Abstract
Proinflammatory cytokines and bacterial products trigger inducible nitric oxide synthase (iNOS) expression and nitric oxide (NO) production in inflammatory and tissue cells. In inflammation, NO acts as an important mediator having both proinflammatory and destructive effects.
Protein kinase C (PKC) is a family of serine–threonine protein kinase isoenzymes involved in signal transduction pathways related to inflammatory responses. The aim of the present study was to investigate the role of classical PKC (cPKC) isoenzymes in the regulation of iNOS expression and NO production in murine J774 macrophages and the mechanisms involved.
RO318220 (inhibits PKCβ, PKCγ and PKCɛ), GÖ6976 (inhibits cPKC isoenzymes PKCα and PKCβ) and LY333531 (inhibits PKCβ) reduced lipopolysaccharide (LPS)-induced NO production and iNOS expression in a dose-dependent manner as did 6 h pretreatment with 1 μM phorbol 12-myristate 13-acetate (PMA) (which was shown to downregulate PKC expression).
PKC inhibitors also reduced LPS-induced iNOS mRNA levels, but they did not affect the half-life of iNOS mRNA. PKC inhibitors did not alter LPS-induced activation of NF-κB as measured by electrophoretic mobility shift assay.
All PKC inhibitors used and pretreatment with 1 μM PMA inhibited signal transducer and activator of transcription 1 (STAT1) activation as measured by the translocation of STAT1α from the cytosol to the nucleus by Western blot. In addition, inhibition of STAT1 activation by AG-490, an inhibitor of JAK-2, also reduced NO production.
These results suggest that cPKC isoenzymes, especially PKCβ, mediate the upregulation of iNOS expression and NO production in activated macrophages in an NF-κB-independent manner, possibly through the activation of transcription factor STAT1.
Keywords: iNOS, LPS, macrophages, nitric oxide, protein kinase C, STAT1
Introduction
Nitric oxide (NO) acts as a signalling molecule in, for example, cardiovascular and neuronal systems. In inflammation, NO is an important mediator having both proinflammatory and destructive effects (Moilanen et al., 1999; Korhonen et al., 2005). High amount of NO is produced by inducible nitric oxide synthase (iNOS) for prolonged time as a response to bacterial products, such as lipopolysaccharide (LPS), and to proinflammatory cytokines (MacMicking et al., 1997; Vallance & Leiper, 2002). NO production in activated macrophages is primarily regulated at the level of iNOS expression (Kleinert et al., 2003; Korhonen et al., 2005).
The protein kinase C (PKC) pathway represents a major signal transduction system in inflammation (Spitaler & Cantrell, 2004). Different tissues seem to have their own characteristic patterns of PKC isoenzyme expression and function (Way et al., 2000). The mammalian PKC family comprises of serine–threonine protein kinase isoenzymes, which are divided into three classes based on their structure and ability to bind cofactors (Newton, 2001). The classical PKC (cPKC) isoenzymes (α, γ and the splice variants βI and βII) are activated by diacylglycerol (DAG), Ca2+ and phosphatidylserine. These isoenzymes are targets of the tumor-promoting phorbol ester PMA (phorbol 12-myristate 13-acetate, also called TPA), a surrogate of DAG. The novel (nPKC) isoenzymes (δ, ɛ, η and θ) are Ca2+-independent and activated by DAG and phosphatidylserine. The third group, atypical PKC (aPKC) isoenzymes (ζ and ι/λ), are Ca2+- and DAG-independent kinases. In contrast to the classical and novel isoenzymes, aPKCs do not respond to phorbol esters (Newton, 2001; Spitaler & Cantrell, 2004). In addition, PKCμ and PKCν are sometimes regarded to form a fourth class of PKC isoenzymes (Newton, 2001).
A role for PKC has been identified in inflammatory diseases, cancer and heart disease, and PKC inhibitors are under development to treat these diseases (Bowling et al., 1999; Chen et al., 2001; Goekjian & Jirousek, 2001; Newton, 2001; Tan & Parker, 2003; Aksoy et al., 2004). Several lines of evidence suggest that cPKC isoenzymes (Fujihara et al., 1994; St-Denis et al., 1998; Giroux & Descoteaux, 2000; Molina-Holgado et al., 2000; Foey & Brennan, 2004), PKCδ (Chen et al., 1998a; Tepperman et al., 2000; Carpenter et al., 2001), PKCη (Chen et al., 1998b; Pham et al., 2003) and PKCɛ (Castrillo et al., 2001; Kang et al., 2001) are involved in the LPS- and cytokine-induced expression of inflammatory genes including iNOS.
The aim of the present study was to investigate the role of cPKC isoenzymes in the regulation of iNOS expression and NO production in activated macrophages and the mechanisms involved. The results suggest that cPKC isoenzymes, probably PKCβ, mediate the upregulation of iNOS expression and NO production in activated macrophages in an NF-κB-independent manner, possibly through the activation of transcription factor signal transducer and activator of transcription 1 (STAT1).
Methods
Materials
Reagents were purchased as follows: RO318220, phorbol 12,13-didecanoate (PDD) and LY333531 were from Alexis Biochemicals (Lausen, Switzerland); GÖ6976, HBDDE and recombinant PKCγ were from Calbiochem (La Jolla, CA, U.S.A.); LPS (Escherichia coli 0111:B4, product number L-4391) was from Sigma Chemical Co. (St Louis, MO, U.S.A.); mouse monoclonal PKCα antibody, rabbit polyclonal iNOS, PKCβI, PKCβII, PKCγ and STAT1α antibodies and goat anti-rabbit HRP-conjugated polyclonal antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, U.S.A.) and goat anti-mouse HRP-conjugated antibody was from Pierce Biotechnology (Rockford, IL, U.S.A.). All other reagents were from Sigma Chemical Co.
Cell culture
J774 macrophages (American Type Culture Collection) were cultured at 37°C in 5% CO2 atmosphere in Dulbecco's modified Eagle's medium with ultraglutamine 1 (Cambrex BioScience, Verviers, Belgium) supplemented with 10% heat-inactivated fetal bovine serum (Cambrex BioScience), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 250 ng ml−1 amphotericin B (Gibco, Paisley, U.K.) and harvested with trypsin-EDTA (Gibco). Cells were seeded on 24-well plates for nitrite measurements and RT–PCR, on six-well plates for Western blot analysis and on 10 cm dishes for translocation studies, preparation of nuclear extracts and electrophoretic mobility shift assay. Cells were grown to confluence prior to the experiments. Toxicity of the tested compounds was ruled out by measuring cell viability using Cell Proliferation Kit II (Roche Diagnostics, Indianapolis, IN, U.S.A.) according to manufacturer's instructions.
