma membrane, the virus switches from a microtubule-based to an actin-based motility and then spreads to neighboring cells (Newsome et al., 2004). The viral membrane protein A36R is required for actin-based motility and kinesin-1 recruitment, which occurs via the TPRs of KLC (Ward and Moss, 2004). Vaccinia virus transport therefore represents an ideal system to test KIM function. To this end, HeLa cells were infected with the vaccinia A36R-YdF strain (Rietdorf et al., 2001) and subsequently transfected with mRFP (Figure 6A), mRFP-KIM (Figure 6B) or mRFP-TPRs (Figure 6C). Extracellular viral particles were then visualized by wide-field microscopy in absence of permeabilization (see Materials and Methods). Strikingly, we found that mRFP-KIM overexpression did not affect the ability of the virus to reach the plasma membrane (Figure 6, compare b and e), suggesting that mRFP-KIM does not perturb its kinesin-1–dependent transport. In contrast, the overexpression of the TPR domain impaired the transport of the viral particles to the cell periphery (Figure 6, compare b and h). Altogether, these experiments indicate that KIM is not a general inhibitor of kinesin-1 function but rather that it blocks the recruitment of Kidins220/ARMS by interfering with a specific binding site on KLC.
Figure 6.
The translocation of vaccinia virus to the plasma membrane is unaffected by KIM. HeLa cells were infected with the A36R-YdF vaccinia virus strain, and 4 h later they were transfected with mRFP (A), mRFP-KIM (B), or mRFP-TPR (C). After a further 4 h, cells were processed for immunofluorescence in absence of permeabilization, and extracellular virus particles were visualized. The distribution of the virus on the plasma membrane is independent of the overexpression of mRFP-KIM (compare b and e), whereas it is reduced by the overexpression of the mRFP-TPR domain (compare b and h). Bars, 5 μm.
KIM Overexpression Reduces MAPK Phosphorylation and the NGF-dependent Differentiation of PC12 Cells
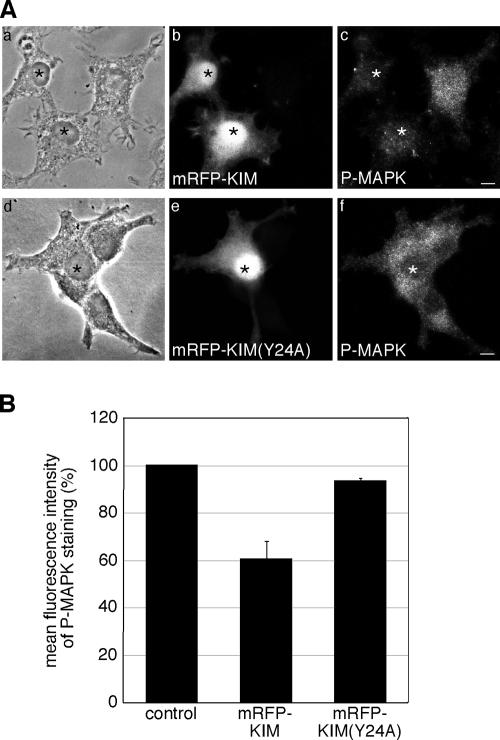
Kidins220/ARMS has been linked to NGF signaling and sustained MAPK activation (Arevalo et al., 2004, 2006), raising the possibility that the kinesin-1–dependent targeting of Kidins220/ARMS plays a role in its signaling function. To test this hypothesis, we examined the effect of KIM overexpression on the NGF-dependent activation of the MAPK pathway. We chose this dominant-negative strategy over a small interfering RNA (siRNA) approach, because down-regulation of Kidins220/ARMS by siRNA has been previously reported to reduce MAPK activation (Arevalo et al., 2004), but it does not directly answer the question of whether a kinesin-1–dependent transport of Kidins220/ARMS is required for NGF signaling. To examine the activation of MAPK, we transfected PC12 cells with either mRFP-KIM or mRFP-KIM(Y24A), and upon short treatment with NGF (see Materials and Methods), cells were stained for the phosphorylated form of MAPK (P-MAPK). As shown in Figure 7A, the P-MAPK labeling was reduced in mRFP-KIM–transfected cells compared with nontransfected cells, whereas the KIM(Y24A) mutant had no significant effect. Quantitative analysis of the mean fluorescence intensity of these samples revealed that mRFP-KIM–transfected cells showed a reduction of 40% in the P-MAPK staining compared with mRFP-KIM(Y24A)–expressing samples or nontransfected cells (Figure 7B). This result suggests that the kinesin-1–dependent transport of Kidins220/ARMS might be required for downstream events in NGF signaling.
