Abstract
In The Institute for Genomic Research Rice Genome Annotation project (http://rice.tigr.org), we have continued to update the rice genome sequence with new data and improve the quality of the annotation. In our current release of annotation (Release 4.0; January 12, 2006), we have identified 42 653 non-transposable element-related genes encoding 49 472 gene models as a result of the detection of alternative splicing. We have refined our identification methods for transposable element-related genes resulting in 13 237 genes that are related to transposable elements. Through incorporation of multiple transcript and proteomic expression data sets, we have been able to annotate 24 799 genes (31 739 gene models), representing ∼50% of the total gene models, as expressed in the rice genome. All structural and functional annotation is viewable through our Rice Genome Browser which currently supports 59 tracks. Enhanced data access is available through web interfaces, FTP downloads and a Data Extractor tool developed in order to support discrete dataset downloads.
INTRODUCTION
Cereal species such as rice (Oryza sativa), maize/corn (Zea mays) and wheat (Triticum aestivum) are utilized throughout the world as major caloric sources (http://faostat.fao.org/site/336/default.aspx). These species, along with other cereal species, share not only sequence conservation but also synteny (1–7), thereby providing an opportunity for leveraging similar cereal sequences. A genome sequence initiative is in the initial stages for wheat (http://www.wheatgenome.org/), in progress for maize (http://www.maizesequence.org/) and multiple projects have been completed for rice (2,8–11). The complete, map-based finished sequence of a japonica subspecies of O.sativa was reported in 2005 by the International Rice Genome Sequencing Project [IRGSP, (11)]. The rice genome sequence will likely serve as a major reference for all cereal genomes as rice is a model cereal (12), the rice genome sequence is of finished quality and most other cereal genome sequences will not be finished to a similar level in the near future. Thus, high-quality, publicly available annotation of the rice genome is imperative for proper interpretation of future cereal genome sequences.
ANNOTATION OF THE RICE GENOME
Annotation of the rice genome by automated methods was previously reported by us and other groups (9–11,13–16). We continue to improve our annotation through a combination of computational advancements and implementation of semi-automation and manual annotation. Prior to Release 4, we modified the annotation pipeline to facilitate and accelerate manual annotation by annotating genes at the pseudomolecule rather than at the bacterial artificial chromosome (BAC) level as described previously (13,14). Pseudomolecule level annotation eliminated the need to resolve models in the overlap region between BACs while streamlining computational processes. The Release 4 pseudomolecule sequence incorporates new sequences and replaces unfinished sequences with finished sequence made available by the IRGSP subsequent to our Release 3 pseudomolecules (January 2004). The length of the 12 pseudomolecules in Release 4 is 372.1 Mb and includes 2.6 Mb of new sequences from the IRGSP [28 BACs/P1 artificial chromosomes clones (PACs)] and 25.1 Mb of updated (217 BACs/PACs) sequences. Thus, 99.1% of the total Release 4 sequence within the pseudomolecules is finished sequence. The number of physical gaps within the rice genome, excluding gaps at the centromeric and telomeric regions, has been reduced from 45 to 38.
In Release 4, at the structural annotation level, we implemented three major modifications and processes that improved the quality of the predicted gene models. First, applying the program to assemble spliced alignments [PASA, (17)] at the pseudomolecule rather than the BAC/PAC level allowed for more robust alignment of expressed sequence tags (ESTs) and full-length cDNAs as the gene models were not truncated at the termini of BAC/PAC clones. Duplicated regions between overlapping BAC/PAC clones were eliminated, thereby facilitating optimal alignment of cognate EST and full-length cDNA sequences to the pseudomolecules. Second, we utilized an improved version of PASA [PASA2, (17)] optimized for the rice genome sequence through manual evaluation of rice gene models. Third, we implemented a manual review step in which 2215 gene models were manually reviewed by human curators. Evaluation of experimental evidence supporting the PASA-proposed models resulted in acceptance of 1920 gene models (87%).
Annotated genes were assigned temporary identifiers in Releases 1 and 2; however, locus identifiers were assigned in Release 3. Thus, we generated a version converter (http://rice.tigr.org/tdb/e2k1/osa1/v_converter/index.shtml) to assist users in tracking genes between releases. With Release 4, some loci were deprecated and became obsolete. Thus, we created an ‘Obsolete Loci Web Page’ to facilitate tracking of loci no longer present in our annotation dataset.