Nitrite assays
Measurement of nitrite accumulation into the culture medium was used to determine NO production. The culture medium was collected at indicated time points and nitrite was measured by Griess reaction (Green et al., 1982).
Preparation of cell lysates for Western blotting
At indicated time points, cells were rapidly washed with ice-cold phosphate-buffered saline (PBS) and solubilized in cold lysis buffer containing 10 mM Tris-base, pH 7.4, 5 mM EDTA, 50 mM NaCl, 1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM sodiumorthovanadate, 20 μg ml−1 leupeptin, 50 μg ml−1 aprotinin, 5 mM NaF, 2 mM sodiumpyrophosphate and 10 μM n-octyl-β-D-glucopyranoside. After incubation for 15 min on ice, lysates were centrifuged (13,400 × g, 4°C, 10 min), supernatants were collected and stored in SDS sample buffer in −20°C. An aliquot of the supernatant was used to determine protein concentration by the Coomassie blue method (Bradford, 1976).
Preparation of cytosolic and particulate fractions for PKC Western blotting
At indicated time points, cells were rapidly washed with ice-cold PBS and solubilized in cold buffer A (20 mM Tris-base, pH 7.4, 10 mM EDTA, 5 mM EGTA, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM sodiumorthovanadate, 10 μg ml−1 leupeptin, 25 μg ml−1 aprotinin and 1.25 mM NaF). After incubation for 15 min on ice, lysates were centrifuged at 100,000 × g for 1 h at 4°C, supernatants were collected and marked as the cytosolic fraction. Pellets were resuspended in cold lysis buffer B (20 mM Tris-base, pH 7.4, 10 mM EDTA, 5 mM EGTA, 1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM sodiumorthovanadate, 10 μg ml−1 leupeptin, 25 μg ml−1 aprotinin, 1.25 mM NaF and 10 μM n-octyl-β-D-glucopyranoside). After incubation for 2 h on ice, lysates were centrifuged at 100,000 × g for 1 h at 4°C, supernatants were collected and marked as the particulate fraction. An aliquot of the supernatant was used to determine protein concentration by the Coomassie blue method (Bradford, 1976).
Preparation of nuclear extracts for electrophoretic mobility shift assay (EMSA) and STAT1α Western blotting
At indicated time points, cells were rapidly washed with ice-cold PBS and solubilized in hypotonic buffer A (10 mM HEPES–KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM sodiumorthovanadate, 10 μg ml−1 leupeptin, 25 μg ml−1 aprotinin, 1 mM NaF and 0.1 mM EGTA). After incubation for 10 min on ice, cells were vortexed for 30 s and the nuclei were separated by centrifugation at 4°C, 21,000 × g for 10 s. Nuclei were resuspended in buffer C (20 mM HEPES–KOH, pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM sodiumorthovanadate, 10 μg ml−1 leupeptin, 25 μg ml−1 aprotinin, 1 mM NaF and 0.1 mM EGTA) and incubated for 20 min on ice. Nuclei were vortexed for 30 s and nuclear extracts were obtained by centrifugation at 4°C, 21,000 × g for 2 min. Protein contents of the nuclear extracts were measured by the Coomassie blue method (Bradford, 1976).
Western blotting
Prior to Western blotting, proteins were boiled for 10 min with SDS sample buffer and 20 μg of protein was used per lane on 8% (iNOS, STAT1α) or 10% (PKC) SDS–polyacrylamide gel and transferred to Hybond ECL™ nitrocellulose membrane (Amersham Biosciences, U.K., Ltd, Little Chalfont, Buckinghamshire, U.K.). After transfer, the membrane was blocked in TBS-T (20 mM Tris-base, pH 7.6, 150 mM NaCl, 0.1% Tween-20) containing 5% nonfat dry milk for 1 h at room temperature and incubated with primary antibody in the blocking solution at 4°C overnight. The membrane was washed with TBS-T and incubated with the secondary antibody in the blocking solution for 30 min at room temperature and washed. Bound antibody was detected using Super Signal® West Pico chemiluminescent substrate (Pierce, Rockford, IL, U.S.A.) and FluorChem™ 8800 imaging system (Alpha Innotech Corporation, San Leandro, CA, U.S.A.). Super Signal® West Dura and Femto (Pierce) were used for the detection of PKC isoenzymes.
Electrophoretic mobility shift assay
EMSA was performed as described previously (Lahti et al., 2002). Briefly, transcription factor consensus oligonucleotides for NF-κB (Promega, Madison, WI, U.S.A.) were 5′-32P-end-labeled with DNA 5′-End Labeling Kit (Roche Diagnostics, Indianapolis, IN, U.S.A.). For binding reactions, 5 μg of nuclear extract was incubated in 20 μl of total reaction volume containing 0.1 mg ml−1 (poly)dI–dC, 1 mM dithiothreitol, 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 200 mM KCl and 10% glycerol for 20 min in room temperature. 32P-labeled oligonucleotide probe (0.2 ng) was added and the reaction mixture was incubated for 10 min. Protein–DNA complexes were separated from DNA probe by electrophoresis on a native 4% polyacrylamide gel. The gel was dried and autoradiographed using intensifying screen at −70°C. The quantitation of densities of specific bands was carried out using FluorChem™ software version 3.1.
RNA extraction and quantitative RT–PCR
Cell homogenization, RNA extraction, reverse transcription of RNA to cDNA and PCR reactions were performed as described previously (Lahti et al., 2003), with the exception that in the reverse transcription reaction, the amount of total RNA reverse transcribed was 100 ng and cDNA used in PCR corresponded to approximately 2.5 ng of total RNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control gene.
The relative mRNA levels were quantified and compared using the relative standard curve method as described in Applied Biosystems User Bulletin number 2. Total RNA was isolated from LPS-stimulated J774 macrophages and reverse transcribed. Standard curves for GAPDH and iNOS were created using dilution series of cDNA corresponding to approximately 1 pg to 10 ng of total RNA in PCR. The threshold cycle values obtained were plotted against dilution factor to create a standard curve. Relative mRNA levels in test samples were then calculated using the standard curve. The relative amount of gene transcript present was calculated and normalized by dividing the calculated value of iNOS by the GAPDH value in each sample.