Figure 7.
Overexpression of KIM reduces the phosphorylation of MAPK in PC12 cells. (A) PC12 cells were transfected with mRFP-KIM (a–c) or mRFP-KIM(Y24A) (d–f) and then fixed and immunostained for P-MAPK (c and f). mRFP-KIM–expressing cells show a reduced P-MAPK staining compared with nontransfected cells and mRFP-KIM(Y24A)–expressing cells. Transfected cells are marked with asterisks. Bars, 5 μm. (B) The mean fluorescence intensities of untransfected cells and cells overexpressing mRFP-KIM or mRFP-KIM(Y24A) were determined as described in Materials and Methods. mRFP-KIM, n = 74 cells; mRFP-KIM(Y24A), n = 63 cells; nontransfected cells, n = 204 derived from three independent experiments. Error bars represent SEM.
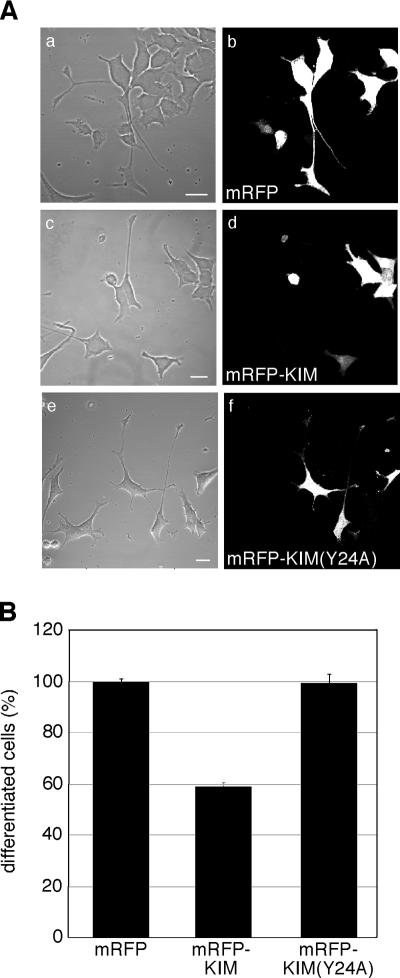
The NGF-mediated activation of the MAPK pathway plays a major role in promoting neuronal differentiation in PC12 cells (Vaudry et al., 2002). In light of the results described above, interfering with Kidins220/ARMS transport by KIM overexpression is predicted to reduce PC12 cells differentiation. To test this hypothesis, we transfected undifferentiated PC12 cells with mRFP, mRFP-KIM, or mRFP-KIM(Y24A) before NGF treatment (Figure 8). Cells were fixed after 3 d, and differentiation was then quantified. KIM overexpression caused a 40% reduction in the percentage of differentiated cells, whereas the transfection of KIM(Y24A) was ineffective (Figure 8B). These results indicate that KIM overexpression interferes with neurite outgrowth in NGF-treated PC12 cells, suggesting that the kinesin-1–dependent transport of Kidins220/ARMS participates in neuronal differentiation.
Figure 8.
KIM overexpression inhibits neurite outgrowth in PC12 cells. (A) Undifferentiated PC12 cells were transfected with mRFP (a and b), mRFP-KIM (c and d), or mRFP-KIM(Y24A) (e and f); treated for 3 d with NGF; and analyzed by confocal microscopy. Bars, 20 μm. (B) Percentage of differentiated cells in each condition is shown. Differentiation in mRFP-expressing cells was set to 100%. Between 50 and 140 cells were counted for each experiment (n = 3). Error bars represent SEM.
