Abstract
The human malaria parasite, Plasmodium falciparum, possesses a broad repertoire of proteins that are proposed to be trafficked to the erythrocyte cytoplasm or surface, based upon the presence within these proteins of a Pexel/VTS erythrocyte-trafficking motif. This catalog includes large families of predicted 2 transmembrane (2TM) proteins, including the Rifin, Stevor and Pfmc-2TM superfamilies, of which each possesses a region of extensive sequence diversity across paralogs and between isolates that is confined to a proposed surface-exposed loop on the infected erythrocyte. Here we express epitope-tagged versions of the 2TM proteins in transgenic NF54 parasites and present evidence that the Stevor and Pfmc-2TM families are exported to the erythrocyte membrane, thus supporting the hypothesis that host immune pressure drives antigenic diversity within the loop. An examination of multiple P.falciparum isolates demonstrates that the hypervariable loop within Stevor and Pfmc-2TM proteins possesses sequence diversity across isolate boundaries. The Pfmc-2TM genes are encoded within large amplified loci that share profound nucleotide identity, which in turn highlight the divergences observed within the hypervariable loop. The majority of Pexel/VTS proteins are organized together within sub-telomeric genome neighborhoods, and a mechanism must therefore exist to differentially generate sequence diversity within select genes, as well as within highly defined regions within these genes.
INTRODUCTION
The bloodstream stages of the human malaria parasite, Plasmodium falciparum, reside enveloped by a parasitophorous vacuole within mature erythrocytes. As the parasite develops within this niche, it extensively modifies the erythrocyte surface and cytoplasm in order to display parasite-encoded receptors on the erythrocyte surface as well as to establish erythrocyte surface solute channels that are involved in nutrient uptake and elimination of metabolic waste products. The requisite ‘extra-parasitic, intra-erythrocytic’ trafficking network is of parasite origin and invention, because the erythrocyte itself is devoid of trafficking machinery that can be co-opted for parasitic means. Little is known regarding the molecular mechanisms of Plasmodium extra-parasitic trafficking, but it likely involves components that are either soluble within the erythrocyte cytoplasm or integral within the parasitophorous vacuolar membrane; the Maurer's clefts that dot the erythrocyte cytoplasm; or possibly small vesicles budding from either of these membrane surfaces [(1–7), reviewed in (8,9)]. Our understanding of protein targeting to the erythrocyte cytoplasm was recently given a revelatory boost by the identification of a trafficking motif that appears to be widely, but not inclusively, present in parasite-encoded, signal peptide-containing proteins that are trafficked to the erythrocyte cytoplasm or surface (10–12). The motif is termed Pexel [Plasmodium export element, (12)] or VTS [vacuolar transport signal, (10)] and has a simple core consensus sequence, ‘RxLxQ/E’. All Pexel/VTS-containing proteins identified to date possess a two exon gene structure in which the signal peptide, oftentimes recessed from the start methionine, is encoded on the short N-terminal exon, whereas the Pexel/VTS motif is encoded on the second exon near the splice junction site [reviewed in (13,14)].
Using the above descriptive features of Pexel/VTS-encoding genes heuristically it is possible to compile a catalog of P.falciparum proteins that are predicted to be trafficked to the erythrocyte. This catalog most notably includes all proteins encoded by the 2 transmembrane (2TM)-containing members of the stevor, rifin and Pfmc-2TM gene families; the knob-associated proteins KAHRP and MESA; the DnaJ domain-containing RESA family; the superfamily of α-helical PHIST domain proteins (9) that contains over 70 members, including the RESA-like proteins; as well as a family of 20 serine/threonine protein kinases of the R45 family that are predicted to be trafficked to the erythrocyte cytoplasm (8,15). In addition, as much as 25% of the total Pexel/VTS catalog is represented by unique proteins. We have determined such a catalog [see also the recent reviews, (9,16)] by combining the catalogs compiled by Marti et al. (12) and Hiller et al. (10); and additionally performing exhaustive BLAST screening of Plasmodium sp. genome sequence databases and conducting extensive genome walking on the P.falciparum genome sequence. The resulting catalog contains over 300 genes that, although daunting in its breadth, can be greatly distilled into families of proteins that are grouped according to paralogous relationships, protein domain architectures and transmembrane topologies. Here we describe the sequence diversity and genomic organizations of genes and extended loci for the Pexel/VTS-containing 2TM families. The rifin, stevor and Pfmc-2TM genes exhibit paralog-specific and isolate-specific divergences that are confined to a hypervariable loop region between the 2TM domains. We present evidence that the 2TM protein families are exported to the erythrocyte membrane, thus supporting the hypothesis that the hypervariable loop is exposed on the erythrocyte surface and that host immune pressure drives antigenic diversity within the loop. The genes encoding 2TM superfamily members are frequently found in sub-telomeric genome neighborhoods that contain Pexel/VTS-encoding genes that do not exhibit sequence diversity across isolates, such as the PHIST domain-containing protein superfamily. Thus it is proposed that the sub-telomeric regions facilitate molecular mechanisms that promote hypervariability within genes, and within discrete regions of these genes, which encode proteins that are exposed to the host immune system; whereas invariant erythrocyte cytoplasmic proteins are shielded from this process.
MATERIALS AND METHODS
Parasite culture and isolate-specific genomic DNA
Asexual and gametocyte NF54 isolate P.falciparum parasites were cultivated in vitro as described (17). To synchronize cultures, schizonts were purified either by magnetic isolation using a MACS depletion column (Miltenyi Biotec) in conjunction with a magnetic separator (18), or by a Percoll-sorbitol gradient; and placed back into culture at 0.5% hematocrit in RPMI 1640 containing 10% human serum. Genomic DNA from P.falciparum isolates GB4, Liberia, 7G8 and Dd2 were the generous gift of Dr Karen Hayton (NIAID, NIH). A cloned progeny parasite line, 3D7B, from a 3D7 × 3D7 self-cross was kindly provided by Dr Thomas Wellems and was cultured for the purification of genomic DNA. Genomic DNA from the parents and progeny of an HB3 × 3D7 cross were the generous gift of Dr Akhil Vaidya.