Statistics
Results are expressed as mean±standard error of mean (s.e.m.). Statistical significance of the results was calculated by the analysis of variances supported by Dunnett adjusted significance levels. Differences were considered significant at P<0.05.
Results
Effects of PKC inhibitors on LPS-induced NO production and iNOS protein expression
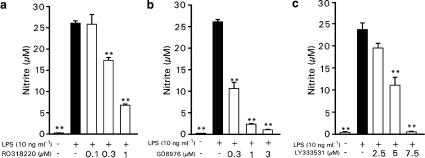
Bacterial endotoxin LPS induced iNOS protein expression and NO production in J774 macrophages. To determine whether PKC activation participated in the upregulation of NO production by LPS, we measured NO production in the presence of PKC inhibitors. RO318220, an inhibitor of PKC isoenzymes β, γ and ɛ (Davis et al., 1992; Wilkinson et al., 1993), and GÖ6976, a selective inhibitor of cPKC isoenzymes (Martiny-Baron et al., 1993), both inhibited LPS-induced NO production in a dose-dependent manner (Figure 1a and b). Exposure to increasing concentration of RO318220 resulted in a 34% (0.3 μM) and 74% (1 μM) inhibition of NO production during 24 h incubation. Exposure to GÖ6976 resulted in 59% (0.3 μM) inhibition of NO production and larger doses (1 and 3 μM) inhibited NO production almost completely (91 and 95%). Since the results with GÖ6976 suggest that the effect of PKC on NO production could be mediated by the cPKC isoenzymes, we studied the effects of LY333531, a selective inhibitor of PKCβ (Jirousek et al., 1996), and HBDDE, an inhibitor of PKC isoenzymes α and γ (Kashiwada et al., 1994), on LPS-induced NO production. LY333531 inhibited NO production in a dose-dependent manner (Figure 1c), but HBDDE did not have any effect on NO production when used up to 100 μM concentrations (at concentrations higher than 100 μM HBDDE started to be toxic to J774 macrophages).
Figure 1.
Effects of PKC inhibitors on LPS-induced NO production in J774 cells. J774 cells were stimulated by LPS (10 ng ml−1) and treated with increasing concentrations of RO318220 (a), GÖ6976 (b) or LY333531 (c). After 24 h incubation, nitrite concentrations in the culture medium were measured as a marker of NO production. Values are mean±s.e.m. (n=6). **P<0.01 as compared with cells treated with LPS only.
In further studies, we investigated the effects of PKC inhibitors on iNOS expression by Western blot. Cells cultured in the absence of LPS did not contain detectable amounts of iNOS protein. Exposure to LPS enhanced iNOS protein expression markedly. RO318220 (1–3 μM), GÖ6976 (0.1–1 μM) and LY333531 (2.5–7.5 μM) inhibited LPS-induced iNOS expression in a dose-dependent manner (Figure 2a–c).
Figure 2.
Effects of PKC inhibitors on LPS-induced iNOS protein expression in J774 cells. J774 cells were stimulated by LPS (10 ng ml−1) and treated with increasing concentrations of RO318220 (a), GÖ6976 (b) or LY333531 (c). After 24 h, incubations were terminated and immunoblots were run using antibody against iNOS. Chemiluminescent signal was quantified as described under the Methods section. Values are mean±s.e.m. (n=3). **P<0.01 as compared with cells treated with LPS only.
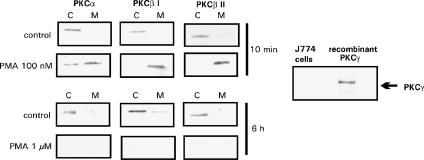
PKC isoenzyme expression in J774 macrophages and the effect of PMA on PKC isoenzyme translocation
Western blot with antibodies specific for cPKC isoenzymes (α, βI, βII and γ) were carried out. Resting J774 cells expressed three cPKC isoenzymes α, βI and βII, but PKCγ was not found (Figure 3). In the further studies, cells were treated with a PKC activator PMA (100 nM), and after 10 min incubation, all three isoenzymes were activated as measured by isoenzyme translocation from the cytosol to the membrane (Figure 3). In addition, incubation with a high concentration of PMA (1 μM) for 6 h resulted in the downregulation of all three PKC isoenzymes (Figure 3). Prolonged exposure to higher concentrations of phorbol esters, such as PMA, is known to cause almost complete downregulation of cPKCs and nPKCs, presumably as a result of proteolysis and it can be used as another means to downregulate PKC activity (Huang et al., 1989; Liu & Heckman, 1998).
Figure 3.
cPKC expression in J774 macrophages and the effects of PMA on PKC isoenzyme translocation. J774 cells were treated with 100 nM PMA or 1 μM PMA as indicated for 10 min or 6 h, respectively. Subsequent to preparation of cell lysates, the expression of individual PKC isoenzymes was assessed by immunoblotting with isoenzyme specific antibodies as outlined in the Methods section. Each experiment is a representative of three others with similar results. C=cytosolic fraction; M=membrane fraction. The expression of PKCγ in resting J774 macrophages was tested by Western blotting using recombinant human PKCγ as a positive control.
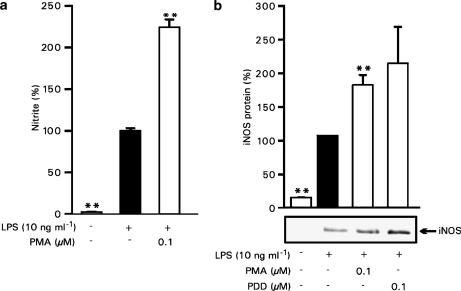
Effects of phorbol esters on LPS-induced NO production and iNOS protein expression
To further determine the participation of PKC in LPS-induced NO production and iNOS expression, we measured the effects of PMA on NO production and iNOS protein expression. When PMA was used at concentrations (100 nM) that activate PKC (Figure 3), it enhanced LPS-induced NO production and iNOS protein expression as shown in Figure 4a and b. Another phorbol ester, PDD, also enhanced iNOS protein expression, when it was used at 100 nM concentration (Figure 4b).
Figure 4.
