Abstract
We report a novel release of the GermOnline knowledgebase covering genes relevant for the cell cycle, gametogenesis and fertility. GermOnline was extended into a cross-species systems browser including information on DNA sequence annotation, gene expression and the function of gene products. The database covers eight model organisms and Homo sapiens, for which complete genome annotation data are available. The database is now built around a sophisticated genome browser (Ensembl), our own microarray information management and annotation system (MIMAS) used to extensively describe experimental data obtained with high-density oligonucleotide microarrays (GeneChips) and a comprehensive system for online editing of database entries (MediaWiki). The RNA data include results from classical microarrays as well as tiling arrays that yield information on RNA expression levels, transcript start sites and lengths as well as exon composition. Members of the research community are solicited to help GermOnline curators keep database entries on genes and gene products complete and accurate. The database is accessible at http://www.germonline.org/.
INTRODUCTION
Molecular biologists and biomedical researchers are confronted with the complex task of processing and interpreting different types of high-throughput genome biological data. Our goal is to facilitate scientific hypothesis building on the basis of comprehensive and accurate knowledgebase entries associated with curated and relevant experimental data. To this end, we have developed the GermOnline cross-species database, an integrated solution for displaying annotated genome sequence information and microarray expression profiling data alongside manually curated knowledge about gene function (1–3). The current release is not a simple update of the previous versions but is based on a novel concept, a different software architecture and an improved graphical user interface. To achieve our aims we harness the extensive capabilities of existing open-source software including the Ensembl genome browser environment (4) with extensions for displaying expression data, the BioMart query system (5) and our own Microarray Information Management and Annotation System (MIMAS) (6). Finally, we implement the MediaWiki online page editing package (http://www.mediawiki.org/) originally developed for the community encyclopedia Wikipedia. Wiki has recently been discussed as a possible approach to biological data management by professional curators together with life scientists (7–9). GermOnline and GermOnline Wiki are available without restrictions at http://www.germonline.org/.
THE SCOPE OF GERMONLINE
GermOnline focuses on information relevant for the cell cycle and gametogenesis in eight key model organisms and Homo sapiens. Since we now include the genome sequences in the database (as opposed to just storing gene identifiers), we selected nine species for which sequenced and fully annotated genomes are available including six (Saccharomyces cerevisiae, Arabidopsis thaliana, Drosophila melanogaster, Rattus norvegicus, Mus musculus and Homo sapiens) that were investigated in high-density oligonucleotide microarray expression profiling studies. The current version of GermOnline therefore lacks three species covered in earlier releases for which complete and fully annotated genome sequence is currently unavailable (Neurospora crassa, Zea mays, Xenopus laevis).
The Ensembl system lends itself well to extension and customization (4). Thus expression data and phylogenetic information could be integrated and displayed while retaining a uniform display. Importantly, moving into a genome-browser environment enabled us to show tiling array data in the context of exon-intron composition at the DNA and RNA level. This is important because tiling arrays contain probes covering the entire genome sequence of a target organism and they are therefore independent of any previous genome annotation. These arrays also detect non-coding RNAs that are difficult or even impossible to trace with classical arrays whose oligonucleotide probes are most often directed against 3′-untranslated sequences or the coding regions of defined genes (10,11).
Lastly, researchers are provided with manually curated database entries describing mechanistic knowledge about genes and gene products important for sexual reproduction. Scientists who work on these genes are solicited to update and extend the entries to help keep them accurate and complete. This process is meant to be carried out in cooperation with our professional scientific curators who establish entries, update them and provide online support for users. The system is based on MediaWiki. Following registration, a pre-requisite for online editing, extensive and convenient text editing options are available for community annotation.
HOW TO RETRIEVE INFORMATION
Search
GermOnline is extensively searchable using gene symbols, standard and systematic names as well as various identifiers from UniProt, Ensembl and GenBank. A complex search form, the GermOnline BioMart, allows users to retrieve gene lists on the basis of functional genomic and expression studies. Recently integrated studies include a comparative mammalian meiotic transcriptome analysis based on rodent and human samples (F. Chalmel, A. Rolland, C. Niederhauser-Wiederkehr, S.S. Chung, P. Demougin, A. Gattiker, J. Moore, J.J. Patard, D.J. Wolgemuth, B. Jégou and M. Primig, submitted). Queries are also possible using experiments with budding yeast based on classical arrays (12,13) as well as high-throughput gene deletion studies (see http://www.germonline.org/Multi/martview) (14–17). This useful feature permits searches across different experiments yielding lists of genes with particular expression patterns and phenotypes.
