Abstract
IntAct is an open source database and software suite for modeling, storing and analyzing molecular interaction data. The data available in the database originates entirely from published literature and is manually annotated by expert biologists to a high level of detail, including experimental methods, conditions and interacting domains. The database features over 126 000 binary interactions extracted from over 2100 scientific publications and makes extensive use of controlled vocabularies. The web site provides tools allowing users to search, visualize and download data from the repository. IntAct supports and encourages local installations as well as direct data submission and curation collaborations. IntAct source code and data are freely available from http://www.ebi.ac.uk/intact.
INTRODUCTION
The understanding of the cell machinery, the characterization of protein function as well as the discovery of new drug targets can be greatly enhanced by studying molecular interactions. We have witnessed in the past few years, a considerable increase of the number of publications reporting molecular interaction, but also the amount of interactions reported in a single publication, scaling from a single to over 22 000 binary interactions (1). The fragmentation of the datasets as well as their lack of formal representation makes it often difficult to reuse the data as the foundation for further research. IntAct addresses these issues by manually annotating published manuscripts reporting molecular interaction data and formalizing it by using a comprehensive set of controlled vocabularies in order to ensure data integrity. The data are made publicly available using the PSI-MI XML Standard (2), providing end users with the highest level of details without compromising the integrity and simplicity of access to the data, thanks to the use of well established standards.
DATA MODEL
The IntAct data model has grown more flexible and detailed over the years in order to cope with the ever evolving level of detail captured by experimentalists (e.g. kinetic data).
We are now going to describe a few key features of the IntAct data model, for a full description please see the annotated UML model on the website.
Molecule types
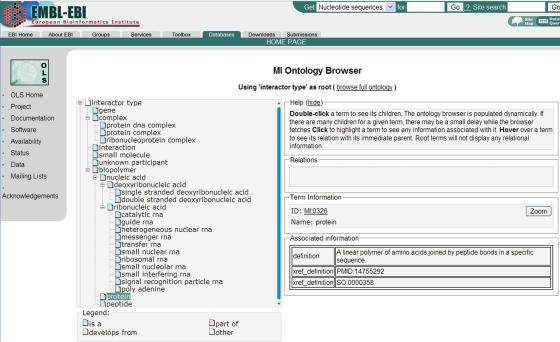
IntAct focuses on the curation of protein–protein interactions, but now also captures a growing number of key studies providing details of DNA, RNA and small molecule interactions. The list of interactor types is still evolving over time and we need our model to encompass these additions without compromising its stability. Thus, we model different molecule types by a generalized ‘interactor’ datatype, which is further specified by a hierarchical controlled vocabulary of the PSI-MI ontology. Hence, should we be adding a new one in the future, the IntAct data model would remain unchanged and a new controlled vocabulary term would be added. You can find more details on the hierarchical structure and the specific terms within this ontology in the Ontology Lookup Service (3) (Figure 1).
Figure 1.
Representation of the hierarchy of interactor type viewed in the Ontology Lookup Service (http://www.ebi.ac.uk/ontology-lookup). Clicking on a controlled vocabulary term displays additional information such as definition, literature references, cross references, etc.
Interacting domains
It is becoming more and more common to find in publications, details such as the relevant domain of an interacting protein. This is known in IntAct as Feature and now allows some lack of clarity by the author in the definition of the domain boundaries of a subsequence, reflecting experimental uncertainties. Here are a few examples of range:
Ser-7,
from 4 to between 10 and 23,
from 78 to <142,
transmembrane region.
One can also use features to represent modifications to the original molecule made by the author, e.g. a C-terminal tag on a protein used in a tandem affinity purification protocol.
Hierarchical build-up
The modeling of a molecular interaction can be convoluted, sometimes requiring the description of complex sub-units that are later assembled to form larger interactions. In order to cope with this requirement, the hierarchical build-up of molecular interactions was introduced. Interactions can be used as an interactor, and thus they can be reused in the context of other interactions.
Negative data
This is reported only to a very strict criteria, e.g. when an author produces contradictory results within a single paper, or when isoforms are shown to behave differently in respect to potential interacting partners. Negative experiments are clearly indicated as such and may be easily filtered out if not required.