Activation of PKC by phorbol esters induces iNOS protein expression and NO production in J774 cells. (a) J774 cells were stimulated by LPS (10 ng ml−1) and treated with PMA (100 nM) or vehicle (DMSO). After 24 h incubation, nitrite concentrations in the culture medium were measured as a marker of NO production. Values are mean±s.e.m. (n=6). (b) J774 cells were stimulated by LPS (10 ng ml−1) and treated with PMA (100 nM), PDD (100 nM) or vehicle. After 24 h, incubations were terminated and immunoblots were run using antibody against iNOS. Chemiluminescent signal was quantified as described under the Methods section. Values are mean±s.e.m. (n=3). **P<0.01 as compared with cells treated with LPS.
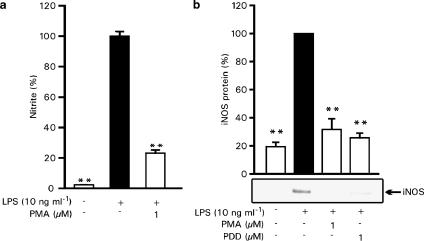
When the cells were pretreated with 1 μM PMA for 6 h before LPS addition (which was shown to downregulate PKC expression, see Figure 3), both LPS-induced NO production and iNOS protein expression were inhibited similarly to the effects of PKC inhibitors (Figure 5a and b). In addition, 6 h pretreatment with PDD (1 μM) had a similar suppressive effect on iNOS expression as 1 μM PMA (Figure 5b). These results further suggest that PKC is involved in the signalling mechanisms mediating LPS-induced iNOS expression and NO production.
Figure 5.
PKC downregulation by 6 h pretreatment with phorbol esters inhibits iNOS protein expression and NO production in J774 cells. (a) J774 cells were pretreated with PMA (1 μM) or vehicle for 6 h before stimulation by LPS (10 ng ml−1). After 24 h incubation, nitrite concentrations in the culture medium were measured as a marker of NO production. Values are mean±s.e.m. (n=6). (b) J774 cells were pretreated with PMA (1 μM), PDD (1 μM) or vehicle for 6 h before stimulation by LPS (10 ng ml−1). After 24 h, incubations were terminated and immunoblots were run using antibody against iNOS. Chemiluminescent signal was quantified as described under the Methods section. Values are mean±s.e.m. (n=3). **P<0.01 as compared with cells treated with LPS.
Effects of PKC inhibitors on iNOS mRNA expression
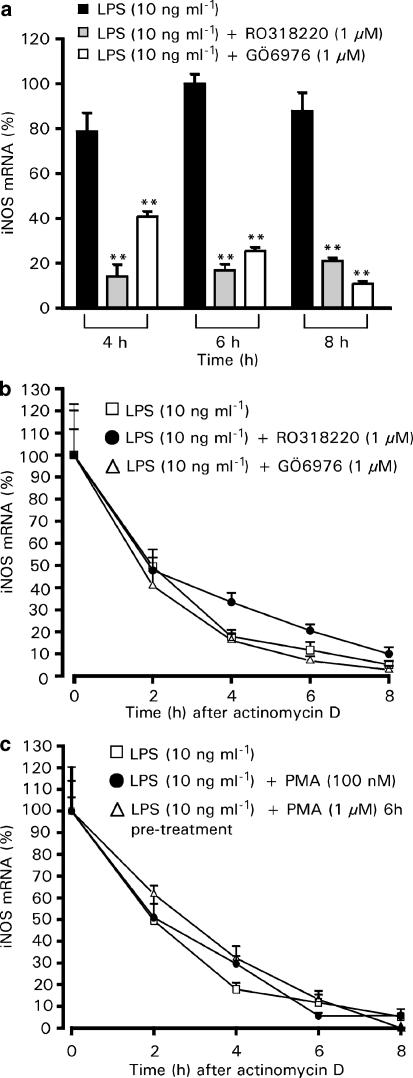
We used RT–PCR to investigate the effects of RO318220 and GÖ6976 on LPS-induced iNOS mRNA expression. In J774 macrophages, LPS-induced transient iNOS mRNA expression which had a 2 h lag phase and peaked at 6 h after the addition of LPS. We chose our time points 2 h before and after the 6 h peak to investigate the effects of PKC inhibitors on iNOS mRNA expression. At all measured time points, 4, 6 and 8 h after LPS induction, the iNOS mRNA levels were reduced in both RO318220- and GÖ6976-treated cells (Figure 6a). To determine whether PKC inhibitors reduce the half-life of iNOS mRNA, the cells were treated with LPS and the tested drugs, and after 6 h, transcription inhibitor actinomycin D (0.1 μg ml−1) was added into the culture. Cells were then further incubated for 0, 2, 4, 6 or 8 h before total RNA was extracted. As shown in Figure 6b, neither of the PKC inhibitors seemed to have any effect on iNOS mRNA half-life, nor did 100 nM PMA or pretreatment with 1 μM PMA (Figure 6c). These results suggest that the effect of cPKC isoenzymes on LPS-induced iNOS protein expression is mediated at the level of iNOS induction rather than at the level of post-transcriptional events.
Figure 6.
Effects of PKC inhibitors on iNOS mRNA expression and stability in J774 cells. (a) J774 cells were stimulated by LPS (10 ng ml−1) and treated with RO318220 (1 μM) or GÖ6976 (1 μM) for 4, 6 or 8 h. At indicated time points, the incubations were terminated and extracted total RNA was subjected to RT–PCR. iNOS mRNA levels were normalized against GAPDH mRNA. (b) Effect of PKC inhibitors on iNOS mRNA degradation. Cells were stimulated by LPS (10 ng ml−1) and treated with RO318220 (1 μM) or GÖ6976 (1 μM) for 6 h before the addition of actinomycin D (0.1 μg ml−1) to inhibit transcription. Incubations were terminated at indicated time points after actinomycin D and extracted total RNA was subjected to RT–PCR. iNOS mRNA levels were normalized against GAPDH mRNA. (c) Effect of PMA on iNOS mRNA degradation. Cells were treated with LPS (10 ng ml−1), with LPS (10 ng ml−1) and PMA (100 nM) to activate PKC or pretreated with PMA (1 μM) for 6 h to downregulate PKC before the addition of LPS (10 ng ml−1). After 6 h incubation with LPS, actinomycin D (0.1 μg ml−1) was added to inhibit transcription. Incubations were terminated at indicated time points after actinomycin D and extracted total RNA was subjected to RT–PCR. iNOS mRNA levels were normalized against GAPDH mRNA. Values are mean±s.e.m. (n=3). **P<0.01 as compared with cells treated with LPS only.
