Abstract
VectorBase (http://www.vectorbase.org/) is a web-accessible data repository for information about invertebrate vectors of human pathogens. VectorBase annotates and maintains vector genomes providing an integrated resource for the research community. Currently, VectorBase contains genome information for two organisms: Anopheles gambiae, a vector for the Plasmodium protozoan agent causing malaria, and Aedes aegypti, a vector for the flaviviral agents causing Yellow fever and Dengue fever.
INTRODUCTION
Even before the completion of the human genome a number of laboratories initiated projects to sequence the genomes of important human pathogens: Plasmodium, Trypanosome and Leishmania species (1–3). The aim of these projects was to better understand the biology of the pathogen through its genome, with the goal of identifying new therapeutics and thus shorten the time from therapeutic lead to marketable product, a notoriously slow process. A more holistic approach to improving our understanding of these pathogens needs to include intermediary vectors where they exist. Over the past few years the cost of genome sequencing has fallen dramatically making it feasible to sequence the genomes of vectors and complete our knowledge of the triumvirate of species involved in many parasitic diseases.
VectorBase is funded by the National Institute of Allergy and Infectious disease (NIAID) as part of a group of Bioinformatics Resource Centres (BRCs) (http://www.brc-central.org/) aiming to provide web-based resources to the scientific community for organisms considered to be causing or transmitting emerging or re-emerging infectious disease. Parallel to this, NIAID has funded a number of genome projects of important vector species that are destined to be housed within the VectorBase system (Table 1).
Table 1.
List of vector species scheduled for inclusion into VectorBase
| Vector | Disease | Status |
|---|---|---|
| Aedes aegyptia | Yellow and Dengue fever | Complete |
| Anopheles gambiae PEST | Malaria | Complete |
| A.gambiae M formb | Malaria | Initiated |
| A.gambiae S formb | Malaria | Initiated |
| Culex pipiens quinquefasciatusa | Lymphatic filariasis | Assembly |
| Glossina morsitansc | Sleeping sickness | Initiated |
| Ixodes scapularisa | Lyme disease | Sequencing |
| Lutzomyia longipalpisd | Leishmaniasis | Planned |
| Pediculus humanusb | Typhus | Initiated |
| Phlebotomus papatasid | Leishmaniasis | Planned |
| Rhodnius prolixusb | Chagas disease | Initiated |
aFunded by NIAID.
bFunded by NHGRI.
cFunded by Wellcome Trust.
dFunded by NHGRI and Wellcome Trust.
VectorBase is involved in all the stages of genome analysis: first-pass annotation of new genome sequences in collaboration with the sequencers, re-annotation of existing genome sequences and submission of these data sets to the public nucleotide databanks.
VectorBase acts as the repository for the genome and predicted gene set providing web access for browsing and data mining capability. VectorBase participates in teaching workshops (supporters include WHO-TDR, MR4, EMBO and BioMalPar) and has undertaken ‘hands-on’ demonstrations at international meetings. VectorBase strives to improve the accuracy and scope of the annotations, expanding controlled vocabularies for the vectors and incorporating new data types (expression, population and variation data).
RESULTS
Data storage
VectorBase uses the Genome Model Organism Database (GMOD) construction set for the storage of genome sequence and annotations. The GMOD CHADO schema facilitates the rapid incorporation of diverse data types, e.g. literature, controlled vocabularies and good inter-operability with the manual annotation effort, using Apollo, as well as the Ensembl system. For web display and access to the genome data and annotation, VectorBase utilises the Ensembl database schema, API and web code (4).
Genome browsing and data mining
The Ensembl system provides a good model for handling genomic data from a number of species in a consistent and unified manner and a highly sophisticated set of interlinked web pages.
Entry points into the genome are through text searches of gene names, symbols and descriptions, pairwise similarity searches and from cross-references in the public nucleotide and protein databanks.
The VectorBase website contains standard Ensembl style gene and transcript pages. Gene pages contain information about the prediction including gene orthologue and protein features (signal peptides, trans-membrane domains, InterPro domains). Gene Ontology (GO) codes and Enzyme Classification (EC) numbers are assigned where possible.
Batch file downloads are available for both the raw sequence data (fasta files of genomic sequence, including repeat masked sequence and ESTs) and the annotation (GFF3 files or a MySQL dump for use with the Ensembl API).
Batch searching capabilities are handled by the powerful data mining tool BioMart (5) and through two spreadsheets, AnoXcel and AegyXcel (6) that contain gene based information about the presence of signal peptides, trans-membrane domains, protein domains and the best similarity with yeast, Drosophila and human.
Annotation
The two genomes currently available through VectorBase are good examples of the multiple roles undertaken by the group.
Anopheles gambiae annotation
The A.gambiae PEST genome was published in 2002 (7) with a genome size of 260 Mb and ∼14 000 genes. The predicted gene set has been reviewed and updated several times since publication. This process involves a blend of automated evidence-based gene prediction (8) and manual approaches. Manual appraisal of gene models is firstly targeted to regions of interest from the community and regions for which we are aware that automated approaches fail. A manual re-annotation of chromosome arm 2L is finished. Manually appraised gene models are highlighted for the user as a separate track on the genome browser.