Plasmid construction and parasite transformation
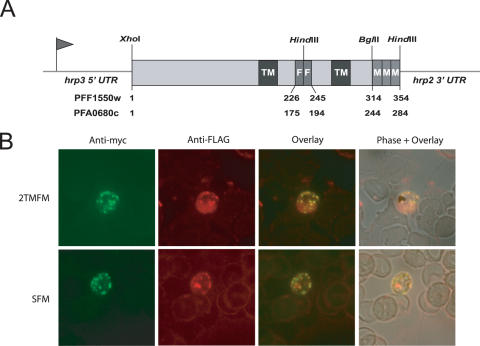
Plasmodium falciparum transfection plasmids were constructed to express stevor (PFF1550w) and Pfmc2TM (PFA0680c) genes which encode tagged proteins having two adjacent FLAG epitopes within the hypervariable loop region plus three adjacent c-myc epitopes at the carboxy termini (Figure 2A). The plasmids were constructed by amplifying the N-terminal portion containing the first TM, and the C-terminal portion encoding the second TM, from P.falciparum NF54 isolate genomic DNA using the following primers: Stevor5′XhoI (5′-CCCTGCAGCTCGAGATGTTATTGTTTAACTTTTTAAT); Stevor3′FLAGHindIII (5′-GGAAGCTTCTTATCGTCGTCATCCTTGTAATCTGTAGTTAATGAACCACCATG); Stevor5′FLAGHindIII (5′-CCAAGCTTGATTACAAGGATGACGACGATAAGGCTCTTAAAGCAACCGAAGCG); Stevor3′ BamHI (5′-GGGGATCCCTTACATAAATGTTTCTTGC); 2TM5′XhoI (5′-CCCTGCAGCTCGAGATGTTTCATTATATTTATAAAAT); 2TM3′FLAGHindIII (5′-GGAAGCTTCTTATCGTCGTCATCCTTGTAATCAATATCAACCGAAAATAAACATAC); 2TM5′FLAGHindIII (5′-CCAAGCTTGATTACAAGGATGACGACGATAAGACATCATCTTCTGCACTTGGC); 2TM3′BglII (5′-GGGGATCCTTTTGTTATTTGCTTTTTGTT). The PCRproducts corresponding to the N-terminal portion and the C-terminal portion of each gene were ligated to reconstitute the full ORF sequences and to introduce two FLAG epitope tags in the middle of the hypervariable loop region (Figure 2A). These full-length products were restriction digested with XhoI and BglII and inserted into the expression vector, pHL-dhfr-3myc, which is a derivative of the pHLH1 vector (19). pHL-dhfr-3myc contains three consecutive c-myc epitopes within an expression cassette that is driven by the P.falciparum 5′hrp3 promoter and the 3′hrp2 terminator (Figure 2A), to ensure a high expression level of stevor and Pfmc-2TM genes; and an hDHFR expression cassette for selection of transformed parasites with pyrimethamine. Expression vectors were transfected into the NF54 P.falciparum isolate by ‘pre-loading’ erythrocytes as described (20). Stable transformant parasites were selected by culturing in media containing 40 ng/ml pyrimethamine. Transfections were validated by performing plasmid rescue experiments; specifically, chemically competent Escherichia coli cells were transformed with 100 ng of purified genomic DNA from selected transformant parasites. Expression of Stevor-FLAG-myc (SFM) in the transformed line, pHL-SFM, and Pfmc2TM-FLAG-myc (2TMFM) in the transformed line, pHL-2TMFM, were confirmed by reverse transcription and real-time PCR using specific primers for PFF1550w (sense: 5′-AAACAAAAGGAAGAGATAAG; antisense: 5′-ATCGCTGCTACATATACAGG) and for PFA0680c (see Supplementary Table 1), following the protocol described below. As a control for expression and localization experiments we used the pHL-dhfr-luciferase line (a gift from K. Deitsch), which expresses the luciferase protein and is a derivative of pHLH1 vector (19).
Immunofluorescence and immunoelectron microscopy
For indirect immunofluorescence microscopy assays, infected erythrocytes were thinly smeared within wells of Teflon coated slides and fixed for 20 min in methanol chilled at −80°C. Slides were blocked in 2% BSA in phosphate-buffered saline (PBS) and then incubated with FITC-conjugated anti-c-myc mAb (1:200; Sigma), or with anti-FLAG polyclonal antibodies (1:200; Sigma) followed by goat anti-rabbit Alexa 594-conjugated IgG (1:2000; Molecular Probes). For electron microscopy, schizonts and late trophozoites from transformant lines were purified on Percoll-sorbitol gradients and the cell pellets were resuspended in 2% paraformaldehyde, 0.1% glutaraldehyde (EM grade) in PBS. The samples were dehydrated through a coldethanol series, infiltrated first with ethanol-LRWhite (London Resin Company, UK), overnight at 4°C, then in pure LRW at 4°C for up to 24 h. The blocks were hardened at 50°C overnight. Sections were cut using a Diatome diamond knife on a Leica Ultracut S ultramicrotome and contrasted with aqueous uranyl acetate for 15 min. Thin sections were incubated with an anti-c-myc mAb (1:25) followed by 6 nM gold-labeled goat anti-mouse IgG (1:100, Aurion). Samples were examined in a JEOL 100 CX-II EM operating at 80 Kv and imaged on Kodak 4489 EM film.
Real-time PCR assays of gene transcription
Gene-specific primers (Supplementary Table 1) were empirically designed for the 13 Pfmc-2TM genes and two control genes, namely, arginyl-tRNA synthetase (PFL0900c) and glutamyl-tRNA synthetase (PF13_0170). The control genes were identified in the P.falciparum 3D7 genome sequence database [(21); http://www.plasmodb.org] as fulfilling transcript expression criteria for ‘housekeeping’ control genes which display consistent transcript expression patterns throughout the parasite life cycle. Real-time PCR was performed using an ABI Prism 7900HT sequence detector (Applied Biosystems). Reactions were prepared in 25 μl volumes using SYBR Green PCR master mix (Applied Biosystems) and 1 μM primers. Specificity of amplification was confirmed by melting-curve analysis for each PCR product. Duplicate PCR were analyzed for each sample. The number of Pfmc-2TM genes were estimated using the 2ddct method (User Bulletin 2, ABI), as well as an adjacent paralogous 2TM gene family, Hyp5 (16), that is present in the genomes of the P.falciparum 3D7 and Dd2 isolates. Real-time PCR experiments were performed on several dilutions of genomic DNA from different isolates using universal primers for Pfmc-2TM and Hyp5 genes, with gene-specific primers for PFL0900c and PF13_0170 used as single copy gene references.