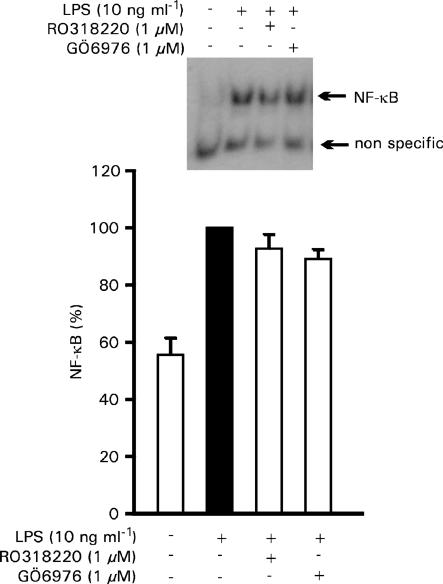
Effects of PKC inhibitors on transcription factors NF-κB and STAT1
To evaluate whether the effect of PKC inhibitors on iNOS mRNA expression levels could be a consequence of their effects on transcription factors, we measured the effects of RO318220 and GÖ6976 on the activation of NF-κB and STAT1, which are essential transcription factors for LPS-induced iNOS expression. The activation of NF-κB was measured by EMSA. RO318220 or GÖ6976 had no effect on NF-κB activation or binding activity (Figure 7). In contrast, when we investigated the effects of PKC inhibitors on STAT1 activation, as measured by the translocation of STAT1α from the cytosol to the nuclei by Western blot, both RO318220 and GÖ6976 inhibited STAT1α translocation (Figure 8a). In addition, the PKCβ-selective inhibitor LY333531 (5 μM), as well as pretreatment for 6 h with 1 μM PMA inhibited STAT1α translocation to the nuclei (Figure 8b and c). These data suggest that the effects of cPKC isoenzymes on LPS-induced iNOS protein expression are NF-κB-independent, but may well be mediated through the activation of transcription factor STAT1.
Figure 7.
Effect of PKC inhibitors on NF-κB activity. J774 cells were stimulated by LPS (10 ng ml−1) and treated with RO318220 (1 μM) or GÖ6976 (1 μM) for 30 min before the preparation of nuclear extracts. NF-κB DNA binding activity was analyzed by EMSA. Densities of specific bands were quantified as described under the Methods section. Values are mean±s.e.m. (n=3).
Figure 8.
The effect of PKC inhibitors and PMA pretreatment on STAT1α translocation. J774 cells were stimulated by LPS (10 ng ml−1) and treated with RO318220 (1 μM), GÖ6976 (1 μM) (a) or LY333531 (5 μM) (b) for 6 h before the preparation of nuclear extracts. STAT1α translocation to the nuclei was determined by Western blotting using specific antibody against STAT1α. (c) Cells were pretreated with PMA (1 μM) or vehicle (DMSO) for 6 h before stimulation with LPS (10 ng ml−1). Cells were further incubated for 6 h before the nuclear extracts were prepared. STAT1α translocation to the nuclei was determined by Western blotting using specific antibody against STAT1α. Chemiluminescent signal was quantified as described under the Methods section. Values are mean±s.e.m. (n=3). *P<0.05, **P<0.01 as compared with cells treated with LPS only.
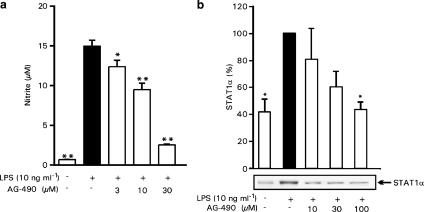
Effects of JAK-2 inhibitor AG-490 on LPS-induced NO production and STAT1 activation
To further investigate the role of STAT1 on LPS-induced NO production, we used JAK-2 inhibitor AG-490. JAK-2 (Janus kinase-2) is an upstream kinase of STAT1 and inhibition of JAK-2 leads to the inhibition of STAT1 (Shuai & Liu, 2003). AG-490 inhibited LPS-induced NO production in a dose-dependent manner (Figure 9a). Same concentrations that downregulated LPS-induced NO production also inhibited LPS-induced STAT1 activation, as measured by the translocation of STAT1α from the cytosol to the nuclei by Western blot (Figure 9b). These results further suggest that the effects of cPKC isoenzymes on iNOS expression and NO production could be mediated through the activation of STAT1.
Figure 9.
Effect of JAK-2 inhibitor AG-490 on LPS-induced NO production in J774 cells. J774 cells were stimulated by LPS (10 ng ml−1) and treated with increasing concentrations of AG-490 (a). After 24 h incubation, nitrite concentrations in the culture medium were measured as a marker of NO production. Values are mean±s.e.m. (n=6). (b) The effect of JAK-2 inhibitor AG-490 on STAT1α nuclear translocation. J774 cells were stimulated by LPS (10 ng ml−1) and treated with increasing concentrations of AG-490 for 4 h before the preparation of nuclear extracts. STAT1α translocation to the nucleus was determined by Western blotting using specific antibody against STAT1α. Chemiluminescent signal was quantified as described under the Methods section. Values are mean±s.e.m. (n=3). *P<0.05, **P<0.01 as compared with cells treated with LPS only.
Discussion
In the present study, we show that inhibition of classical isoenzymes, especially PKCβ, inhibits LPS-induced iNOS expression and NO production in activated J774 macrophages, and that this effect is possibly mediated through the inhibition of transcription factor STAT1.
Distribution of PKC isoenzymes is cell type- and tissue-specific. PKCα, βI, βII, δ, ɛ and ζ seem to be ubiquitous isoenzymes, and are found in most tissues (Liu & Heckman, 1998). Classical isoenzyme PKCγ is largely restricted to the central nervous system and spinal cord (Liu & Heckman, 1998; Way et al., 2000). In the present study, we focused on cPKCs and found that PKCα, PKCβI and PKCβII are expressed in macrophage cultures used. PKC regulates various inflammatory functions in an isoenzyme-specific manner (Tan & Parker, 2003). The regulation of cell signalling events by single PKC isoenzymes have also been shown to differ between cell types (Paul et al., 1997).