DISCUSSION
The diverse outcomes triggered by trophic stimuli are achieved through the precise compartmentalization of effector molecules, which ensure the activation of different signaling cascades according to the temporal and spatial pattern of stimulation (Smith and Scott, 2002). Molecular motors play a key role in determining the appropriate response to growth factors by delivering neurotrophin receptor complexes to specific domains of the plasma membrane as well as by targeting activated receptors to local and long-range trafficking routes (Verhey and Rapoport, 2001; Yano and Chao, 2004; Howe and Mobley, 2005; Zweifel et al., 2005). The kinesin-1 family of molecular motors is implicated in the transport of an increasing number of proteins, which include cytoskeletal components (Xia et al., 2003; Kimura et al., 2005; Utton et al., 2005); receptor-associated factors, such as GRIP and GRIF (Setou et al., 2002; Brickley et al., 2005; Hoogenraad et al., 2005); and the synaptosome-associated protein of 25 kDa (SNAP25) and SNAP23 (Diefenbach et al., 2002). Kinesin-1 has also been shown to play a crucial role in the transport of organelles, such as synaptic vesicles (Su et al., 2004) and lysosomes (Nakata and Hirokawa, 1995). Furthermore, kinesin-1 localization has been recently shown to be a marker of axonal specification (Jacobson et al., 2006). Based on these observations, the general principle that signaling pathways are linked to the activity of molecular motors, and in particular, to kinesin-1–mediated processes, has recently emerged. The JNK pathway, which relies on the binding of JIPs and related factors to the TPR motifs of KLC (Bowman et al., 2000; Verhey et al., 2001), represents probably the most compelling example in support to this model. Our findings provide new evidence for the link between molecular motors and signaling networks, suggesting that the kinesin-1–mediated transport of Kidins220/ARMS plays a role in the NGF-triggered activation of the MAPK pathway.
Following up our initial observation that the C-terminal portion of Kidins220/ARMS interacts with KLC in a yeast two-hybrid screen, we established that this protein interacts with kinesin-1 both in PC12 cells and in brain tissue. In addition, Kidins220/ARMS and kinesin-1 display an overlapping distribution along the neurites of NGF-treated PC12 cells and primary neurons. To map the binding determinants between Kidins220/ARMS and KLC1, we generated a series of mutants of both proteins. Interestingly, we found that the region of KLC containing the TPR repeats, which in most cases mediates the interaction with specific cargoes (Karcher et al., 2002; Hirokawa and Takemura, 2005), was not sufficient for optimal binding to Kidins220/ARMS. A stretch of amino acids extending to the N-terminus of KLC, which spans the linker region and about half of the HR repeats (aa 83-199), is required in addition to the first two TPR motifs (aa 199-296). This binding mode is also distinct from that recently described for rootletin, a component of ciliar rootlets, which involves only the HR domain of KLC (Yang and Li, 2005). The engagement of a portion of the HR domain in Kidins220/ARMS binding does not preclude its interaction with KHC and the recruitment of a functional kinesin-1 motor complex (Figure 1D and 5). Furthermore, this novel binding interface would potentially allow the simultaneous interaction of other cargoes with the remaining TPR motifs. Kidins220/ARMS may therefore mediate the recruitment of components of the neurotrophin signaling pathway onto neurite transport carriers. Altogether, these results are indicative of the unique nature of the interface used by Kidins220/ARMS for its binding to kinesin-1. As a consequence, interfering with this interaction is unlikely to affect other kinesin-1–dependent processes. Interestingly, the removal of the region 1–63 of KLC1 results in an increased binding efficiency of Kidins220/ARMS, suggesting that the N-terminus of KLC1 might act as a regulatory domain. These findings offer new insights on novel aspects of KLC regulation, which will be analyzed in future studies.
Using a similar approach, we were able to restrict the region responsible for the binding of Kidins220/ARMS to KLC to a small sequence within its C-terminus, which is sufficient for this interaction. This region, termed KIM (aa 1356-1395), represents a novel kinesin-1 binding motif, because it does not bear any similarities with other proteins reported to interact with KLC. Crucially, both targeting to the neurite tips and colocalization with KHC are lost in a Kidins220/ARMS mutant lacking the KIM domain. These findings confirm the novelty of the recognition interface between Kidins220/ARMS and kinesin-1, and further our understanding on how different cargoes interact independently with this multifunctional motor complex.