Sequence analyses
The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda, MD) and the P.falciparum genome nucleotide sequence database [(21); www.plasmodb.org] were searched using the BLASTP or TBLASTN programs (22). Iterative database searches were conducted using the PSI-BLAST program with either a single sequence or an alignment used as the query, with the PSSM inclusion expectation E-value threshold of 0.01 (unless specified otherwise); the searches were iterated until convergence. Multiple alignments were constructed using the T_Coffee and MUSCLE programs, followed by manual correction based on the PSI-BLAST results (23,24). Potential transmembrane regions were predicted using TopPred (25) and TMHMM-2 (26). The Ghanaian P.falciparum isolate sequence information was used by permission and was accessed via the BLAST server at The Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/p_falciparum). The P.falciparum HB3 isolate raw sequence reads (The Broad Institute) were obtained from the NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi) and formatted for database searches using the TBLASTN program. Nucleotide sequence alignments of extended loci containing Pfmc-2TM genes were performed using ClustalW (27).
RESULTS AND DISCUSSION
Classification of Pexel/VTS-containing proteins
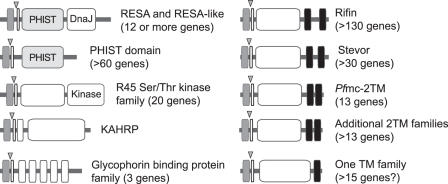
To determine a representative catalog of Pexel/VTS-containing proteins we have combined the catalogs compiled by Marti et al. (12) and Hiller et al. (10); performed exhaustive BLAST screening of Plasmodium sp. genome sequence databases using all identified Pexel/VTS-containing proteins and position-specific score matrix (PSSM) profiles as queries; and conducted extensive genome walking to identify ORFs within sub-telomeric regions of the P.falciparum genome sequence. A similar analysis was recently described (9,16) and appears to correlate well with our resulting catalog, which contains over 300 Pexel/VTS-containing proteins that are predicted to be trafficked to the erythrocyte. These proteins can be grouped into several families of proteins according to their paralogous relationships, protein domain architectures and transmembrane topologies (Figure 1). These groups include two superfamilies, the 2TM proteins and the PHIST domain-containing proteins, which together represent >75% of all the Pexel/VTS-containing proteins. The remaining proteins include the knob-associated proteins KAHRP and MESA; a family of 20 serine/threonine protein kinases of the R45 family as well as a family of 15 uncharacterized predicted 1TM proteins. The 1TM proteins are grouped together based upon their similarity in architectures; namely, overall size, C-terminal TM domain, the presence of signal peptide sequences plus Pexel/VTS motifs at their N-termini, two exon gene structure and, in some instances, paralogous relationships. The 2TM superfamily is encoded by seven or more paralogous gene families, that have a shared architecture of ∼300 amino acid lengths and a 2TM topology. Both the Rifin proteins, encoded by over 130 genes, and the related Stevor proteins, with >35 genes, are placed within this topological superfamily as well as the recently described Pfmc-2TM family (28).
Figure 1.
Schematic representation of the catalog of P.falciparum Pexel/VTS-containing proteins. Signal peptide sequences are represented by gray boxes; transmembrane domains are represented by black boxes. All proteins are encoded by two exon genes; the location of the intron is represented by a gray triangle. The Pexel/VTS motif is encoded near the 5′ end of the second exon and is represented by a narrow open box. The locations of signal peptide cleavage sites have not been determined, and the N-termini of mature proteins are not known. Predicted globular domains are indicated by open boxes.
Protein domain architecture of the 2TM superfamily members
Alignment of 13 members of the Pfmc-2TM family reveals a high degree of amino acid conservation throughout the protein, including the TM and signal peptide domains [described in Ref. (28); see also Supplementary Figure 1]. Notably, the overall conservation within the Pfmc-2TM family highlights a marked divergence within the ∼17 amino acid long hypervariable region between the two TM domains (boxed regions in Supplementary Figure 1). Three members of the family (PF11_0014, PFC1080c and MAL7P1.58) differ within the C-terminal-most 30 amino acids due to a shared single nucleotide deletion and resulting frameshift. It is not known if this mutation results in non-functional or non-expressed proteins, or if it indicates alternate protein functions. Alignments of full-length amino acid sequences from 23 Stevor (Supplementary Figure 2) and 73 Rifin (Supplementary Figure 3) proteins also support a 2TM topology and conserved architectures consisting of: (i) highly conserved amino termini regions within the respective families, as well as similarity between Rifins and Stevors near the amino terminus; (ii) a hypervariable region between two conserved TM domains and (iii) a highly conserved, positively charged, short C-terminal domain (29). In contrast to the Stevor proteins, the Rifin N-terminal-most TM domain is atypical and contains an abundance of glycine and alanine residues, as well as a central proline residue that is conserved in a predominant subset of Rifins and is suggestive of a relationship with ion channels (15).
Cellular localization of the 2TM superfamilies
For a given 2TM family, the pronounced sequence variability within the region between the TM domains, when contrasted with the relative sequence conservation in the N-terminal and C-terminal regions, suggests that this region defines a loop that is exposed to antibody-mediated immune pressure at the external surface of the erythrocyte. As a logical conclusion, it has been proposed that the erythrocyte surface is the final trafficking destination for the 2TM proteins and that they conduct their principal function at this site (28,29). However, the cellular localization of the 2TM superfamily remains speculative and only the Rifin proteins have been shown to be exposed at the erythrocyte surface (30), whereas the Stevor and Pfmc-2TM proteins have been described as Maurer's clefts proteins (28,31,32). To determine the cellular localization of these proteins we generated transgenic NF54 parasites that abundantly express either Stevor (SFM, Stevor-FLAG-myc transgenic line) or Pfmc-2TM (2TMFM, Pfmc2TM-FLAG-myc transgenic line) proteins, each of which were tagged with three c-myc epitopes at the carboxyl terminus plus two FLAG epitopes within the hypervariable loop (Figure 2A). IFA microscopy studies using anti-c-myc and anti-FLAG antibodies showed that the tagged proteins were efficiently expressed within the cytoplasm of infected erythrocytes (Figure 2B); and both antibodies gave staining patterns that likely correspond to Maurer's cleft localization, as described (28,31).
Figure 2.
Immunolocalization of epitope-tagged proteins. (A) Schematic representation of the SFM (Stevor-FLAG-myc) and 2TMFM (Pfmc-2TM-FLAG-myc) recombinant proteins. For the respective genes, two FLAG epitopes (F) were placed between the 2TM domains and three myc epitopes (M) were inserted at the carboxy terminus. The lengths, in amino acids, of the chimeric proteins are indicated at the bottom. The restriction sites specific for the insertion of tags are indicated. (B) Immunofluorescence assays (IFA) showing the localization of SFM and 2TMFM in the transformed lines, pHL-SFM and pHL-2TMFM. IFA studies were performed on air-dried P.falciparum transformed parasites. Infected erythrocytes were stained with FITC-conjugated anti-c-myc mAb and with anti-FLAG polyclonal antibodies followed by goat anti-rabbit Alexa 594-conjugated IgG. As a negative control, air-dried pHL-dhfr-luc P.falciparum parasites gave no signal (data not shown).