In the present study, four PKC inhibitors with different PKC isoenzyme profiles were used to study the role of PKC and its classical isoenzymes in LPS-induced iNOS protein expression and NO production in macrophages. PKC inhibitors RO318220, GÖ6976 and LY333531 inhibited LPS-induced iNOS expression and NO production in a dose-dependent manner. RO318220 is a PKC inhibitor, which inhibits isoenzymes β, γ and ɛ (Davis et al., 1992; Wilkinson et al., 1993), GÖ6976 is more selective to cPKC isoenzymes (Martiny-Baron et al., 1993) and LY333531 is a selective inhibitor of PKCβ (Jirousek et al., 1996). HBDDE, which is a relatively selective inhibitor of PKCα and PKCγ (Kashiwada et al., 1994), did not have any effect on LPS-induced NO production. These results suggest that PKC, probably its isoenzymes PKCβI and PKCβII, mediate the LPS-induced upregulation of iNOS expression and NO production in macrophages.
The results may be complicated by the fact that RO318220 and GÖ6976 have been reported to inhibit some other kinases in addition to PKC (Davies et al., 2000). Therefore, we also studied the effects of phorbol esters PMA and PDD on LPS-induced iNOS expression and NO production. Phorbol esters are known to activate cPKC isoenzymes (Castagna et al., 1982), whereas a longer pretreatment with a higher concentration of phorbol esters have been shown to result in the downregulation of cPKCs, presumably due to proteolysis (Huang et al., 1989; Liu & Heckman, 1998). The bidirectional effect of PMA and PDD on cPKCs was seen also in the present study. PMA and PDD at 100 nM concentration enhanced cPKC activation, while 6 h pretreatment with 1 μM concentration of PMA or PDD suppressed cPKC expression. When used at cPKC-activating concentrations, PMA and PDD enhanced iNOS expression and NO production. In contrast, cPKC downregulation due to 6 h pretreatment with PMA or PDD (1 μM) resulted in the suppression of iNOS expression and NO production, similarly as inhibition of cPKCs by pharmacological means. These results further support the role of cPKC isoenzymes in the regulation of iNOS expression and NO production in macrophages.
To determine the mechanisms by which the regulation by PKC is mediated, we studied the effects of PKC inhibitors RO318220 and GÖ6976, and PMA on LPS-induced iNOS mRNA expression and mRNA stability. Our results show that inhibition of cPKCs does not effect the stability of LPS-induced iNOS mRNA. The effect of PKC isoenzymes is rather at the level of iNOS transcription, since PKC inhibitors decreased the expression of iNOS mRNA already at the early time points after addition of LPS.
NF-κB and STAT1 appear to be important transcription factors for the enhanced iNOS gene expression in macrophages exposed to LPS (Lowenstein et al., 1993; Chartrain et al., 1994; Xie et al., 1994; Gao et al., 1998; Jacobs & Ignarro, 2001). In the present study, all three PKC inhibitors used (RO318220, GÖ6976 and LY333531) inhibited STAT1 activation as measured by translocation of the transcription factor from the cytosol to nuclei as did pretreatment with 1 μM PMA. In contrast, none of the treatments did inhibit NF-κB activation as measured by EMSA. These results suggest that the regulation of LPS-induced iNOS protein expression by PKC is NF-κB-independent and is most likely mediated through the activation of transcription factor STAT1.
RO318220 and GÖ6976 have earlier been reported to inhibit LPS and IFN-γ-induced NO production in macrophages (Paul et al., 1997; Chen, B.C. et al., 1998; Chen et al., 1998a). In addition, PKCδ and PKCη have been suggested to regulate NO production and iNOS expression in activated macrophages and some other cell types (Chen et al., 1998a, 1998b; Carpenter et al., 2001; Banan et al., 2003; Pham et al., 2003). The present study extends the earlier data by providing a cellular mechanism for the inhibitory effects of RO318220 and GÖ6976 on LPS-induced iNOS expression and NO production in macrophages. Our results show that inhibition of cPKC isoenzymes results in the suppression of STAT1 activation, which may well explain the inhibitory effect on iNOS expression and NO production. In addition, we were able to show that LY333531, a selective inhibitor of PKCβ, also suppressed STAT1 activation and iNOS expression, supporting the role of PKCβ in the regulation of iNOS expression in activated macrophages.
In conclusion, the present results show that inhibition of cPKC isoenzymes, especially PKCβ, inhibits the LPS-induced activation of transcription factor STAT1, iNOS expression and NO production in macrophages. The results suggest that inhibition of cPKC isoenzymes provides a way to prevent iNOS protein expression and NO production in inflammation, offering a novel target for the development of anti-inflammatory drugs.
Acknowledgments
We wish to thank Mrs Niina Ikonen for her technical assistance and Mrs Heli Määttä for her secretarial help. The study was supported by grants from the Academy of Finland, from the National Technology Agency of Finland and from the Medical Research Fund of Tampere University Hospital.
