Abstract
The microbial genome database for comparative analysis (MBGD) is a comprehensive platform for microbial comparative genomics. The central function of MBGD is to create orthologous groups among multiple genomes from precomputed all-against-all similarity relationships using the DomClust algorithm. The database now contains >300 published genomes and the number continues to grow. For researchers who are interested in ongoing genome projects, we have now started a new service called ‘My MBGD,’ which allows users to add their own genome sequences to MBGD for the purpose of identifying orthologs among both the new and the existing genomes. Furthermore, in order to make available the rapidly accumulating information on closely related genome sequences, we enhanced the interface for pairwise genome comparisons using the CGAT interface, which allows users to see nucleotide sequence alignments of non-coding as well as coding regions. MBGD is available at http://mbgd.genome.ad.jp/.
INTRODUCTION
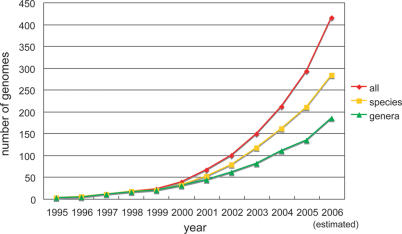
More than 300 genomic sequences have been determined to date, and the number of completed sequences continues to grow. Extracting useful information from such a growing number of genomes is a major challenge in comparative genomics. Interestingly, many of the completed genomic sequences are closely related to each other; of the 293 genomic sequences available at the end of 2005, the number of unique species (for which at least one genome sequence was determined) is 211, and the number of unique genera is only 135 (Figure 1). It is important to conduct comparative analyses not only of distantly related genomes, but also of closely related genomes, since we can extract different types of information about biological functions and evolutionary processes from comparisons of genomes at different evolutionary distances.
Figure 1.
Increase in the number of complete microbial (bacteria and archaea) genome sequences. The number of genomes published between January and June 2006 was doubled for the purpose of estimating total number of genomes newly determined during the year 2006. The data are taken from the GOLD database (13).
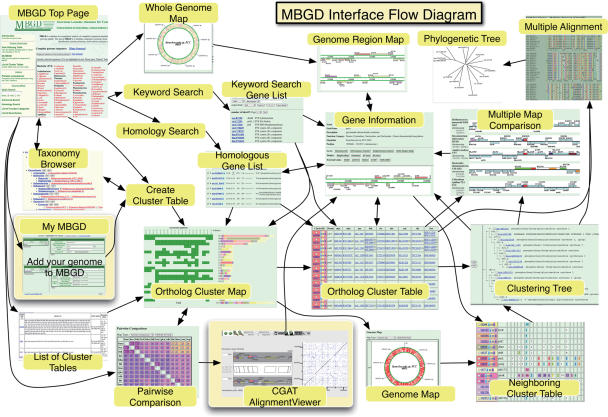
MBGD is a microbial genome database that provides a platform for large-scale comparative genome analysis based on comprehensive ortholog classification (1) (Figure 2). Unlike COG (2), TIGRFAMs (3) and other databases of orthologous groups constructed with curation processes, MBGD is comprehensive and routinely updated. Unlike OrthoMCL-DB (4), IMG (5) and other databases of orthologous groups constructed by automated procedures, MBGD allows users to classify genes dynamically. The key features of MBGD derive from an efficient clustering algorithm named DomClust (6), which is a hierarchical clustering algorithm for constructing ortholog groups at the domain (rather than gene) level from precomputed all-against-all similarity relationships. With this algorithm, MBGD not only provides the orthologous groups among the latest genomic data available, but also allows users to create their own ortholog groups using a specified set of organisms. The latter feature is especially useful when the user's interest is focused on some taxonomically related organisms (1); in fact, MBGD is most effectively used when an appropriate number of genomes are selected. However, in the previous version, users could only choose published genomes whose sequences were already available in MBGD.
Figure 2.
The user interface flow diagram of MBGD. The arrows represent possible transitions between pages. The new functionalities, My MBGD and CGAT AlignmentViewer, are highlighted.
With the growing amount of the microbial genomic information in mind, we have started a new service called ‘My MBGD,’ which allows users to add their own genome sequences to MBGD for the purpose of finding orthologous relationships among the newly added genomes and the existing genomes. Furthermore, in order to facilitate comparisons of closely related genomes, we have also enhanced the interface of pairwise comparison using the CGAT interface (7), which is a Java applet for displaying genome alignment on both dotplot and alignment viewers.