The locus report
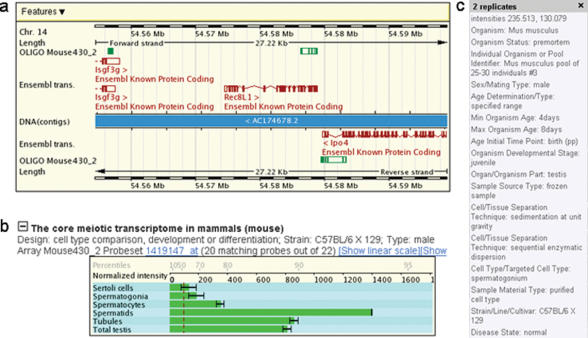
Users can call up information on a given locus by using various queries including gene symbols and identification numbers. The report page covers the exon-intron structure, GeneChip probe target sequences as well as extensive graphical display of relevant microarray expression data. Examples of genomic DNA annotation data and GeneChip target sequences (for yeast S98) as well as GeneChip expression data are shown for REC8 from S.cerevisiae (Figure 1) and Rec8L1 from M.musculus (Figure 2). The expression data for the orthologues can be directly compared because they were obtained in budding yeast cells (12) and mammalian germ cells undergoing meiotic differentiation (F. Chalmel, A. Rolland, C. Niederhauser-Wiederkehr, S.S. Chung, P. Demougin, A. Gattiker, J. Moore, J.J. Patard, D.J. Wolgemuth, B. Jégou and M. Primig, submitted; http://www.biozentrum.unibas.ch/primig/mam_sperm). In addition, a substantial number of new profiling studies using mouse and rat (18–21) testicular samples as well as sporulating yeast cultures (22) are available. The array experiments are extensively described in a MIAME-compliant manner and users can call up more detailed information by clicking on the sample name (Figure 2,c). The database report page contains links to PubMed, GEO and ArrayExpress, as well as to supporting web portals whenever available, to provide information about the source of array data on display.
Figure 1.
Genome annotation and S98 GeneChip expression profiling data for REC8 in budding yeast. (a) The chromosome number is indicated. On a genomic map, REC8 and its neighbouring genes as annotated by SGD, and the gene-specific S98 array probe target sequences (OLIGO YG-S98) are shown in red and green, respectively. (b) Normalized linear (or log2 transformed) expression signals obtained with S98 arrays and samples from budding yeast growing in rich medium with acetate as the sole carbon source (YPA) and sporulation medium at different time points as indicated, are shown in blue and green, respectively, in a bar diagram. Expression signals of individual genes are correlated with all other signals on the array via the percentile scale. The dotted red line indicates an empirical signal threshold level for background noise.
Figure 2.
Genome annotation and MG 430 2.0 GeneChip expression profiling data for Rec8L1 in mouse. The chromosome number is indicated. (a) On a genomic map, Rec8L1 and its neighbouring genes as annotated by Ensembl, and the gene-specific MG 430 2.0 array probe target sequences (OLIGO Mouse430_2) are shown in red and green, respectively. (b) Normalized linear (or log2 transformed) expression signals obtained with MG430 2.0 arrays and testicular mouse samples as indicated are shown in a bar diagram. Expression signals of individual genes are correlated with all other signals on the array via the percentile scale. The dotted red line indicates an empirical signal threshold level for background noise.
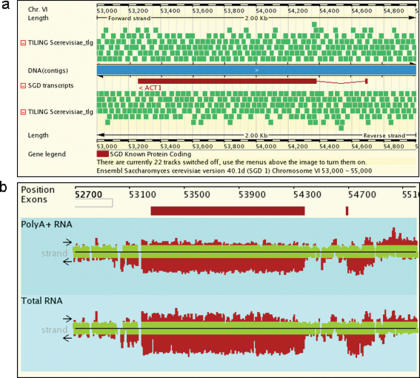
The report page now includes data from novel tiling arrays whose probes cover the entire genome sequence of a target organism such as budding yeast. A typical signal from mitotically growing yeast is shown for ACT1 whose exons are detected by the array with remarkable accuracy (Figure 3) (10).
Figure 3.
Genome annotation and tiling array expression profiling data for ACT1 in budding yeast. The chromosome number is indicated. (a) On a genomic map, the two exons in ACT1 and the tiling array target sequences (TILING Scerevisiae_tlg) are shown in red and light green, respectively. Normalized and transformed expression signals obtained with tiling arrays and samples from budding yeast growing in rich medium (YPD) are shown for two different RNA isolation protocols as indicated (25).