CURATION PROCESS AND QUALITY ASSURANCE
IntAct strives to provide users with data featuring a high level of structured annotation (i.e. as far as available in the publication). The ways to achieve this goal are manifold:
Controlled vocabularies
IntAct makes extensive use of ontologies to represent experimental conditions as well as general concepts such as databases or interactor types and thus enforces data integrity and provides a powerful means for searching data. IntAct mainly uses the ontologies of the PSI-MI standard for molecular interactions (Table 1).
Table 1.
Major categories of controlled vocabulary in IntAct
| Ontology type | Number of terms |
|---|---|
| Interaction detection method | 175 |
| Interaction type | 62 |
| Participant detection method | 30 |
| Interactor type | 28 |
| Sequence feature type | 174 |
| Sequence feature detection | 31 |
Mapping of biological objects
Interacting molecules are systematically mapped to stable identifiers from public databases such as UniProtKB (4) for proteins, ChEBI (5) and the DDBJ/EMBL/GenBank (6) nucleotide databases for nucleic acids for small molecules. This is a highly time consuming part of the curation process but it is also crucial to ensure precision and comparability of the data. In cases where the authors give sequence information when describing a feature such as an interacting residue or binding site, this is mapped back to the parent sequence (or, when possible, the appropriate isoform) in UniProtKB. In cases where sequence information is not given, e.g. when identification is made by antibody detection, it is assumed that the authors annotation is correct however maintaining within IntAct an association between both the interaction and the corresponding descriptions of both the interaction and participant detection methods allows the user to make their own assessment of the accuracy of this data. When mapping high throughput datasets, there is often a small proportion of participants which cannot be traced due to the instability of the identifier used. Protein are remapped to UniProtKB, to allow use of their versioning and archiving services to maintain mappings and author identifiers are retained and revisited to attempt to improve coverage upto 100%.
Curation manual
Over the years, we have written and maintained a very detailed curation manual explaining how IntAct records are being curated. This manual is publicly available from the IntAct home page.
Expert curation
All records are manually annotated by domain experts, using the curation manual as a reference guide. Every record is then cross-checked by a second curator.
Software checking
By studying the record produced over time, a set of recurrent data consistency issues has been identified. Computational checking for these cases is performed on a nightly basis. Curators are sent reports and requested to amend the records concerned.
Direct submissions
Authors of publications reporting molecular interaction data are encouraged to submit the interaction data to IntAct prior to publication. On finalization of the record, we will issue a public accession number that can be referred to in the manuscript. However, the data will only be released on publication of the manuscript or on explicit request of the data submitter. For details of submission methods and formats, please refer to the deposition page of the International Molecular intraction Exchange (IMEx) consortium of molecular interaction databases at http://imex.sf.net.
Curation collaborations
IntAct increasingly collaborates with partners on specific curation topics, either performing targeted curation for collaborators, or providing a private instance of IntAct as well as infrastructure and support for curation project by external partners. If you are interested in either of these, please contact intact-help@ebi.ac.uk. IntAct data is released on a weekly basis and is available on the web site as well as for download in PSI-MI 1.0 and 2.5 XML format (classified by organism and publication).
APPLICATIONS
In addition to its publicly available data, IntAct provide several web applications allowing users to browse, visualize and perform analyzes of the data stored in the repository (be it their own local instance of IntAct or the EBI public repository). We are now going to describe a few enhancements made on existing applications as well as new applications made available to the community.
Easy data download
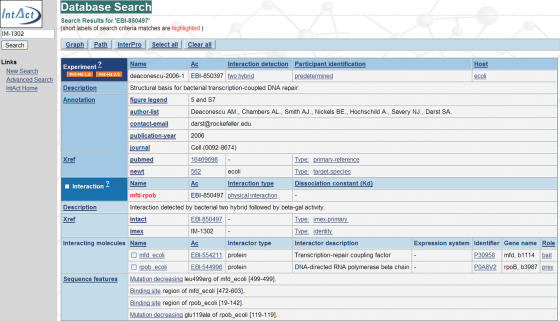
The experiment view (Figure 2) allows users to download the publication currently being viewed by simply clicking on one of the icons in the upper left corner, two formats are currently available: PSI-MI XML 1.0 and 2.5.
Figure 2.