Abbreviations
- EMSA
electrophoretic mobility shift assay
- iNOS
inducible nitric oxide synthase
- LPS
lipopolysaccharide
- NO
nitric oxide
- PDD
phorbol 12,13-didecanoate
- PKC
protein kinase C
- PMA
phorbol 12-myristate 13-acetate
- STAT1
signal transducer and activator of transcription 1
References
- AKSOY E., GOLDMAN M., WILLEMS F. Protein kinase C epsilon: a new target to control inflammation and immune-mediated disorders. Int. J. Biochem. Cell Biol. 2004;36:183–188. doi: 10.1016/s1357-2725(03)00210-3. [DOI] [PubMed] [Google Scholar]
- BANAN A., FARHADI A., FIELDS J.Z., ZHANG L.J., SHAIKH M., KESHAVARZIAN A. The δ-isoform of protein kinase C causes inducible nitric-oxide synthase and nitric oxide up-regulation: key mechanism for oxidant-induced carbonylation, nitration, and disassembly of the microtubule cytoskeleton and hyperpermeability of barrier of intestinal epithelia. J. Pharmacol. Exp. Ther. 2003;305:482–494. doi: 10.1124/jpet.102.047308. [DOI] [PubMed] [Google Scholar]
- BOWLING N., WALSH R.A., SONG G., ESTRIDGE T., SANDUSKY G.E., FOUTS R.L., MINTZE K., PICKARD T., RODEN R., BRISTOW M.R., SABBAH H.N., MIZRAHI J.L., GROMO G., KING G.L., VLAHOS C.J. Increased protein kinase C activity and expression of Ca2+-sensitive isoforms in the failing human heart. Circulation. 1999;99:384–391. doi: 10.1161/01.cir.99.3.384. [DOI] [PubMed] [Google Scholar]
- BRADFORD M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- CARPENTER L., CORDERY D., BIDEN T.J. Protein kinase Cδ activation by interleukin-1β stabilizes inducible nitric-oxide synthase mRNA in pancreatic β-cells. J. Biol. Chem. 2001;276:5368–5374. doi: 10.1074/jbc.M010036200. [DOI] [PubMed] [Google Scholar]
- CASTAGNA M., TAKAI Y., KAIBUCHI K., SANO K., KIKKAWA U., NISHIZUKA Y. Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J. Biol. Chem. 1982;257:7847–7851. [PubMed] [Google Scholar]
- CASTRILLO A., PENNINGTON D.J., OTTO F., PARKER P.J., OWEN M.J., BOSCA L. Protein kinase Cɛ is required for macrophage activation and defense against bacterial infection. J. Exp. Med. 2001;194:1231–1242. doi: 10.1084/jem.194.9.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHARTRAIN N.A., GELLER D.A., KOTY P.P., SITRIN N.F., NUSSLER A.K., HOFFMAN E.P., BILLIAR T.R., HUTCHINSON N.I., MUDGETT J.S. Molecular cloning, structure, and chromosomal localization of the human inducible nitric oxide synthase gene. J. Biol. Chem. 1994;269:6765–6772. [PubMed] [Google Scholar]
- CHEN B.C., CHOU C.F., LIN W.W. Pyrimidinoceptor-mediated potentiation of inducible nitric-oxide synthase induction in J774 macrophages. Role of intracellular calcium. J. Biol. Chem. 1998;273:29754–29763. doi: 10.1074/jbc.273.45.29754. [DOI] [PubMed] [Google Scholar]
- CHEN C.C., WANG J.K., LIN S.B. Antisense oligonucleotides targeting protein kinase C-α, -βI, or -δ but not -η inhibit lipopolysaccharide-induced nitric oxide synthase expression in RAW 264.7 macrophages: involvement of a nuclear factor κB-dependent mechanism. J. Immunol. 1998a;161:6206–6214. [PubMed] [Google Scholar]
- CHEN C.C., WANG J.K., CHEN W.C., LIN S.B. Protein kinase Cη mediates lipopolysaccharide-induced nitric-oxide synthase expression in primary astrocytes. J. Biol. Chem. 1998b;273:19424–19430. doi: 10.1074/jbc.273.31.19424. [DOI] [PubMed] [Google Scholar]
- CHEN L., HAHN H., WU G., CHEN C.H., LIRON T., SCHECHTMAN D., CAVALLARO G., BANCI L., GUO Y., BOLLI R., DORN G.W., II, MOCHLY-ROSEN D. Opposing cardioprotective actions and parallel hypertrophic effects of δPKC and ɛPKC. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11114–11119. doi: 10.1073/pnas.191369098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIES S.P., REDDY H., CAIVANO M., COHEN P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS P.D., HILL C.H., LAWTON G., NIXON J.S., WILKINSON S.E., HURST S.A., KEECH E., TURNER S.E. Inhibitors of protein kinase C. 1. 2, 3-bisarylmaleimides. J. Med. Chem. 1992;35:177–184. doi: 10.1021/jm00079a024. [DOI] [PubMed] [Google Scholar]
- FOEY A.D., BRENNAN F.M. Conventional protein kinase C and atypical protein kinase Cζ differentially regulate macrophage production of tumour necrosis factor-α and interleukin-10. Immunology. 2004;112:44–53. doi: 10.1111/j.1365-2567.2004.01852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUJIHARA M., CONNOLLY N., ITO N., SUZUKI T. Properties of protein kinase C isoforms (βII, ɛ, and ζ) in a macrophage cell line (J774) and their roles in LPS-induced nitric oxide production. J. Immunol. 1994;152:1898–1906. [PubMed] [Google Scholar]
- GAO J.J., FILLA M.B., FULTZ M.J., VOGEL S.N., RUSSELL S.W., MURPHY W.J. Autocrine/paracrine IFN-αβ mediates the lipopolysaccharide-induced activation of transcription factor Stat1α in mouse macrophages: pivotal role of Stat1α in induction of the inducible nitric oxide synthase gene. J. Immunol. 1998;161:4803–4810. [PubMed] [Google Scholar]
- GIROUX M., DESCOTEAUX A. Cyclooxygenase-2 expression in macrophages: modulation by protein kinase C-α. J. Immunol. 2000;165:3985–3991. doi: 10.4049/jimmunol.165.7.3985. [DOI] [PubMed] [Google Scholar]
- GOEKJIAN P.G., JIROUSEK M.R. Protein kinase C inhibitors as novel anticancer drugs. Exp. Opin. Invest. Drugs. 2001;10:2117–2140. doi: 10.1517/13543784.10.12.2117. [DOI] [PubMed] [Google Scholar]
- GREEN L.C., WAGNER D.A., GLOGOWSKI J., SKIPPER P.L., WISHNOK J.S., TANNENBAUM S.R. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal. Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- HUANG F.L., YOSHIDA Y., CUNHA-MELO J.R., BEAVEN M.A., HUANG K.P. Differential down-regulation of protein kinase C isozymes. J. Biol. Chem. 1989;264:4238–4243. [PubMed] [Google Scholar]
- JACOBS A.T., IGNARRO L.J. Lipopolysaccharide-induced expression of interferon-β mediates the timing of inducible nitric-oxide synthase induction in RAW 264.7 macrophages. J. Biol. Chem. 2001;276:47950–47957. doi: 10.1074/jbc.M106639200. [DOI] [PubMed] [Google Scholar]