GermOnline report pages now also include phylogenetic data from the Ensembl Compara database, and from the Orthologous Matrix Project (OMA, http://www.cbrg.ethz.ch/research/orthologous/) that aims at building phylogenetic trees of species using a conservative approach to identify ‘true’ orthologs (23,24). This feature provides efficient cross-connection between locus report pages of conserved genes from different species represented in the database. Users can therefore quickly call up genome annotation information, expression data and (where available), manually curated data on related genes that may well fulfill similar functions across species.
ONLINE ESTABLISHING AND EDITING OF DATABASE ENTRIES
Establishing a new entry
The GermOnline Wiki provides the research communities working on the cell cycle, gametogenesis and gamete function with a comprehensive online editing system to write database entries that cover relevant genes across species (http://wiki.germonline.org/). Our goal is to build entries that are complete and accurate and that can be easily updated. Scientists are solicited to edit, amend and correct pages as they deem appropriate, using free text and references to appropriate peer-reviewed publications. We provide researchers with the MediaWiki system that is also used by the highly successful online information source Wikipedia (http://www.wikipedia.org/). This community resource was recently found by experts to be as reliable as Encyclopaedia Britannica as far as scientific topics are concerned (8). Moreover, Wiki-like approaches to knowledge management in biomedical sciences are currently being discussed as a way of motivating the research community to participate in genome annotation (7,9). Registered users of the GermOnline Wiki can establish new pages for genes represented in the database for which no community annotation is available as yet. This is possible via the locus report page by following the link ‘contribute your knowledge to GermOnline’ located in the ‘Community annotation’ section. GermOnline Wiki prompts users to register via an extremely brief process and to log in, leading them to an empty text field. The information content of a page consists of free text and images or figures that can be copied from published papers. Most journals do not own the copyright and allow re-publication of images as long as they are appropriately referenced. In other cases, GermOnline has obtained permission to re-publish images.
Editing an existing entry
The GermOnline report page includes a prominent community annotation window enabling users to browse through the corresponding GermOnline Wiki entry. Scientists can edit the entry by following the ‘enlarge window’ link. Examples for REC8 from S.cerevisiae (Figure 4, a and b) and Rec8L1 from M.musculus are given (Figure 4c). Several hundred entries on genes that were written using the previous submission/curation system, were migrated into the new environment and can be accessed and updated. A history page enables users to trace changes and authors who made them and extensive online help is available via the Wiki pages. Any edited page is visited by GermOnline curators within one business day for verification purposes and to prevent vandalism.
Figure 4.
Gene annotation using GermOnline Wiki. (a) Examples for an insert into the locus report page and (b) full-size Wiki pages covering REC8 from yeast and (c) Rec8L1 from mouse are shown.
FUTURE DEVELOPMENT
We intend to integrate relevant classical and tiling microarray data as they get published for all species represented in the database. Furthermore, in coordination with the ongoing development of MIMAS, we will include genome-wide protein-DNA interaction data (ChIP-Chip assays) and, where available, high-throughput data on protein structures and protein interaction networks as well as pathways.
CONCLUSION
We report the development of an innovative approach to knowledge management that combines the advanced capabilities of specific solutions for genome browsing (Ensembl), phylogenetic analysis (OMA), microarray data annotation (MIMAS) and online editing of database entries (MediaWiki). Based on our previous experience with GermOnline we conclude that any significant input from life scientists requires an efficient and flexible system that permits researchers to do what they are trained to do: provide a concise and precise summary of mechanistic knowledge established for a given gene product using free text, figures and original references. Since time is a critical factor it is crucial that any online editing system allows easy access and does not require extensive usage of complex forms. MediaWiki as implemented in GermOnline is an ideal solution for this purpose. The time for large-scale community annotation involving regular contributions by most, if not all scientists may not yet have come. However, we believe that it is important to provide appropriate prototype tools early on to help change the way biological and biomedical researchers produce and disseminate knowledge in the not too distant future.
Acknowledgments
We thank R. Poehlmann for systems administration and the Ensembl team members for providing outstanding services. We gratefully acknowledge the guidance and support of the members of the International Board. A.G. and J.M. are supported by the Swiss Institute of Bioinformatics. Funding to pay the Open Access publication charges for this article was provided by the Swiss Institute of Bioinformatics.
Conflict of interest statement. None declared.