At the functional annotation level, we provided more thorough identification of transposable element (TE)-related gene models by updating the TIGR Oryza Repeat Database (18). A total of 201 new rice repetitive sequences from GenBank were added to the TIGR Oryza Repeat Database (S. Ouyang, unpublished data). Automated putative function assignment, which incorporates similarity search results at the protein level and the Pfam domain annotation, was also improved in Release 4 through implementation of an iterative set of reviews and updates such that inconsistencies and uninformative names were replaced with more appropriate names. Implementation of this semi-automated process generated more consistent and accurate gene function assignments. Annotation statistics (Release 4) are available on the project website (http://rice.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml) and reflect improvements in both structural and functional annotation methods. We have also now made available via the Genome Browser and FTP site, InterPro search results.
Expression data
A variety of rice expression data types are available. They include a large collection of ESTs and full-length cDNAs (19) which provide a powerful resource not only for empirical support of transcript expression but also for improvement and validation of gene structure through the PASA component of our annotation pipeline. Quantitative transcript data are available through a massively parallel signature sequencing project [MPSS (20)] and a serial analysis of gene expression (SAGE) project [(21), http://www.mgosdb.org]. A limited set of proteomic data are also available (22). In Release 4, we used both transcript and proteomic expression data types to enhance gene model functional annotation and to generate an expression matrix and tool for the rice genome. For genes containing identifiable transcript (EST, full-length cDNA, MPSS, SAGE) or proteomic data, we have appended the functional annotation to indicate that the gene is expressed. At least one type of expression evidence was found for a total of 31 739 (50.5%) gene models. Of these, 22 804 (36.3%) models were completely supported by a PASA assembly (Table 1). Expression support for each gene model is available through a web interface (http://www.tigr.org/tdb/e2k1/osa1/locus_expression_evidence.shtml) in which the availability of each data type is summarized and linked to the Genome browser (see below). A tissue breakdown of EST evidence for the gene models is available through the Rice Gene Expression Anatomy Viewer (http://www.tigr.org/tdb/e2k1/osa1/dnav.shtml). The frequency of PASA-mapped ESTs in a given gene model can be queried on a tissue basis thereby allowing a graphical expression display. A virtual northern hybridization showing the expression level on an EST library basis is also available. For gene models with MPSS data, transcript frequency on a library basis is available through the University of Delaware Rice MPSS website (http://mpss.udel.edu/rice/).
Table 1.
Summary of annotated rice genes with expression evidence
| Evidence | Locus | Model | ||
|---|---|---|---|---|
| Number | % | Number | % | |
| Any expression evidence | 24 799 | 44.37 | 31 739 | 50.52 |
| PASA fully supporteda | 17 252 | 30.87 | 22 804 | 36.30 |
| FL-cDNA | 16 875 | 30.19 | 18 862 | 30.02 |
| EST | 19 394 | 34.70 | 26 002 | 41.39 |
| MPSS | 20 211 | 36.16 | 26 223 | 41.74 |
| SAGE | 9 593 | 17.16 | 13 736 | 21.86 |
| Proteomics | 1 952 | 3.49 | 2 654 | 4.22 |
Percentage refers to the portion of loci or models supported by a type of expression evidence.
aEvery exon of the gene model is covered by a PASA assembly.
Genome Browser display of structural, functional and comparative annotation
With the increased genomic sequencing activity not only in rice but also in other cereals, we have expanded and displayed our functional rice annotation in our Rice Genome Browser which is based on the Generic Genome Browser (23). There are 59 tracks available in the Genome Browser and related tracks are grouped into 10 categories. A separate web page briefly explains the data, methods and parameters for the generation of each track (http://www.tigr.org/tigr-scripts/osa1_web/gbrowse/rice/?help=citations). Track expansion reflects increased availability of sequence and functional genomic data for the cereal species, including rice. A notable addition is inclusion of Transcript Assemblies rather than Gene Indices for transcript support of the gene models. Previously, we had used the TIGR Gene Indices (24) for display of clustered, assembled transcript evidence. For rice and other species with genome annotation data, this was problematic as the TIGR Gene Indices are constructed using not only ESTs and full-length cDNAs but also any annotated genes (from the genome sequencing projects) available in GenBank in the build. Thus, the TIGR Gene Indices contain non-experimentally based sequences and are a possible source of annotation error propagation. To address this issue, we built ‘Transcript Assemblies’ for >200 plant species using a similar but more stringent clustering/assembly process that includes ESTs, full-length cDNAs and mRNAs exclusively and excludes non-experimental sequences such as genome annotations [(25), http://plantta.tigr.org]. These assemblies represent true transcripts derived empirically and can be used in structural and functional annotation of gene models. Transcript Assemblies were aligned to the rice genome and are viewable in the Genome Browser.