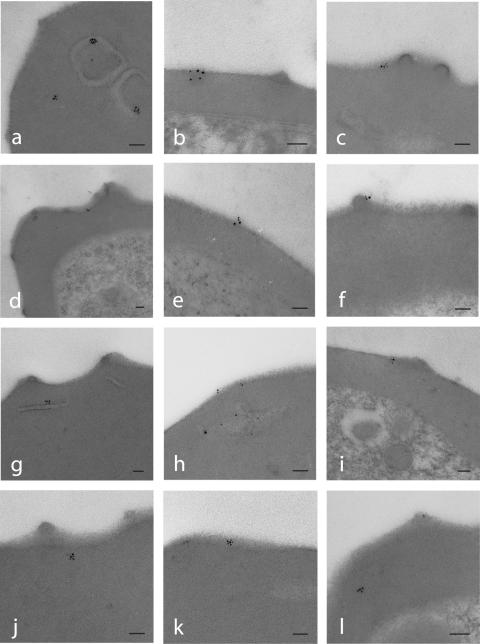
The IFA studies of the chimeric transgenic proteins did not have sufficient resolution to determine if these proteins are transported beyond the Maurer's clefts to the erythrocyte surface; therefore, we used immunoelectron microscopy with anti-c-myc antibodies, which gave a stronger and more specific staining by IFA than anti-FLAG antibodies, to examine the cellular localization of the epitope-tagged proteins. In both the SFM and 2TMFM transgenic lines numerous gold particles were associated with membranous structures that likely correspond to Maurer's clefts, thus confirming the above immunofluorescence microscopy, as well as published data [(Figure 3a,g,h); (28,31,32)]. In addition, gold particles were associated with the erythrocyte membrane, and in a few instances associated with knobs, supporting the hypothesis that the 2TM proteins pass through Maurer's clefts en route to their final destination, the erythrocyte surface. However, a small amount of protein (2–3 gold particles or clusters per thin section of erythrocyte) was observed in association with the surface; and we were not able to detect the tagged proteins by IFA on live cells using anti-FLAG antibodies (data not shown), although these proteins were abundantly expressed in both the SFM and 2TMFM transgenic lines. The relative abundance of gold particles, as well as of IFA fluorescence, in Maurer's clefts versus the erythrocyte membrane suggests that Maurer's clefts might function to regulate the level of surface expression, in addition to a role in trafficking. Moreover, it might explain the lack of surface expression in studies that describe the Stevor and Pfmc-2TM proteins as Maurer's cleft residents (28,31,32). It is currently being addressed if the inefficiency of surface transport is partially a phenotype of the NF54 isolate, and if greater erythrocyte surface display can be achieved using other transgenic P.falciparum isolates, such as IT4. Nonetheless, we show that the 2TM proteins are trafficked to the erythrocyte membrane and that the hypervariable region between two conserved TM domains likely defines a loop that is exposed on the erythrocyte surface.
Figure 3.
Immunogold localization of SFM and 2TMFM in the transformed lines pHL-SFM and pHL-2TMFM. Thin sections of late trophozoites and schizonts from pHL-SFM line (a–f) and pHL-2TMFM line (g–l) were probed with an anti-c-myc mAb and revealed by 6 nm gold-labeled anti-mouse IgG. Gold particles were associated with Maurer's clefts (a, g and h) or the erythrocyte surface (b–f, h, i, k and l). Some particles were associated with knobs (c, f, i and l). Bar, 0.25 μm.
Diversity within the hypervariable loop of the 2TM superfamily members
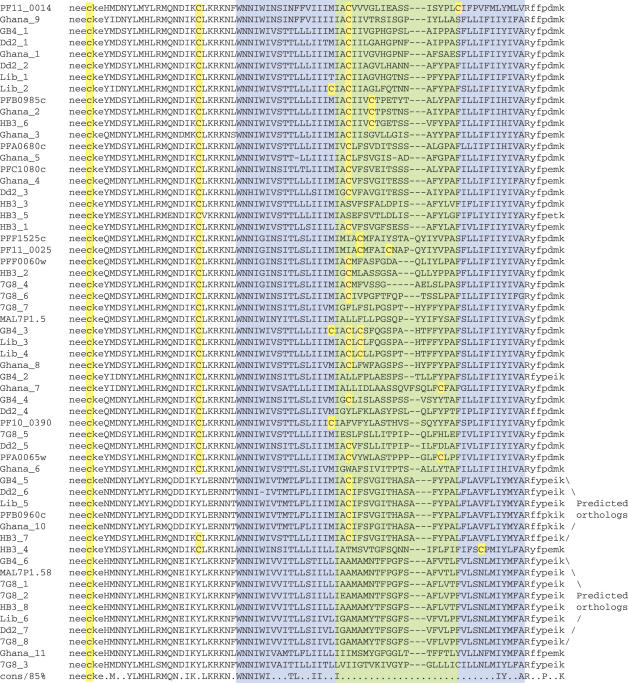
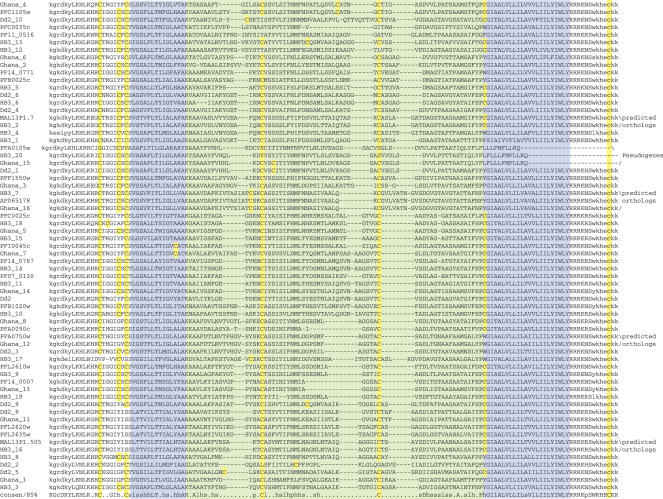
To implicate if immune pressure selects for divergence within the loop region and shapes an antigenically-diverse gene repertoire, we determined the sequence diversity between paralogs and across isolates for the Pfmc-2TM and Stevor families. Genomic DNA PCR products were isolated and sequenced for a region that spans the hypervariable loop of Pfmc-2TM genes from the P.falciparum African isolates, Liberia and GB4; the Southeast Asian isolate, Dd2; and the South American isolate, 7G8. The PCR products were amplified using universal PCR primers specific to the Pfmc-2TM gene family and flanking the loop region (see primer locations in Supplementary Figure 1). It was not attempted to exhaustively clone the entire repertoire of Pfmc-2TM genes from each isolate; rather, representative numbers of genes were obtained. In addition, 11 Pfmc-2TM gene sequences were obtained from the P.falciparum Ghanaian isolate genome sequence project database (The Wellcome Trust Sanger Institute), as well as 11 genes identified by TBLASTN screening of raw sequences from the genome sequence project for the P.falciparum HB3 Honduran isolate (The Broad Institute). The translated amino acid sequences within the loop region are highly divergent in all Pfmc-2TM versions (Figure 4), supporting the hypothesis that the divergence is selected for by immune pressure. Two exceptions to this rule were observed: the gene MAL7P1.58 in the 3D7 isolate, which is highly conserved across isolates and is located within the Chr7 internal cluster of var genes in the 3D7 genome sequence; and the gene fragment PFB0960c in the 3D7 isolate, which is predicted to be a pseudogene because the 3D7, Ghanaian and Dd2 versions possess frameshift mutations and the 3D7 and Ghanaian versions additionally lack a start methionine.