The observation that Kidins220/ARMS accumulates at the neurite tips of NGF-differentiated PC12 cells (Iglesias et al., 2000) suggests that its targeting to these structures might be mediated by kinesin-1. By imaging EGFP-Kidins220/ARMS in PC12 neurites, we were able to visualize Kidins220/ARMS carriers, which moved bidirectionally over long distances with an average speed compatible with a kinesin-1–mediated transport (Woehlke and Schliwa, 2000). Overexpression of the isolated KIM domain reduces both the total displacement and the average speed of the Kidins220/ARMS carriers, suggesting that this domain acts as a dominant-negative inhibitor for the trafficking of the Kidins220/ARMS-KLC1 complex. This inhibition was specific for Kidins220/ARMS trafficking, because other kinesin-1–dependent processes, such as vaccinia virus targeting to the plasma membrane, were unaffected. The KIM peptide therefore represents a unique and specific tool to dissect the dynamics of Kidins220/ARMS transport in vivo.
Scaffolding proteins have a crucial role in the regulation of signaling pathways, because they channel the flow of information toward proper cellular responses by holding together preassembled signaling complexes (Pawson and Nash, 2003). In PC12 cells, neurotrophins can trigger both proliferation and differentiation. In this system, the transient activation of MAPK signaling is associated with a proliferative outcome, whereas the sustained activation of the same pathway leads to neuronal differentiation (Marshall, 1995; Vaudry et al., 2002). Kidins220/ARMS has been recently shown to connect activated Trk receptors to the downstream CrkL–C3G–Rap1 complex by providing a scaffold where receptors and downstream modulators come into contact with each other. The recruitment of these factors on Kidins220/ARMS and the formation of a stable signaling platform lead to a prolonged activation of the MAPK pathway, which ultimately contributes to neurite outgrowth and differentiation (Arevalo et al., 2004, 2006). Here, we show that KIM overexpression reduces the activation of the MAPK pathway and PC12 cell differentiation in response to NGF. On this basis, we suggest that the cellular responsiveness to neurotrophins depends on the correct delivery of Kidins220/ARMS-positive carriers to the neurite tips, which is driven by the recruitment of the kinesin-1 motor complex to these transport organelles. However, the possibility that KIM overexpression affects not only the interaction with kinesin-1 but also other signaling events linked to neurite outgrowth cannot be dismissed. This is particularly relevant in light of the multiple binding partners and phosphorylation sites identified for Kidins220/ARMS (Iglesias et al., 2000; Kong et al., 2001; Arevalo et al., 2004). Likewise, other molecular motors might also be involved in the transport of Kidins220/ARMS at specific stages of development and/or in different cell types.
In conclusion, the formation of a complex with the KLC subunit of kinesin-1 is necessary for the transport and intracellular localization of Kidins220/ARMS. We propose a model in which the kinesin-1–mediated transport of Kidins220/ARMS is required for the correct targeting of this scaffolding protein to neurite tips and might regulate the cellular response to neurotrophic stimuli.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. L. Thomas for the preparation of the KC construct and advice, K. J. Verhey for the pcDNA3-HA-KLC-HR and pcDNA3-HA-KLC-TPR plasmids, D. Badford and J. Yang for the Pp5 construct, and V. Cavalli for the SyD/JIP-3 cDNA and antibody. We thank D. Worth for the cloning of the KLC1 constructs, A. Nicol for the Mathematica workbook used for tracking data analysis, and P. Fitzjohn for the structure prediction analysis of KIM and KLC. We are grateful to S. Tooze, A. Behrens, and members of the Molecular Neuropathobiology laboratory for critical reading of the manuscript. This work was supported by Cancer Research UK.