The IntAct experiment view. This view provides a very deep level of detail, including publication reference, experimental conditions and details of the interaction and the participants. In the example shown above, the specific binding region is described as an amino acid sequence range. In addition, there are details of point mutation leading to a decrease of the participants' binding affinity. This interaction was found by searching for its IMEx accession number (i.e. IM-1302). It appears in the record as a cross reference (Xref) of the interaction. Should users be wishing to download this data, a list of available formats is available on the top left corner of the screen. The download encompasses the whole dataset related to the publication currently being displayed.
Textual browsing
In order to respond to the increasing amount of data being stored in the database as well as the number of interactions that can be extracted from a single publication, we have developed ways to easily browse through large collections of data while keeping usability and performance at their best. Whenever a user request matches a large amount of data, a paging mechanism splits the dataset in smaller chunks that the user can navigate at will.
Data search
A simple, yet versatile, search engine is available and allow to search for a broad variety of criteria such as publication ID, InterPro domain, UniProtKB ID, gene names, IMEx ID. When searching through large amount of data, search criteria combination and filtering become crucial features. In order to give users more freedom, a Lucene-based (http://lucene.apache.org) search module was integrated, thus, giving more flexibility when building queries as well as the opportunity to apply controlled vocabulary filters. For instance, one can search all experiments using the interaction detection method ‘fluorescent resonance energy transfer’. Hierarchical controlled vocabularies can be displayed graphically in order to simplify term's selection.
Visualisation
Previously we reported on a functionality which allowed the user to display common Gene Ontology (GO) (7) annotation shared by a cluster of interacting molecules (8). We now allow users to highlight interactor sharing the same InterPro (9) annotation. Furthermore, the currently displayed interaction network can be saved in either PSI-MI XML 1.0 or 2.5. Doing so allow users to import their data into third party tools, such as Cytoscape (10) or Proviz (11) to enable further analysis.
Over the past two years, we have introduced new applications allowing users to perform analysis of interaction networks. MiNe (Minimal connecting Interaction Network) enables the understanding of how a given set of proteins relate to each other by looking for the shortest path connecting them in the underlying interaction network. The result of such query is displayed graphically using our visualization engine. The resulting interaction network can be downloaded in PSI-MI XML.
Statistics
The number of interactions curated in IntAct has almost tripled in the past two years, you can find more details such as the species' coverage and the representation of interaction detection methods on our statistics page available online: http://www.ebi.ac.uk/intact/statisticView.
There is an increasing number of scientific publication reporting on large scale interactome analysis based on pull-down of complexes. A crucial step in planning large scale experiments is the bait selection. IntAct provides an experimental tool that aims at assisting scientist by computing a prioritized list of ‘best bait’ which are expected to yield the highest return in experimental effort. These lists are generated using the Pay-As-You-Go strategy (12) which detects and prioritizes those proteins which have the highest likelihood of being hubs based on the current data within IntAct for various species. Using this strategy would save a large amount of experimental effort. This of course relies on the timely deposition of experimental data into the IntAct database in order that the Pay-As-You-Go algorithm remains up-to-date and effective.
FUTURE DEVELOPMENT
IntAct is constantly being improved and new services are made available regularly. Here are a few upcoming services:
Tabular data
Though PSI-MI XML provides a very detailed representation of molecular interaction data, many users are seeking a simplified representation of it. IntAct is going to provide tabular data files in the new PSI-MI tabular format containing binary interactions, reference to the originating publications as well as a minimal details about experimental conditions.
Datasets
As a result of an increasing number of external collaborations, we have increased the level of topic specific annotations, for example interactions believed to be involved in disease states such as cancer and Alzheimer's and organism specific sets such as cyanobacteria. We are developing extensions of our textual browsing tools for displaying these dataset more comprehensively, including statistics and improved access to data downloads in various formats.
Confidence score
Molecular interaction data originating from large scale experiments can be of varying quality. False positives can result from many causes: interactions that are identified in an experiment but actually never take place in the cell or inaccurate interpretation/breakdown of a complex into binary interactions. We are currently developing a statistical method that will allow the identification of interactions that are more likely to be biologically relevant.