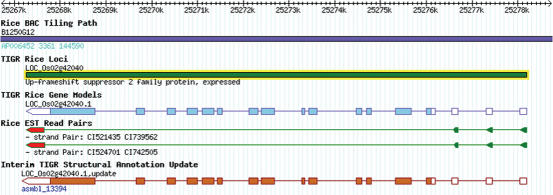
A set of ∼780 000 rice ESTs with read-pair information is available from the Rice Full-Length cDNA Sequencing Consortium. There are 400 534 sequences from 200 267 clones that are ‘read paired’. The read-pair information provides a valuable resource for improving structural annotation as it clearly defines the 5′ and 3′ transcript boundaries. We extracted read-pair information from GenBank EST records and aligned the read-paired ESTs to the rice genome, which is viewable as a separate track in the Rice Genome Browser with the direction of the EST read pairs shown graphically. Owing to the depth of the EST coverage, we can provide strong evidence for the boundaries of the transcript as well as alternative splice forms (Figure 1). Furthermore, with access to >1.2 M ESTs for rice, we have updated our structural annotation using the PASA2 program and have added an Interim TIGR Structural Annotation Update track on the browser so that any PASA-structural annotation updates occurring between versions is immediately user accessible.
Figure 1.
Display of EST read pairs in the TIGR Rice Genome Browser. Shown is a rice locus (LOC_Os02g42040) which lacks full-length cDNA support yet has empirical support for the 5′ and 3′ termini through multiple EST read pairs. The direction of the EST read pairs is indicated by an arrow and the 5′-termini is colored in green while the 3′-termini in red.
We mapped probes from five publicly available rice microarray platforms to the rice genome. Using a stringent cut-off of 100% identity and 100% coverage, 19 853 (98.1%) NSF Rice 20K Array probes (www.ricearray.org), 42 703 (98.6%) NSF Rice 45K Array probes (www.ricearray.org), 19 516 (90.8%) Agilent rice array probes (http://www.chem.agilent.com/cag/bsp/oligoGL/012106_D_GeneList_20050601.htm), 585 705 (92.8%) Affymetrix Rice Genome Array probes (http://www.affymetrix.com/products/arrays/specific/rice.affx) and 40 215 (68.9%) Yale/BGI rice oligoarray probes (26) have been mapped on to the genome. We updated the Generic Genome Browser to version 1.64 and upgraded the Gbrowse server to MySQL 5, thereby increasing performance.
Segmental duplication
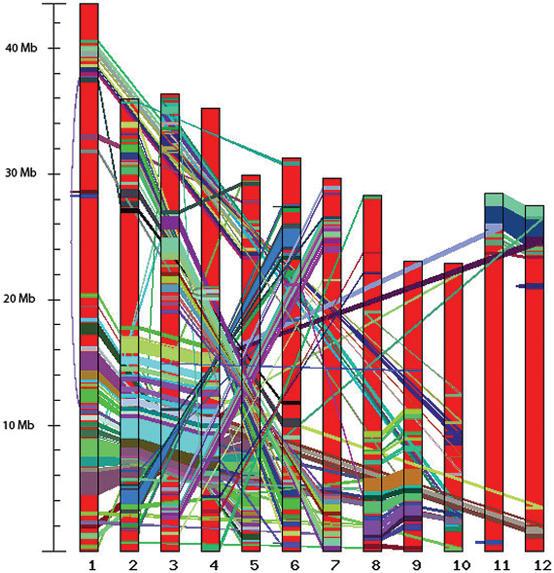
Segmental duplication in the rice genome can account for 15–62% of the sequence depending on the methods and parameters employed (27–31). To provide researchers with a resource for gene and genome evolution studies, we have provided an analysis of segmental duplication in Release 4. This analysis utilized the DAGChainer program (30,32) with parameters 100 and 500 kb for maximum distance allowed between two collinear gene pairs in order to identify segmentally duplicated genes. At 100 and 500 kb maximum distance allowed between two collinear gene pairs, 100 Mb (27.0%) and 194 Mb (52.3%), respectively, of the rice genome was located within the duplicated blocks which comprise 2403 and 3468 collinear gene pairs (100 and 500 kb maximum distance, respectively). A graphical representation of the segmentally duplicated regions can be viewed through a web interface (http://rice.tigr.org/tdb/e2k1/osa1/segmental_dup/index.shtml, Figure 2) with linkages to lists of genes located within the duplicated blocks.