Abbreviations used:
- EGFP
enhanced green fluorescent protein
- GST
glutathione S-transferase
- HR
heptad repeat
- KC
C-terminal part (aa 1209-1762) of Kidins220/ARMS
- Kidins220/ARMS
kinase D-interacting substrate of 220 kDa/ankyrin repeat-rich membrane spanning
- KLC
kinesin light chain
- KHC
kinesin heavy chain
- kinesin-3/KIF1A
kinesin family member 1A
- KIM
KLC-interacting motif
- MAPK
mitogen-activated protein kinase
- MN
motor neuron
- p75NTR
p75 neurotrophin receptor
- mRFP
monomeric red fluorescent protein
- NGF
nerve growth factor
- PKD
protein kinase D
- P-MAPK
phosphorylated mitogen-activated protein kinase
- Pp5
protein phosphatase 5
- SyD/JIP-3
Sunday Driver/c-Jun NH2-terminal kinase JNK interacting protein-3
- Syt
synaptotagmin I
- TPR
tetratricopeptide repeat.
Footnotes

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0453) on November 1, 2006.
REFERENCES
- Aravind L., Iyer L. M., Leipe D. D., Koonin E. V. A novel family of P-loop NTPases with an unusual phyletic distribution and transmembrane segments inserted within the NTPase domain. Genome Biol. 2004;5:R30. doi: 10.1186/gb-2004-5-5-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arevalo J. C., Pereira D. B., Yano H., Teng K. K., Chao M. V. Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation. J. Biol. Chem. 2006;281:1001–1007. doi: 10.1074/jbc.M504163200. [DOI] [PubMed] [Google Scholar]
- Arevalo J. C., Yano H., Teng K. K., Chao M. V. A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein. EMBO J. 2004;23:2358–2368. doi: 10.1038/sj.emboj.7600253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M., Barde Y. A. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 2000;14:2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- Bohnert S., Schiavo G. Tetanus toxin is transported in a novel neuronal compartment characterized by a specialized pH regulation. J. Biol. Chem. 2005;280:42336–42344. doi: 10.1074/jbc.M506750200. [DOI] [PubMed] [Google Scholar]
- Bowman A. B., Kamal A., Ritchings B. W., Philp A. V., McGrail M., Gindhart J. G., Goldstein L. S. Kinesin-dependent axonal transport is mediated by the Sunday driver (SYD) protein. Cell. 2000;103:583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- Brickley K., Smith M. J., Beck M., Stephenson F. A. GRIF-1 and OIP106, members of a novel gene family of coiled-coil domain proteins: association in vivo and in vitro with kinesin. J. Biol. Chem. 2005;280:14723–14732. doi: 10.1074/jbc.M409095200. [DOI] [PubMed] [Google Scholar]
- Cabrera-Poch N., Sanchez-Ruiloba L., Rodriguez-Martinez M., Iglesias T. Lipid raft disruption triggers protein kinase C and Src-dependent protein kinase D activation and Kidins220 phosphorylation in neuronal cells. J. Biol. Chem. 2004;279:28592–28602. doi: 10.1074/jbc.M312242200. [DOI] [PubMed] [Google Scholar]
- Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Cavalli V., Kujala P., Klumperman J., Goldstein L. S. Sunday Driver links axonal transport to damage signaling. J. Cell Biol. 2005;168:775–787. doi: 10.1083/jcb.200410136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M. S., Arevalo J. C., Chao M. V. Ternary complex with Trk, p75, and an ankyrin-rich membrane spanning protein. J. Neurosci. Res. 2004;78:186–192. doi: 10.1002/jnr.20262. [DOI] [PubMed] [Google Scholar]
- Chao M. V. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat. Rev. Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Cyr J. L., Pfister K. K., Bloom G. S., Slaughter C. A., Brady S. T. Molecular genetics of kinesin light chains: generation of isoforms by alternative splicing. Proc. Natl. Acad. Sci. USA. 1991;88:10114–10118. doi: 10.1073/pnas.88.22.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. K., Cohen P. W., Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5, implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach R. J., Diefenbach E., Douglas M. W., Cunningham A. L. The heavy chain of conventional kinesin interacts with the SNARE proteins SNAP25 and SNAP23. Biochemistry. 2002;41:14906–14915. doi: 10.1021/bi026417u. [DOI] [PubMed] [Google Scholar]