Curated complexes
Many protein complexes can be isolated as a functional unit and their role in the cell, and the processes in which they are involved in, are thought to be well understood. However, the actual protein composition of such complexes can vary under different physiological conditions and the importance of such changes are far from fully comprehended. Authors often do not make clear the actual protein content of a complex when describing its activity and the nomenclature may often be misleading—the transcription factor AP-1, e.g. is in fact 16 different combinations of homo- and heterodimers. IntAct is developing a dictionary of complex nomenclature, with protein content clearly defined and linked to experimental evidence, with each variation of a complex given a distinct name and a separate entry. This information is being linked to the corresponding entry in the pathway database, Reactome (13), to give contextual information.
DISCUSSION
IntAct has initially been developed to support local installation and has now instances running around the world. Pharmaceutical companies, research laboratories as well as interaction databases have chosen to adopt our open source database and toolkit and whenever the need arise add novel or adapt existing functionality. If you are interested in a collaboration or a local IntAct installation, please contact us at intact-help@ebi.ac.uk or simply use the freely available source code.
Working toward giving fully inclusive access to the ever growing amount of molecular interaction data is a vast task, likely to be beyond the reach of any single interaction data resource. To share the curation workload, avoid redundant curation and ensure consistency in annotation policies, five public databases, BIND (14), MINT (15), DIP (16), MPact (17) and IntAct, have formed the IMEx consortium (IMEx—http://imex.sourceforge.net) to exchange molecular interaction records between partners. This cumulative effort should result in an overarching repository that is broader in scope and deeper in information than any individual efforts and one that scientists can use to better understand issues of health and disease or in the development of new drugs and therapeutics. To assist IMEx partners in capturing as much of published interaction data as possible, please refer to the IMEx website and submit your data pre-publication to one of the IMEx partners. To aid this process, and to ensure minimum data loss on submission due to the use of ambiguous or unstable identifiers, it is suggested that such data be compliant with the recently published Minimum Information required for reporting a Molecular Interaction Experiment (MIMIx) standard compliant (18).
Acknowledgments
Funded by EU grant number QLRI-CT-2001-00015 under the RTD programme ‘Quality of Life and Management of Living Resources’ and EU contract no. 21902 ‘Felics—Free European Life-Science Information and Computational Services’. Funding to pay the Open Access publication charges for this article was provided by Felics.
Conflict of interest statement. There are no conflict of interest.
REFERENCES
- 1.Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 2.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 3.Cote R.G., Jones P., Apweiler R., Hermjakob H. The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics. 2006;7:97. doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galperin M.Y. The Molecular Biology Database Collection: 2006 update. Nucleic Acids Res. 2006;34:D3–D5. doi: 10.1093/nar/gkj162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochrane G., Aldebert P., Althorpe N., Andersson M., Baker W., Baldwin A., Bates K., Bhattacharyya S., Browne P., van den Broek A., et al. EMBL Nucleotide Sequence Database: developments in 2005. Nucleic Acids Res. 2006;34:D10–D15. doi: 10.1093/nar/gkj130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Bateman A., Binns D., Bradley P., Bork P., Bucher P., Cerutti L., et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–D205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iragne F., Nikolski M., Mathieu B., Auber D., Sherman D. ProViz: protein interaction visualization and exploration. Bioinformatics. 2005;21:272–274. doi: 10.1093/bioinformatics/bth494. [DOI] [PubMed] [Google Scholar]
- 12.Lappe M., Holm L. Unraveling protein interaction networks with near-optimal efficiency. Nat. Biotechnol. 2004;22:98–103. doi: 10.1038/nbt921. [DOI] [PubMed] [Google Scholar]
- 13.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfarano C., Andrade C.E., Anthony K., Bahroos N., Bajec M., Bantoft K., Betel D., Bobechko B., Boutilier K., Burgess E., et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 16.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guldener U., Munsterkotter M., Oesterheld M., Pagel P., Ruepp A., Mewes H.W., Stumpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orchard S., Salwinski L., Kerrien S., Montecchi-Palazzi L., Oesterheld M., Stümpflen V., Ceol A., Chatr-aryamontri A., Armstrong J., Woollard P., et al. The Minimum Information required for reporting a Molecular Interaction Experiment (MIMIx) Nat. Biotechnol. 2006 doi: 10.1038/nbt1324. (In Press) [DOI] [PubMed] [Google Scholar]