Figure 4.
Alignments of the divergent loop and flanking sequences for Pfmc-2TM family members from isolates 3D7, GB4, Liberia, Dd2, 7G8, HB3 and Ghana. 3D7 isolate genes are denoted by their protein names; genes from other isolates are numbered in the order of their discovery. The TM domains are indicated by blue boxes and a green box indicates the hypervariable loop. Translated sequences corresponding to the sense and antisense PCR primers are indicated in lowercase font. The bottom line reflects 85% consensus. Cysteine residues are highlighted in yellow.
Isolate-specific differences were also examined for the hypervariable loop within the Stevor protein family. Using ‘universal’ primers within the highly conserved regions flanking the 2TM domains, 11 stevor gene sequences were cloned from the Dd2 isolate, supplemented with 20 genes and 14 genes, respectively from the HB3 and Ghanaian isolate sequence databases (Figure 5). The overwhelming majority of the sequences, both across paralogs and between isolates, showed extensive diversity that is localized to the loop region. As observed with the Pfmc-2TM genes, the most conserved stevor gene is a pseudogene fragment that is present in the 3D7 (PFA0105w), HB3, and Ghana isolates, and four additional predicted orthologs were identified.
Figure 5.
Alignments of the divergent loop and flanking sequences for Stevor family members from isolates 3D7 Dd2, HB3 and Ghana. 3D7 isolate genes are denoted by their protein names; genes from other isolates are numbered in the order of their discovery. The TM domains are indicated by blue boxes and a green box indicates the hypervariable loop. Translated sequences corresponding to the sense and antisense PCR primers are indicated in lowercase font. The bottom line reflects 85% consensus. Cysteine residues are highlighted in yellow.
In summary, the hypervariable loop examined for Pfmc-2TM and Stevor sequences from multiple isolates, as well as within 3D7 paralogs for the Rifin family (Supplementary Figure 3), is highly divergent within a precise region encoding the erythrocyte surface-exposed loop of the 2TM superfamily genes, and this diversity is likely in response to host immune pressure that is encountered due to exposure at the erythrocyte surface. Following, a general theme is described of variable versus invariant genes, and this distinction is correlated with gene location within the chromosomes; status as intact or pseudogenes; and the ability of genes to undergo positive selection for antigenic variants via antibody-mediated immune pressure.
Amplification and nucleotide conservation of extended Pfmc-2TM loci containing Pexel/VTS-encoding genes
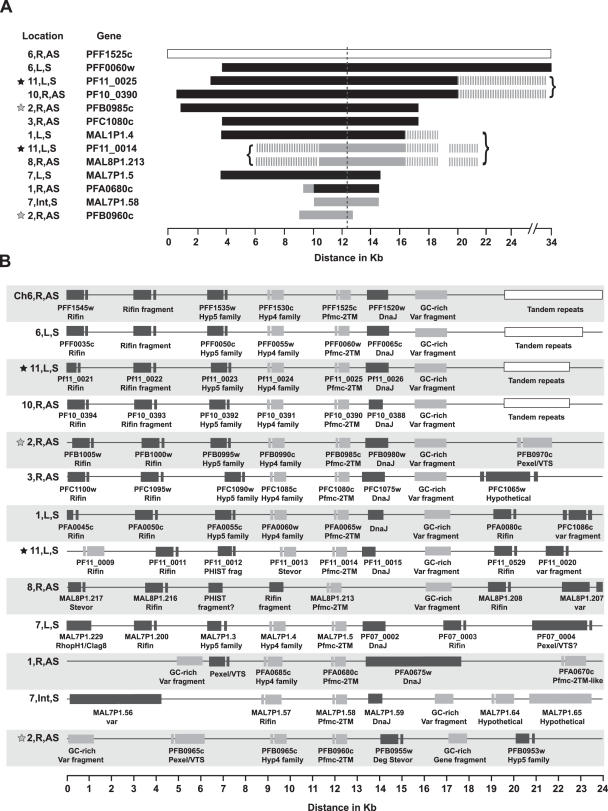
In the course of determining if the high degree of nucleotide conservation within Pfmc-2TM paralogs extends to adjacent non-coding regions, it was noticed that the genes reside within a locus that is present, as either extended or fragmented versions, as 13 copies in the 3D7 isolate genome. The extended versions share 36 Kb or more of sequence similarity, including >95% nt sequence identity spanning 15 Kb of sequence (Figure 6A); and almost 100% identity within numerous stretches >1 Kb, particularly for an alignment of the extended loci of the Pfmc-2TM genes PFF1525c, PFF0060w, PF11_0025 and PF10_0390 (the topmost four loci in Figure 6A). The loci identified as truncated versions (e.g. the loci containing PFA0680c, MAL7P1.58 or PFB0960c) might have additional sequence identities that were not recognized by the ClustalW alignment program, perhaps due to large deletions or insertions. It is possible that the amplified sub-telomeric loci are identical to one or more ‘sub-telomeric blocks’ described in the P.falciparum genome sequence manuscript [e.g. SB-4 or SB-5; see Figure 2 of Ref. (33)]. The conservation of sequences flanking Pfmc-2TM genes is reminiscent of the subsets of var genes which share substantial conserved flanking nucleotide sequences (34), although the var gene loci do not extend to the length, nor to the degree of the nucleotide conservation of the Pfmc-2TM loci.
Figure 6.