- JIROUSEK M.R., GILLIG J.R., GONZALEZ C.M., HEATH W.F., MCDONALD J.H., III, NEEL D.A., RITO C.J., SINGH U., STRAMM L.E., MELIKIAN-BADALIAN A., BAEVSKY M., BALLAS L.M., HALL S.E., WINNEROSKI L.L., FAUL M.M. (S)-13-[(dimethylamino)methyl]-10, 11, 14, 15-tetrahydro-4, 9:16, 21-dimetheno-1H, 13H-dibenzo[e, k]pyrrolo[3, 4-h][1, 4, 13]oxadiazacyclohexadecene-1, 3(2H)-dione ( LY333531) and related analogues: isozyme selective inhibitors of protein kinase Cβ. J. Med. Chem. 1996;39:2664–2671. doi: 10.1021/jm950588y. [DOI] [PubMed] [Google Scholar]
- KANG J., YANG M., JOU I., JOE E. Identification of protein kinase C isoforms involved in interferon-gamma-induced expression of inducible nitric oxide synthase in murine BV2 microglia. Neurosci. Lett. 2001;299:205–208. doi: 10.1016/s0304-3940(01)01515-4. [DOI] [PubMed] [Google Scholar]
- KASHIWADA Y., HUANG L., BALLAS L.M., JIANG J.B., JANZEN W.P., LEE K.H. New hexahydroxybiphenyl derivatives as inhibitors of protein kinase C. J. Med. Chem. 1994;37:195–200. doi: 10.1021/jm00027a025. [DOI] [PubMed] [Google Scholar]
- KLEINERT H., SCHWARZ P.M., FÖRSTERMANN U. Regulation of the expression of inducible nitric oxide synthase. Biol. Chem. 2003;384:1343–1364. doi: 10.1515/BC.2003.152. [DOI] [PubMed] [Google Scholar]
- KORHONEN R., LAHTI A., KANKAANRANTA H., MOILANEN E. Nitric oxide production and signalling in inflammation. Curr. Drug Targets. 2005;4:471–479. doi: 10.2174/1568010054526359. [DOI] [PubMed] [Google Scholar]
- LAHTI A., JALONEN U., KANKAANRANTA H., MOILANEN E. c-Jun NH2-terminal kinase inhibitor anthra(1, 9-cd)pyrazol-6(2H)-one reduces inducible nitric-oxide synthase expression by destabilizing mRNA in activated macrophages. Mol. Pharmacol. 2003;64:308–315. doi: 10.1124/mol.64.2.308. [DOI] [PubMed] [Google Scholar]
- LAHTI A., KANKAANRANTA H., MOILANEN E. p38 mitogen-activated protein kinase inhibitor SB203580 has a bi-directional effect on iNOS expression and NO production. Eur. J. Pharmacol. 2002;454:115–123. doi: 10.1016/s0014-2999(02)02490-1. [DOI] [PubMed] [Google Scholar]
- LIU W.S., HECKMAN C.A. The sevenfold way of PKC regulation. Cell Signal. 1998;10:529–542. doi: 10.1016/s0898-6568(98)00012-6. [DOI] [PubMed] [Google Scholar]
- LOWENSTEIN C.J., ALLEY E.W., RAVAL P., SNOWMAN A.M., SNYDER S.H., RUSSELL S.W., MURPHY W.J. Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferon γ and lipopolysaccharide. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9730–9734. doi: 10.1073/pnas.90.20.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACMICKING J., XIE Q.W., NATHAN C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- MARTINY-BARON G., KAZANIETZ M.G., MISCHAK H., BLUMBERG P.M., KOCHS G., HUG H., MARME D., SCHÄCHTELE C. Selective inhibition of protein kinase C isozymes by the indolocarbazole Gö 6976. J. Biol. Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- MOILANEN E., WHITTLE B.J.R., MONCADA S.Nitric oxide as a factor in inflammation Inflammation: Basic principles and Clinical Correlates 1999Philadelphia: Lippincott, Williams & Wilkins; 787–800.ed. Gallin, J.I. & Snyderman, R. pp [Google Scholar]
- MOLINA-HOLGADO E., ORTIZ S., MOLINA-HOLGADO F., GUAZA C. Induction of COX-2 and PGE2 biosynthesis by IL-1β is mediated by PKC and mitogen-activated protein kinases in murine astrocytes. Br. J. Pharmacol. 2000;131:152–159. doi: 10.1038/sj.bjp.0703557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEWTON A.C. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- PAUL A., DOHERTY K., PLEVIN R. Differential regulation by protein kinase C isoforms of nitric oxide synthase induction in RAW 264.7 macrophages and rat aortic smooth muscle cells. Br. J. Pharmacol. 1997;120:940–946. doi: 10.1038/sj.bjp.0700976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHAM T.N.Q., BROWN B.L., DOBSON P.R.M., RICHARDSON V.J. Protein kinase C-eta (PKC-η) is required for the development of inducible nitric oxide synthase (iNOS) positive phenotype in human monocytic cells. Nitric Oxide. 2003;9:123–134. doi: 10.1016/j.niox.2003.09.006. [DOI] [PubMed] [Google Scholar]
- SHUAI K., LIU B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 2003;3:900–911. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- SPITALER M., CANTRELL D.A. Protein kinase C and beyond. Nat. Immunol. 2004;5:785–790. doi: 10.1038/ni1097. [DOI] [PubMed] [Google Scholar]
- ST-DENIS A., CHANO F., TREMBLAY P., ST-PIERRE Y., DESCOTEAUX A. Protein kinase C-α modulates lipopolysaccharide-induced functions in a murine macrophage cell line. J. Biol. Chem. 1998;273:32787–32792. doi: 10.1074/jbc.273.49.32787. [DOI] [PubMed] [Google Scholar]
- TAN S.L., PARKER P.J. Emerging and diverse roles of protein kinase C in immune cell signalling. Biochem. J. 2003;376:545–552. doi: 10.1042/BJ20031406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEPPERMAN B.L., CHANG Q., SOPER B.D. Protein kinase C mediates lipopolysaccharide- and phorbol-induced nitric-oxide synthase activity and cellular injury in the rat colon. J. Pharmacol. Exp. Ther. 2000;295:1249–1257. [PubMed] [Google Scholar]
- VALLANCE P., LEIPER J. Blocking NO synthesis: how, where and why. Nat. Rev. Drug. Discov. 2002;1:939–950. doi: 10.1038/nrd960. [DOI] [PubMed] [Google Scholar]
- WAY K.J., CHOU E., KING G.L. Identification of PKC-isoform-specific biological actions using pharmacological approaches. Trends Pharmacol. Sci. 2000;21:181–187. doi: 10.1016/s0165-6147(00)01468-1. [DOI] [PubMed] [Google Scholar]
- WILKINSON S.E., PARKER P.J., NIXON J.S. Isoenzyme specificity of bisindolylmaleimides, selective inhibitors of protein kinase C. Biochem. J. 1993;294:335–337. doi: 10.1042/bj2940335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XIE Q.W., KASHIWABARA Y., NATHAN C. Role of transcription factor NF-κB/Rel in induction of nitric oxide synthase. J. Biol. Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]