- Frischknecht F., Moreau V., Rottger S., Gonfloni S., Reckmann I., Superti-Furga G., Way M. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401:926–929. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- Gatto G. J., Jr, Geisbrecht B. V., Gould S. J., Berg J. M. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- Ghanekar Y., Lowe M. Protein kinase D: activation for Golgi carrier formation. Trends Cell Biol. 2005;15:511–514. doi: 10.1016/j.tcb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Gindhart J. G., Jr, Goldstein L. S. Tetratrico peptide repeats are present in the kinesin light chain. Trends Biochem. Sci. 1996;21:52–53. [PubMed] [Google Scholar]
- Herreros J., Ng T., Schiavo G. Lipid rafts act as specialized domains for tetanus toxin binding and internalization into neurons. Mol. Biol. Cell. 2001;12:2947–2960. doi: 10.1091/mbc.12.10.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- Hoogenraad C. C., Milstein A. D., Ethell I. M., Henkemeyer M., Sheng M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat. Neurosci. 2005;8:906–915. doi: 10.1038/nn1487. [DOI] [PubMed] [Google Scholar]
- Howe C. L., Mobley W. C. Long-distance retrograde neurotrophic signaling. Curr. Opin. Neurobiol. 2005;15:40–48. doi: 10.1016/j.conb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Huang E. J., Reichardt L. F. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Iglesias T., Cabrera-Poch N., Mitchell M. P., Naven T. J., Rozengurt E., Schiavo G. Identification and cloning of Kidins220, a novel neuronal substrate of protein kinase D. J. Biol. Chem. 2000;275:40048–40056. doi: 10.1074/jbc.M005261200. [DOI] [PubMed] [Google Scholar]
- Jacobson C., Schnapp B., Banker G. A. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 2006;49:797–804. doi: 10.1016/j.neuron.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Junco A., Bhullar B., Tarnasky H. A., van der Hoorn F. A. Kinesin light-chain KLC3 expression in testis is restricted to spermatids. Biol. Reprod. 2001;64:1320–1330. doi: 10.1095/biolreprod64.5.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher R. L., Deacon S. W., Gelfand V. I. Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 2002;12:21–27. doi: 10.1016/s0962-8924(01)02184-5. [DOI] [PubMed] [Google Scholar]
- Kimura T., Watanabe H., Iwamatsu A., Kaibuchi K. Tubulin and CRMP–2 complex is transported via kinesin-1. J. Neurochem. 2005;93:1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- Kong H., Boulter J., Weber J. L., Lai C., Chao M. V. An evolutionarily conserved transmembrane protein that is a novel downstream target of neurotrophin and ephrin receptors. J. Neurosci. 2001;21:176–185. doi: 10.1523/JNEUROSCI.21-01-00176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli G., Herreros J., Osborne S. L., Montecucco C., Rossetto R., Schiavo G. Functional characterisation of tetanus and botulinum neurotoxins binding domains. J. Cell Sci. 1999;112:2715–2724. doi: 10.1242/jcs.112.16.2715. [DOI] [PubMed] [Google Scholar]
- Marshall C. J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Nakata T., Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J. Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome T. P., Scaplehorn N., Way M. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science. 2004;306:124–129. doi: 10.1126/science.1101509. [DOI] [PubMed] [Google Scholar]
- Okada Y., Yamazaki H., Sekine-Aizawa Y., Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Pawson T., Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Pfister K. K., Wagner M. C., Stenoien D. L., Brady S. T., Bloom G. S. Monoclonal antibodies to kinesin heavy and light chains stain vesicle-like structures, but not microtubules, in cultured cells. J. Cell Biol. 1989;108:1453–1463. doi: 10.1083/jcb.108.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Friedman D. S., Goldstein L. S. Two kinesin light chain genes in mice. Identification and characterization of the encoded proteins. J. Biol. Chem. 1998;273:15395–15403. doi: 10.1074/jbc.273.25.15395. [DOI] [PubMed] [Google Scholar]