A locus containing a central Pfmc-2TM gene that is duplicated 13 times in the P.falciparum 3D7 isolate genome sequence. (A) The predicted length (Kb) of locus duplication, determined as nucleotide identities shared with the extended PFF1525c locus (upper row, indicated by an open rectangle). The left-hand columns indicate for each locus the predicted chromosome and telomere location (left, L; right, R); orientation (sense, S; antisense, AS); and Pfmc-2TM gene identifier. Adjacent loci are indicated by a gray star (Chr2, R) and black star (Chr11, L). A vertical dashed line, at approximately the 12.4 Kb mark, indicates the location of the Pfmc-2TM gene within each locus. Gray horizontal bars indicate >85% nt identity with the PFF1525c locus, and black horizontal bars indicate >95% identity. The curly brackets unify additional sequence similarities (dashed gray bars) that are specific to PF11_0025 and PF10_0390; PF11_0014 and MAL8P1.213; and PFA0065w, PF11_0014 and MAL8P1.213. (B) Predicted ORFs within the 13 loci extending ∼12 Kb upstream and 20 Kb downstream of each centralized Pfmc-2TM gene. ORFs on the sense strand are shown in gray, and ORFs on the antisense strand are in black. All ORFs encode Pexel/VTS-containing genes, var genes or predicted pseudogenes.
To rule out the trivial possibility that the amplification of the highly conserved extended Pfmc-2TM locus is the result of genome sequence assembly or sequencing errors within the 3D7 genome sequence project database [accessed at www.plasmodb.org; (21)], the presence of 13 Pfmc-2TM loci was confirmed by sequencing of PCR products amplified using gene-specific, rather than universal Pfmc-2TM primers (data not shown). In addition, real-time genomic DNA PCR estimations of gene copy number indicated at least 10 copies, by comparing amplification rates using universal versus gene-specific Pfmc-2TM primers and single copy gene-specific control primers (data not shown). Moreover, the genome sequences for the Ghanaian and HB3 isolates each contain at least 11 Pfmc-2TM genes (Figure 4), as well as extended regions of nucleotide conservation in the Ghanaian and HB3 isolates with untranslated region (UTR) flanking regions of 3D7 isolate Pfmc-2TM loci, indicating the conservation of extended loci across isolates (data not shown).
All amplified loci are found on multiple chromosomes in sub-telomeric regions, in association with rifin genes, and have conserved orientations with respect to the telomeres (chromosomal locations and orientations are indicated in the left-hand column, Figure 6A). An exception to this rule is the truncated locus containing the Pfmc-2TM gene, MAL7P1.58, which is located within the internal cluster of var and rifin genes on Chr7. Chr2 and Chr11 each harbor tandem copies of the locus (denoted by gray and black stars, respectively, in Figure 6A) and, in fact, the PFB0960c locus on Chr2 is truncated immediately downstream of the Pfmc-2TM gene due to fusion with the locus containing PFB0985c. The pairs of loci indicated by brackets in Figure 6A show similarities that may indicate relationships in the ancestral progression of the locus duplications. The ORFs within the extended loci predominantly encode Pexel/VTS-containing proteins, PfEMP1 proteins or apparent pseudogenes (Figure 6B). The full-length versions of the amplified locus each contain one Pfmc-2TM gene; one gene from the 2TM paralogous family Hyp5 (e.g. PFF1535w); a gene encoding a Pexel/VTS-containing protein that may not possess a TM domain (e.g. PFF1530c); and one or more rifin gene. Additionally, a DnaJ domain-containing gene (e.g. PFF1520w) lies immediately downstream, and on the opposite strand, of each Pfmc-2TM gene. In contrast to the DnaJ domain-containing Pexel/VTS genes, these versions appear to be pseudogenes and lack an N-terminal exon and start methionine, with the notable exception of a predicted two exon, signal peptide/Pexel/VTS-containing version that has an insertion encoding a pronounced repeat region (PFA0675w).
Although the loop region within the Pfmc-2TM proteins is variable across paralogs and across P.falciparum isolates, the ORFs adjacent these genes within the extended locus are relatively invariant across paralogs and in predicted orthologs within the HB3 isolate genome sequence (e.g. predicted orthologs for PFF1535w, PFF1530c and PFF1520w). The rifin gene (e.g. PFF1540w) is highly conserved in the duplicated versions, including a nearly identical copy on Chr11 that is also a predicted pseudogene. A predicted ortholog of PFF1540w is absent from the HB3 isolate genome sequence database, perhaps due to the incompleteness of the sequence coverage, but the predicted PFF1540w paralog within the Chr2 locus (containing PFB0895c; Figure 6A), itself a pseudogene, identifies a virtually identical predicted ortholog and pseudogene in the HB3 isolate. Thus, additional support is provided for the theme that sub-telomeric pseudogenes are excluded from mechanisms generating divergence, or selection for antigenic diversity, despite their inclusion in highly variant gene families. As a result, pseudogenes are found conserved across isolate boundaries.
Generation of diversity within the hypervariable loop of 2TM proteins
The stark discrepancy of the sequence diversity within the Pfmc-2TM loop, versus the highly conserved flanking regions, suggests that the parasite possesses a mechanism for creating hypervariability within a discrete region, perhaps in concert with immune pressure that selects for antigenic variants. Extrapolating this logic to the rifin and stevor genes, the parasite may also have a mechanism for generating diversity within the larger loops contained within these genes. The sub-telomeric location of variable gene families, such as var, rifin and stevor, likely confers an enhanced capacity for gene diversification and amplification through mechanisms of recombination, including that between non-homologous chromosomal ends (35). The clustering of telomeres at the nuclear periphery is thought to play a role in facilitating recombination (35,36). The relative roles of single crossover recombination events versus gene conversion mechanisms involving replacements of short fragments of sequence information remain unclear. It is possible that the DNA segments encoding the loop regions are prone to strand breaks, and the repair mechanism requires extensive base-pair alignments mediated by the nucleotide conservation within the extended loci and a gene conversion event involving donor sequences. In addition, single nucleotide mutations might accumulate within the loop regions during this process, if introduced by a mutagenic DNA polymerase that functions in translesion DNA repair, such as Plasmodium orthologs of Rev1p and Rev3p (37). The P.falciparum var family has been shown to undergo recombination-mediated variation during meiosis (36,38), which contributes to great sequence diversity between paralogs and across isolates for the erythrocyte surface membrane protein 1 (PfEMP1). Analysis of progeny of a genetic cross between HB3 × 3D7 and HB3 × Dd2 demonstrated the generation of non-parental var genotypes (36); and these genotypes appeared more frequently than expected from the calculated meiotic crossover rate of P.falciparum (39). In addition, an HB3 × HB3 self-cross led to the appearance of a new var form as detected by an additional restriction fragment.