Figure 2.
Segmental duplication within rice. Segmentally duplicated blocks among the 12 rice chromosomes are hyperlinked to gene lists, thereby allowing the users to navigate between a genome level view of the segmental duplication and annotation at the gene level annotation. The figure displays the segmental duplication within the rice genome generated using 100 kb as the maximum distance allowed between two collinear gene pairs.
Community annotation
We initiated a Community Annotation project in which gene family experts contribute structural and functional annotation. We have developed a web-based interface for Community Annotators to identify gene(s) for annotation through either sequence-based or text-based selection tools. Once selected, the Community Annotator can annotate gene structure using a simple web interface and then add functional annotation including putative function, gene name and evidence support. Annotators subsequently validate the Community Annotation and identify TIGR-generated models requiring updates in future releases. To date, we have 30 Community Annotators registered, have released 454 genes (representing 23 gene families) with Community Annotation and have another 142 genes (representing 9 gene families) in the queue for validation.
DATA AVAILABILITY
All data from our project are available through a series of web interfaces, FTP downloads and a Data Extractor developed for bulk downloads. Although FTP provides all rice genomic data available at TIGR, the Data Extractor (http://rice.tigr.org/tdb/e2k1/osa1/data_download.shtml) allows users to obtain many data types within a user-specified coordinate range. By using the Batch Download Tool (http://rice.tigr.org/tdb/e2k1/osa1/batch_download.shtml), through submission of a list of gene identifiers (loci or feat_names), users can batch download data from a specified list of features, including sequence, function and Gene Ontology assignment of gene models.
RELEASE 5 ACTIVITIES
We are currently preparing an updated release (Release 5). Owing to the limited amount of new or upgraded sequence available from the IRGSP (0.4 Mb in total, seven BACs/fosmid clones), we have elected to retain our pseudomolecules from Release 4 and update the annotation only. A major component of our annotation improvements will be the incorporation of ∼780 000 ESTs from GenBank as new transcript evidence using the PASA2 program. Currently, we have made available as a track on our Rice Genome Browser the PASA2-proposed gene models using ∼1.2 million ESTs, including the newly released ∼780 000 EST, as an Interim Structural Annotation update. For Release 5, we are in the process of manually reviewing the structural annotation of ∼2000 genes.
CONCLUSIONS
The rice genome will serve as a major resource for the sequence and annotation of other cereal genomes making it critical that high quality, uniform and publicly accessible annotation is available. We have improved the quality of annotation of the rice genome in our project and continued to make these data more accessible to the scientific community. We plan additional quality improvements in our annotation in the future which will maximize the incorporation of all evidence into the most accurate structural and functional annotation of the rice genome possible.
Acknowledgments
The efforts of the TIGR Bioinformatics department in generating a suite of tools and resources for eukaryotic sequence and annotation are appreciated. Work on rice genome annotation is funded by a grant (to C.R.B.) from the National Science Foundation (DBI-0321538). Funding to pay the Open Access publication charges for this article was provided by was provided by NSF DBI-0321538.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gale M.D., Devos K.M. Comparative genetics in the grasses. Proc. Natl Acad. Sci. USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 3.Sorrells M.E., La Rota M., Bermudez-Kandianis C.E., Greene R.A., Kantety R., Munkvold J.D., Miftahudin Mahmoud A., Ma X., Gustafson P.J., et al. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 2003;13:1818–1827. doi: 10.1101/gr.1113003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ware D., Stein L. Comparison of genes among cereals. Curr. Opin. Plant Biol. 2003;6:121–127. doi: 10.1016/s1369-5266(03)00012-8. [DOI] [PubMed] [Google Scholar]
- 5.The Rice Chromosome3 Sequencing Consortium. Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res. 2005;15:1284–1291. doi: 10.1101/gr.3869505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Rice Chromosome 10 Sequencing Consortium. In-depth view of structure, activity, and evolution of rice chromosome 10. Science. 2003;300:1566–1569. doi: 10.1126/science.1083523. [DOI] [PubMed] [Google Scholar]