- Rietdorf J., Ploubidou A., Reckmann I., Holmstrom A., Frischknecht F., Zettl M., Zimmermann T., Way M. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 2001;3:992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruiloba L., Cabrera-Poch N., Rodriguez-Martinez M., Lopez-Menendez C., Martin Jean-Mairet R., Higuero A. M., Iglesias T. Protein kinase D intracellular localization and activity control kidins220 traffic through a PDZ-binding motif. J. Biol. Chem. 2006;281:18888–18900. doi: 10.1074/jbc.M603044200. [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Schmelz M., Sodeik B., Ericsson M., Wolffe E. J., Shida H., Hiller G., Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal R. A. Selectivity in neurotrophin signaling: theme and variations. Annu. Rev. Neurosci. 2003;26:299–330. doi: 10.1146/annurev.neuro.26.041002.131421. [DOI] [PubMed] [Google Scholar]
- Setou M., Seog D. H., Tanaka Y., Kanai Y., Takei Y., Kawagishi M., Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- Smith F. D., Scott J. D. Signaling complexes: junctions on the intracellular information super highway. Curr. Biol. 2002;12:R32–R40. doi: 10.1016/s0960-9822(01)00646-7. [DOI] [PubMed] [Google Scholar]
- Stenoien D. L., Brady S. T. Immunochemical analysis of kinesin light chain function. Mol. Biol. Cell. 1997;8:675–689. doi: 10.1091/mbc.8.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q., Cai Q., Gerwin C., Smith C. L., Sheng Z. H. Syntabulin is a microtubule-associated protein implicated in syntaxin transport in neurons. Nat. Cell Biol. 2004;6:941–953. doi: 10.1038/ncb1169. [DOI] [PubMed] [Google Scholar]
- Terlecky S. R., Nuttley W. M., McCollum D., Sock E., Subramani S. The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J. 1995;14:3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utton M. A., Noble W. J., Hill J. E., Anderton B. H., Hanger D. P. Molecular motors implicated in the axonal transport of tau and alpha-synuclein. J. Cell Sci. 2005;118:4645–4654. doi: 10.1242/jcs.02558. [DOI] [PubMed] [Google Scholar]
- Vaudry D., Stork P. J., Lazarovici P., Eiden L. E. Signaling pathways for PC12 cell differentiation: making the right connections. Science. 2002;296:1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- Verhey K. J., Lizotte D. L., Abramson T., Barenboim L., Schnapp B. J., Rapoport T. A. Light chain-dependent regulation of kinesin's interaction with microtubules. J. Cell Biol. 1998;143:1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K. J., Meyer D., Deehan R., Blenis J., Schnapp B. J., Rapoport T. A., Margolis B. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K. J., Rapoport T. A. Kinesin carries the signal. Trends Biochem. Sci. 2001;26:545–550. doi: 10.1016/s0968-0004(01)01931-4. [DOI] [PubMed] [Google Scholar]
- Ward B. M., Moss B. Vaccinia virus A36R membrane protein provides a direct link between intracellular enveloped virions and the microtubule motor kinesin. J. Virol. 2004;78:2486–2493. doi: 10.1128/JVI.78.5.2486-2493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke G., Schliwa M. Walking on two heads: the many talents of kinesin. Nat. Rev. Mol. Cell Biol. 2000;1:50–58. doi: 10.1038/35036069. [DOI] [PubMed] [Google Scholar]
- Xia C. H., Roberts E. A., Her L. S., Liu X., Williams D. S., Cleveland D. W., Goldstein L. S. Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J. Cell Biol. 2003;161:55–66. doi: 10.1083/jcb.200301026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Li T. The ciliary rootlet interacts with kinesin light chains and may provide a scaffold for kinesin-1 vesicular cargos. Exp. Cell Res. 2005;309:379–389. doi: 10.1016/j.yexcr.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Yano H., Chao M. V. Mechanisms of neurotrophin receptor vesicular transport. J. Neurobiol. 2004;58:244–257. doi: 10.1002/neu.10321. [DOI] [PubMed] [Google Scholar]
- Zweifel L. S., Kuruvilla R., Ginty D. D. Functions and mechanisms of retrograde neurotrophin signalling. Nat. Rev. Neurosci. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.