The high conservation of the extended Pfmc-2TM and stevor loci in P.falciparum could contribute to gene conversion events by allowing for the alignment of chromosome ends as a basis for ectopic recombination events between paralogous genes. To seek examples of recombination or gene conversion, we studied a cloned progeny of a 3D7 × 3D7 self-cross and three cloned progeny from a 3D7 × HB3 cross. Sequencing of the hypervariable loop for all 13 Pfmc-2TM and 39 stevor genes in the progeny of the 3D7 × 3D7 self-cross did not reveal any new sequences (data not shown). Although a lack of new Pfmc-2TM or stevor forms implies that there were no ectopic recombination events between paralogous genes, allelic recombination events cannot be ruled out. PCR amplification of the 3D7 Pfmc-2TM genes of all three progeny cloned from the 3D7 × HB3 cross and sequencing of genomic DNA from two of these progeny also revealed a complete conservation of the inherited members of the Pfmc-2TM family (the inherited genes in the progeny were traced through the mapping of chromosomal recombination events). Thus, no new Pfmc-2TM genes were seen following a non-self cross. Moreover, a partial survey of stevor genes isolated using ‘universal’ PCR primers from a single 3D7 × HB3 progeny also failed to show novel sequences. These results indicate that diversity within the hypervariable loop of the Pfmc-2tm and stevor gene families is not mediated through meiotic recombination events to the extent seen in vars; although it is possible that the much larger size of var genes increases the chances of intragenic recombination events. Because cultured 3D7 parasites do not develop novel Pfmc-2TM or stevor genes, it is possible that mitotic recombination events are not responsible for the sequence diversity that was observed in the proposed loop. However, proper determination of mitotic recombination events would require cloning of individual parasites, since single mutation events would be difficult to detect from a bulk culture unless those parasites represented a significant portion of the population. It is possible that the large diversity seen in specific regions of the 2TM gene families may be acquired through a slower mechanism such as genetic drift. It is also plausible that in the absence of immune pressure, recombination events within these gene families do not occur more frequently than the natural recombination rate determined for P.falciparum. In addition, a possible pathway for the accumulation of single mutations within the loop regions should be considered. Nevertheless, the wide variation seen within the loop of the 2TM proteins implies a parasite-mediated mechanism for generation of antigenic diversity, and perhaps a mechanism of antigenic variation.
CONCLUSIONS
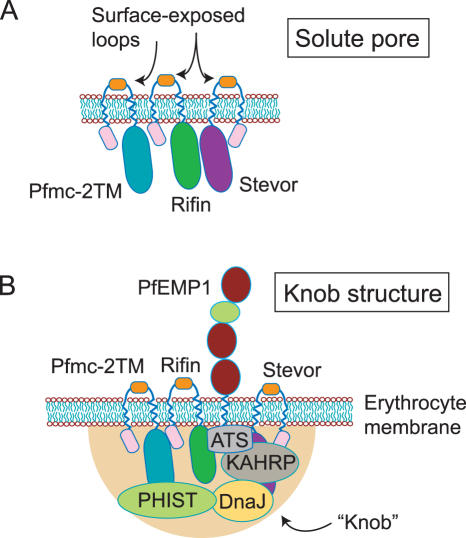
The shared architecture of a 2TM topology and hypervariable loop unites the Pfmc-2TM, Rifin and Stevor proteins and indicates similar functions and likely erythrocyte membrane localization as their final trafficking and functional destination. The amplification of the 2TM protein-encoding genes into large families, and the profound divergence within each family and across isolates within a region confined to a proposed membrane surface loop, combine to suggest that antibody-mediated immune pressure has expanded their repertoire and shaped their topology. The Pfmc-2TM genes are contained within a locus that is amplified, in varying lengths of conservation, 13 times in the P.falciparum genome. The striking nucleotide conservation across the length of this locus, when contrasted with the high nucleotide diversity confined to the hypervariable loop within the Pfmc-2TM genes, suggests that the parasite possesses a mechanism of creating divergence within a discrete, bounded region. The investment of over 200 genes to encode the catalog of 2TM proteins suggests that these proteins participate in the formation of a critical erythrocyte surface structure, perhaps forming a transmembrane anchor for the knob that underpins PfEMP1-mediated cytoadherance, or 2TM subunits of a multi-subunit, multi-transmembrane erythrocyte surface solute channel (Figure 7). Our observations from immunoelectron localization studies of chimeric 2TM proteins suggest that erythrocyte surface expression is not relegated solely to knobs (Figure 3), albeit these studies were performed following the overexpression of transgenic epitope-tagged 2TM proteins.
Figure 7.
Two models for the function of the 2TM proteins. In the upper panel (A) the proteins participate in formation of a multi-subunit, multi-TM solute pore. An alternative model is proposed in which the 2TM proteins modulate endogenous erythrocyte solute channels. In the lower panel (B) the proteins form the knob structure containing PfEMP1 proteins and mediating cytoadherence. A third model might integrate the knob and solute pore and knobs into a single erythrocyte surface structure.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online
Supplementary Material
Acknowledgments
We are grateful to Stephane Egee for his considerable help in the construction of expression plasmids and for valuable discussions. Dr Karen Hayton (NIAID/NIH) is thanked for the generous gift of genomic DNA from P.falciparum isolates. Dr Matthew Berriman (The Wellcome Trust Sanger Institute) is thanked for allowing use of the genome sequence data for the P.falciparum Ghanaian isolate. The Broad Institute is thanked for providing to the scientific community the genome sequence for the P.falciparum HB3 isolate. T.J.T. would like to acknowledge the support of the William Randolph Hearst Foundation. Funding to pay the Open Access publication charges for this article was provided by the William Randolph Hearst Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Haeggstrom M., Kironde F., Berzins K., Chen Q., Wahlgren M., Fernandez V. Common trafficking pathway for variant antigens destined for the surface of the Plasmodium falciparum-infected erythrocyte. Mol. Biochem. Parasitol. 2004;133:1–14. doi: 10.1016/j.molbiopara.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Estrano C., Bhattacharjee S., Harrison T., Haldar K. Cooperative domains define a unique host cell-targeting signal in Plasmodium falciparum-infected erythrocytes. Proc. Natl Acad. Sci. USA. 2003;100:12402–12407. doi: 10.1073/pnas.2133080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papakrivos J., Newbold C.I., Lingelbach K. A potential novel mechanism for the insertion of a membrane protein revealed by a biochemical analysis of the Plasmodium falciparum cytoadherence molecule PfEMP-1. Mol. Microbiol. 2005;55:1272–1284. doi: 10.1111/j.1365-2958.2004.04468.x. [DOI] [PubMed] [Google Scholar]