Aedes aegypti annotation
The A.aegypti Liverpool strain was sequenced and assembled by The Institute of Genomic Research (TIGR) and the Broad Institute. Aedes has a large genome size of 1.3 Gb and a predicted gene complement of ∼16 000 genes. This represents the first-pass annotation of the genome using automated approaches. Improvements in quality will be achieved by manual efforts and enhancements to the evidence-based automated gene predictions.
Sequence comparisons between Anopheles and Aedes
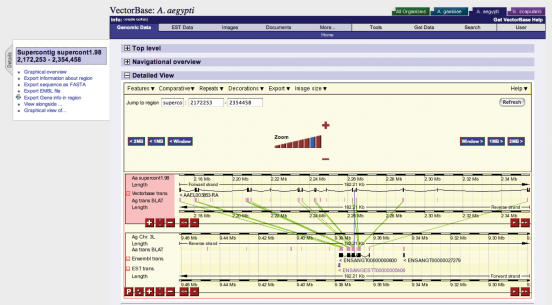
The presence of two related mosquito genomes in VectorBase allows for comparative analysis to identify conserved regions between the genomes. This can be useful in verifying, and correcting, gene models and for studying gene family expansions. Translated BLAT (9) similarities between the two mosquito species and with Drosophila have been identified. Figure 1 shows a view of an orthologous locus between the two mosquito genomes and highlights the expanded intron size in A.aegypti and related higher repeat content.
Figure 1.
Comparative display of Aedes aegypti gene AAEL003853 (upper panel) with the Anopheles gambiae orthologue (lower panel). Green lines join blocks of similarity between the two genomes and highlights the expansion of intron size due to an increased frequency of repeat sequences.
Use of Distributed Annotation System (DAS) in VectorBase
The DAS protocol (10) allows community researchers to integrate and display their data sets in the genome browser window. This is especially powerful with alternative sets of gene predictions. As an example, the Anopheles browser contains DAS tracks for alternate EST based gene predictions from AnoEST (11), an independent re-annotation effort by Li et al. (12) and predictions based on mass spectrometry data (13).
Microarray data
Microarray data exists for both Anopheles and Aedes and array probes from both species are mapped to the genome. These alignments are displayed in the browser and queries can be made against these via BioMart. Concise expression summaries for probes and genes are made available as experiments are published.
FUTURE DIRECTIONS
At least four new arthropod vector genomes will soon be incorporated into VectorBase: the mosquito Culex pipiens quinquefasciatus, the tick Ixodes scapularis, the kissing bug Rhodnius prolixus and the human body louse, Pediculus humanus (see Table 1 for more details). Furthermore, the genomes of two molecular forms of A.gambiae (the S and M forms which are considered to be incipient or possibly distinct species) will soon be completed and integrated into VectorBase. We will continue to re-annotate the existing mosquito genomes to improve gene prediction drawing more on manual/community annotation and comparative analysis with additional arthropod genomes.
The increased importance of manual/community annotation is being addressed by the development of a CHADO-based database for tracking internal VectorBase manual annotation and submissions from the community.
Other material of interest to the vector community is being incorporated, including the newly developed controlled vocabulary of mosquito anatomy (http://obo.sourceforge.net/detail.cgi?mosquito_anatomy) and other vector-related ontologies.
VectorBase is an ongoing project and the scope and usability of the site are improving rapidly. The coming year will see a significant expansion in the number of vector genomes housed.
Acknowledgments
The core VectorBase project is funded by contract HHSN266200400039C from the NIAID, and supported, in part, by the BioMalPar network of excellence. We would like to thank the reviewers for their helpful comments and insights. Funding to pay the Open Access publication charges for this article was provided by NIH-NIAID.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C., Lennard N.J., Caler E., Hamlin N.E., Haas B., et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 3.Ivens A.C., Peacock C.S., Worthey E.A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M.A., Adlem E., Aert R., et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birney E., Andrews D., Caccamo M., Chen Y., Clarke L., Coates G., Cox T., Cunningham F., Curwen V., Cutts T., et al. Ensembl 2006. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krasprzyk A., Keefe D., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E. Ensmart: A generic system for fast and flexible acces to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribeiro J.M.C., Topalis P., Louis C. AnoXcel: an Anopheles gambiae protein database. Insect Mol. Biol. 2004;13:449–457. doi: 10.1111/j.0962-1075.2004.00503.x. [DOI] [PubMed] [Google Scholar]
- 7.Holt R.A., Subramanian G.M., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 8.Curwen V., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M., Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowell R.D., Jokerst R.M., Day A., Eddy S.R., Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kriventseva E.V., Koutsos A.C., Blass C., Kafatos F.C., Christophides G.K., Zdobnov E.M. Toward A.gambiae functional genomics. Genome Res. 2005;15:893–899. doi: 10.1101/gr.3756405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Riehle M.M., Zhang Y., Xu J., Oduol F., Gomez S.M., Eigelmeier K., Ueberheide B.M., Shabanowitz J., Hunt D.F., et al. Anopheles gambiae genome reannotation through synthesis of ab initio and comparative gene prediction algorithms. Genome Biol. 2006;7:R24. doi: 10.1186/gb-2006-7-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalume D.E., Peri S., Reddy R., Zhong J., Okulae M., Kumar N., Pandey A. Genome annotation of Anopheles gambiae using mass spectrometry-derived data. BMC Genomics. 2005;19:128. doi: 10.1186/1471-2164-6-128. [DOI] [PMC free article] [PubMed] [Google Scholar]