REFERENCES
- 1.Primig M., Wiederkehr C., Basavaraj R., Sarrauste de Menthiere C., Hermida L., Koch R., Schlecht U., Dickinson H.G., Fellous M., Grootegoed J.A., et al. GermOnline, a new cross-species community annotation database on germ-line development and gametogenesis. Nature Genet. 2003;35:291–292. doi: 10.1038/ng1203-291. [DOI] [PubMed] [Google Scholar]
- 2.Wiederkehr C., Basavaraj R., Sarrauste de Menthiere C., Hermida L., Koch R., Schlecht U., Amon A., Brachat S., Breitenbach M., Briza P., et al. GermOnline, a cross-species community knowledgebase on germ cell differentiation. Nucleic Acids Res. 2004;32:D560–D567. doi: 10.1093/nar/gkh055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiederkehr C., Basavaraj R., Sarrauste de Menthiere C., Koch R., Schlecht U., Hermida L., Masdoua B., Ishii R., Cassen V., Yamamoto M., et al. Database model and specification of GermOnline Release 2.0, a cross-species community annotation knowledgebase on germ cell differentiation. Bioinformatics. 2004;20:808–811. doi: 10.1093/bioinformatics/bth030. [DOI] [PubMed] [Google Scholar]
- 4.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasprzyk A., Keefe D., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermida L., Schaad O., Demougin P., Descombes P., Primig M. MIMAS: an innovative tool for network-based high density oligonucleotide microarray data management and annotation. BMC Bioinformatics. 2006;7:190. doi: 10.1186/1471-2105-7-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caddick S. Wiki and other ways to share learning online. Nature. 2006;442:744. doi: 10.1038/442744c. [DOI] [PubMed] [Google Scholar]
- 8.Giles J. Internet encyclopaedias go head to head. Nature. 2005;438:900–901. doi: 10.1038/438900a. [DOI] [PubMed] [Google Scholar]
- 9.Yager K. Wiki ware could harness the Internet for science. Nature. 2006;440:278. doi: 10.1038/440278a. [DOI] [PubMed] [Google Scholar]
- 10.David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M. A high-resolution map of transcription in the yeast genome. Proc. Natl Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 12.Primig M., Williams R.M., Winzeler E.A., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E. The core meiotic transcriptome in budding yeasts. Nature Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 13.Williams R.M., Primig M., Washburn B.K., Winzeler E.A., Bellis M., Sarrauste de Menthiere C., Davis R.W., Esposito R.E. The Ume6 regulon coordinates metabolic and meiotic gene expression in yeast. Proc. Natl Acad. Sci. USA. 2002;99:13431–13436. doi: 10.1073/pnas.202495299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briza P., Bogengruber E., Thur A., Rutzler M., Munsterkotter M., Dawes I.W., Breitenbach M. Systematic analysis of sporulation phenotypes in 624 non-lethal homozygous deletion strains of Saccharomyces cerevisiae. Yeast. 2002;19:403–422. doi: 10.1002/yea.843. [DOI] [PubMed] [Google Scholar]
- 15.Deutschbauer A.M., Williams R.M., Chu A.M., Davis R.W. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enyenihi A.H., Saunders W.S. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabitsch K.P., Toth A., Galova M., Schleiffer A., Schaffner G., Aigner E., Rupp C., Penkner A.M., Moreno-Borchart A.C., Primig M., et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- 18.Iguchi N., Tobias J.W., Hecht N.B. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc. Natl Acad. Sci. USA. 2006;103:7712–7717. doi: 10.1073/pnas.0510999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLean D.J., Friel P.J., Pouchnik D., Griswold M.D. Oligonucleotide microarray analysis of gene expression in follicle-stimulating hormone-treated rat Sertoli cells. Mol. Endocrinol. 2002;16:2780–2792. doi: 10.1210/me.2002-0059. [DOI] [PubMed] [Google Scholar]
- 20.Namekawa S.H., Park P.J., Zhang L.F., Shima J.E., McCarrey J.R., Griswold M.D., Lee J.T. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 21.Shima J.E., McLean D.J., McCarrey J.R., Griswold M.D. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 22.Hochwagen A., Wrobel G., Cartron M., Demougin P., Niederhauser-Wiederkehr C., Boselli M.G., Primig M., Amon A. Novel response to microtubule perturbation in meiosis. Mol. Cell. Biol. 2005;25:4767–4781. doi: 10.1128/MCB.25.11.4767-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dessimoz C., Canarozzi G., Gil M., Marghadant D., Roth A., Schneider A., Gonnet G.H. Lecture Notes in Computer Science. Berlin/Heidelberg: Springer; 2005. Vol. 3678/2005. [Google Scholar]
- 24.Gil M., Dessimoz C., Gonnet G.H. A dimensionless fit measure for phylogenetic distance trees. J. Bioinform. Comput. Biol. 2005;3:1429–1440. doi: 10.1142/s0219720005001636. [DOI] [PubMed] [Google Scholar]
- 25.Huber W., Toedling J., Steinmetz L.M. Transcript mapping with high-density oligonucleotide tiling arrays. Bioinformatics. 2006;22:1963–1970. doi: 10.1093/bioinformatics/btl289. [DOI] [PubMed] [Google Scholar]