- 4.Taraschi T.F., O'Donnell M., Martinez S., Schneider T., Trelka D., Fowler V.M., Tilley L., Moriyama Y. Generation of an erythrocyte vesicle transport system by Plasmodium falciparum malaria parasites. Blood. 2003;102:3420–3426. doi: 10.1182/blood-2003-05-1448. [DOI] [PubMed] [Google Scholar]
- 5.Trelka D.P., Schneider T.G., Reeder J.C., Taraschi T.F. Evidence for vesicle-mediated trafficking of parasite proteins to the host cell cytosol and erythrocyte surface membrane in Plasmodium falciparum infected erythrocytes. Mol. Biochem. Parasitol. 2000;106:131–145. doi: 10.1016/s0166-6851(99)00207-8. [DOI] [PubMed] [Google Scholar]
- 6.Wickert H., Gottler W., Krohne G., Lanzer M. Maurer's cleft organization in the cytoplasm of Plasmodium falciparum-infected erythrocytes: new insights from three-dimensional reconstruction of serial ultrathin sections. Eur. J. Cell Biol. 2004;83:567–582. doi: 10.1078/0171-9335-00432. [DOI] [PubMed] [Google Scholar]
- 7.Wickham M.E., Rug M., Ralph S.A., Klonis N., McFadden G.I., Tilley L., Cowman A.F. Trafficking and assembly of the cytoadherence complex in Plasmodium falciparum-infected human erythrocytes. EMBO J. 2001;20:5636–5649. doi: 10.1093/emboj/20.20.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnefoy S., Guillotte M., Langsley G., Mercereau-Puijalon O. Plasmodium falciparum: characterization of gene R45 encoding a trophozoite antigen containing a central block of six amino acid repeats. Exp. Parasitol. 1992;74:441–451. doi: 10.1016/0014-4894(92)90206-p. [DOI] [PubMed] [Google Scholar]
- 9.Marti M., Baum J., Rug M., Tilley L., Cowman A.F. Signal-mediated export of proteins from the malaria parasite to the host erythrocyte. J. Cell Biol. 2005;171:587–592. doi: 10.1083/jcb.200508051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiller N.L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estrano C., Haldar K. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 11.Knuepfer E., Rug M., Klonis N., Tilley L., Cowman A.F. Trafficking of the major virulence factor to the surface of transfected P.falciparum-infected erythrocytes. Blood. 2005;105:4078–4087. doi: 10.1182/blood-2004-12-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marti M., Good R.T., Rug M., Knuepfer E., Cowman A.F. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 13.Horrocks P., Muhia D. Pexel/VTS: a protein-export motif in erythrocytes infected with malaria parasites. Trends Parasitol. 2005;21:396–399. doi: 10.1016/j.pt.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Templeton T.J., Deitsch K.W. Targeting malaria parasite proteins to the erythrocyte. Trends Parasitol. 2005;21:399–402. doi: 10.1016/j.pt.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Schneider A.G., Mercereau-Puijalon O. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics. 2005;6:30. doi: 10.1186/1471-2164-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sargeant T.J., Marti M., Caler E., Carlton J.M., Simpson K., Speed T.P., Cowman A.F. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ifediba T., Vanderberg J.P. Complete in vitro maturation of Plasmodium falciparum gametocytes. Nature. 1981;294:364–366. doi: 10.1038/294364a0. [DOI] [PubMed] [Google Scholar]
- 18.Taylor H.M., Grainger M., Holder A.A. Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect. Immun. 2002;70:5779–5789. doi: 10.1128/IAI.70.10.5779-5789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y., Sifri C.D., Lei H.H., Su X.Z., Wellems T.E. Transfection of Plasmodium falciparum within human red blood cells. Proc. Natl Acad. Sci. USA. 1995;92:973–977. doi: 10.1073/pnas.92.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deitsch K., Driskill C., Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahl A., Brunk B., Crabtree J., Fraunholz M.J., Gajria B., Grant G.R., Ginsburg H., Gupta D., Kissinger J.C., Labo P., et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Notredame C., Higgins D.G., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 24.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claros M.G., von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 26.Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 27.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sam-Yellowe T.Y., Florens L., Johnson J.R., Wang T., Drazba J.A., Le Roch K.G., Zhou Y., Batalov S., Carucci D.J., Winzeler E.A., et al. A Plasmodium gene family encoding Maurer's cleft membrane proteins: structural properties and expression profiling. Genome Res. 2004;14:1052–1059. doi: 10.1101/gr.2126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng Q., Cloonan N., Fischer K., Thompson J., Waine G., Lanzer M., Saul A. stevor and rif are Plasmodium falciparum multicopy gene families which potentially encode variant antigens. Mol. Biochem. Parasitol. 1998;97:161–176. doi: 10.1016/s0166-6851(98)00144-3. [DOI] [PubMed] [Google Scholar]
- 30.Kyes S.A., Rowe J.A., Kriek N., Newbold C.I. Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc. Natl Acad. Sci. USA. 1999;96:9333–9338. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaviratne M., Khan S.M., Jarra W., Preiser P.R. Small variant STEVOR antigen is uniquely located within Maurer's clefts in Plasmodium falciparum-infected red blood cells. Eukaryot. Cell. 2002;1:926–935. doi: 10.1128/EC.1.6.926-935.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Przyborski J.M., Miller S.K., Pfahler J.M., Henrich P.P., Rohrbach P., Crabb B.S., Lanzer M. Trafficking of STEVOR to the Maurer's clefts in Plasmodium falciparum-infected erythrocytes. EMBO J. 2005;24:2306–2317. doi: 10.1038/sj.emboj.7600720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kraemer S.M., Smith J.D. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol. Microbiol. 2003;50:1527–1538. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 35.Kooij T.W., Carlton J.M., Bidwell S.L., Hall N., Ramesar J., Janse C.J., Waters A.P. A Plasmodium whole-genome synteny map: Indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 2005;1:e44. doi: 10.1371/journal.ppat.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freitas-Junior L.H., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P.falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 37.Aravind L., Iyer L.M., Wellems T.E., Miller L.H. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 38.Taylor H.M., Kyes S.A., Newbold C.I. Var gene diversity in Plasmodium falciparum is generated by frequent recombination events. Mol. Biochem. Parasitol. 2000;110:391–397. doi: 10.1016/s0166-6851(00)00286-3. [DOI] [PubMed] [Google Scholar]
- 39.Su X., Ferdig M.T., Huang Y., Huynh C.Q., Liu A., You J., Wootton J.C., Wellems T.E. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.