- 7.The Rice Chromosomes 11 and 12 Sequencing Consortia. The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol. 2005;3:20. doi: 10.1186/1741-7007-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barry G.F. The use of the Monsanto draft rice genome sequence in research. Plant Physiol. 2001;125:1164–1165. doi: 10.1104/pp.125.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 10.Yu J., Wang J., Lin W., Li S., Li H., Zhou J., Ni P., Dong W., Hu S., Zeng C., et al. The Genomes of Oryza sativa: a history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 12.Goff S.A. Rice as a model for cereal genomics. Curr. Opin. Plant Biol. 1999;2:86–89. doi: 10.1016/S1369-5266(99)80018-1. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Q., Ouyang S., Liu J., Suh B., Cheung F., Sultana R., Lee D., Quackenbush J., Buell C.R. The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 2003;31:229–233. doi: 10.1093/nar/gkg059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Q., Ouyang S., Wang A., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., et al. The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiol. 2005;138:18–26. doi: 10.1104/pp.104.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito Y., Arikawa K., Antonio B.A., Ohta I., Naito S., Mukai Y., Shimano A., Masukawa M., Shibata M., Yamamoto M., et al. Rice Annotation Database (RAD): a contig-oriented database for map-based rice genomics. Nucleic Acids Res. 2005;33:D651–D655. doi: 10.1093/nar/gki083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohyanagi H., Tanaka T., Sakai H., Shigemoto Y., Yamaguchi K., Habara T., Fujii Y., Antonio B.A., Nagamura Y., Imanishi T., et al. The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Jr, Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouyang S., Buell C.R. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi S., Satoh K., Nagata T., Kawagashira N., Doi K., Kishimoto N., Yazaki J., Ishikawa M., Yamada H., Ooka H., et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- 20.Nakano M., Nobuta K., Vemaraju K., Tej S.S., Skogen J.W., Meyers B.C. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gowda M., Jantasuriyarat C., Dean R.A., Wang G.L. Robust-LongSAGE (RL-SAGE): a substantially improved LongSAGE method for gene discovery and transcriptome analysis. Plant Physiol. 2004;134:890–897. doi: 10.1104/pp.103.034496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koller A., Washburn M.P., Lange B.M., Andon N.L., Deciu C., Haynes P.A., Hays L., Schieltz D., Ulaszek R., Wei J., et al. Proteomic survey of metabolic pathways in rice. Proc. Natl Acad. Sci. USA. 2002;99:11969–11974. doi: 10.1073/pnas.172183199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quackenbush J., Cho J., Lee D., Liang F., Holt I., Karamycheva S., Parvizi B., Pertea G., Sultana R., White J. The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001;29:159–164. doi: 10.1093/nar/29.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Childs K., Hamilton J., Zhu W., Ly E., Cheung F., Wu H., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P. Nucleic Acids Res. 2007. The TIGR Plant Transcript Assemblies Database. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao W., Wang J., He X., Huang X., Jiao Y., Dai M., Wei S., Fu J., Chen Y., Ren X., et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32:D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandepoele K., Simillion C., Van de Peer Y. Evidence that rice and other cereals are ancient aneuploids. Plant Cell. 2003;15:2192–2202. doi: 10.1105/tpc.014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guyot R., Keller B. Ancestral genome duplication in rice. Genome. 2004;47:610–614. doi: 10.1139/g04-016. [DOI] [PubMed] [Google Scholar]
- 29.Paterson A.H., Bowers J.E., Chapman B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin H., Zhu W., Silva J.C., Gu X., Buell C.R. Intron gain and loss in segmentally duplicated genes in rice. Genome Biol. 2006;7:R41. doi: 10.1186/gb-2006-7-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X., Shi X., Hao B., Ge S., Luo J. Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol. 2005;165:937–946. doi: 10.1111/j.1469-8137.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- 32.Haas B.J., Delcher A.L., Wortman J.R., Salzberg S.L. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data from our project are available through a series of web interfaces, FTP downloads and a Data Extractor developed for bulk downloads. Although FTP provides all rice genomic data available at TIGR, the Data Extractor (http://rice.tigr.org/tdb/e2k1/osa1/data_download.shtml) allows users to obtain many data types within a user-specified coordinate range. By using the Batch Download Tool (http://rice.tigr.org/tdb/e2k1/osa1/batch_download.shtml), through submission of a list of gene identifiers (loci or feat_names), users can batch download data from a specified list of features, including sequence, function and Gene Ontology assignment of gene models.