MY MBGD: ADD YOUR OWN GENOME DATA TO MBGD
The data-building protocol of MBGD was previously described (1), and is summarized in Figure 3. In MBGD, similarity relationships among all protein-coding genes are identified by all-against-all BLAST searches (8) with adjusted E-values ≤ 0.01, and for each of them, an optimal local alignment is calculated by the dynamic programming (DP) algorithm (9) using the JTT PAM250 scoring matrix (10); similarity scores, percentage identities, PAM distances and alignment positions are then taken from these alignments and stored in the database. Conserved domains are also searched in each sequence by the RPS–BLAST program against the CDD database (11). The clustering algorithm, DomClust, is then invoked to create orthologous groups among genomes using these similarity data, and a function category is assigned to each gene based on this clustering result.
Figure 3.
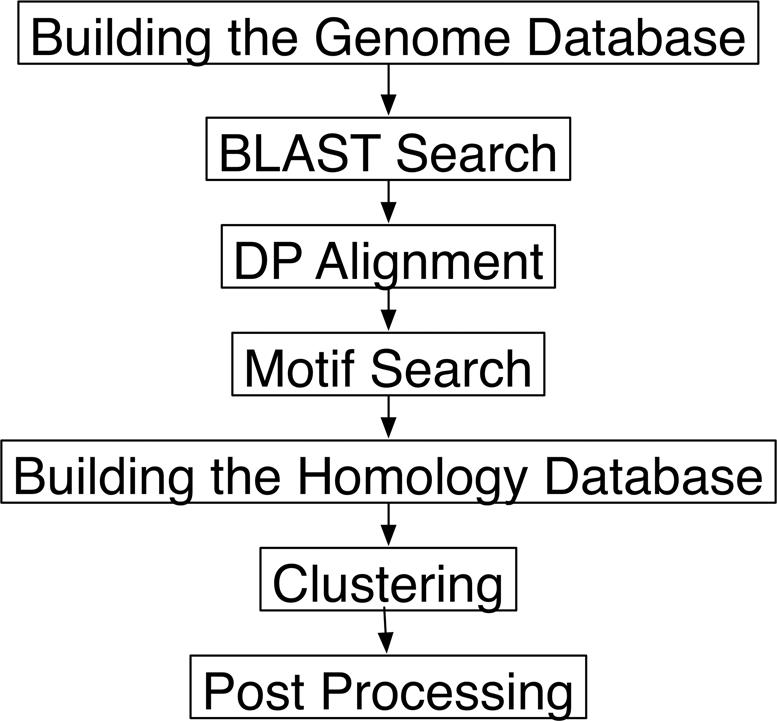
The protocol for database construction in MBGD.
My MBGD accepts genome data submitted by users and processes them according to this protocol. Currently, users must prepare translated sequences of all genes identified in the genome before submission. Sequence data are accepted in either GenBank format or FASTA format; in the latter case, users are also requested to provide a tab-delimited gene annotation table containing the information on the location and direction of each gene. Submitted data are stored in the database as separate tables. In MBGD, the private data submitted by the user and the public data already stored are logically merged using the ‘merge table’ feature implemented in the MySQL database management system. This mechanism enables us to allow users to use almost every function of MBGD without noticing the differences between My MBGD and the usual MBGD, except that the number of genome sequences that users are allowed to compare simultaneously is strictly limited in My MBGD: users must choose a set of organisms to be compared before starting the data construction process.
The all-against-all similarity search is the most time-consuming process. Actually, similarities are calculated between the query genomes and all the selected genomes. Depending on the server load, it typically takes a few hours to complete the calculation on our PC cluster system. The system notifies the user by e-mail after the data construction process is completed. After that, users can use every function just as with the usual MBGD; here the user's genomes are, by default, named ‘ug1’, ‘ug2’ and so on.
Users can create several ortholog cluster tables by choosing different sets of organisms to be compared, and switch these tables to conduct different types of analyses in the ‘List of Custer Tables’ page (Figure 2). For example, one can create two cluster tables, one of which is created from a set of genomes that are closely related to the target genome and the other from a set of genomes that are distantly related to each other.
Basically, the My MBGD functionality can be used without registration. In this case, however, a user cannot access the same data from different browsers due to the HTTP cookie mechanism that My MBGD uses to distinguish users. On the other hand, a registered user can see the same data from any browser on any machine.
CGAT: AN ALIGNMENT VIEWER FOR PAIRWISE COMPARISONS OF CLOSELY RELATED GENOMES
Although many microbial genome sequences exhibit a high sequence similarity, those that are too closely related cannot be usefully compared via MBGD, e.g. for the purpose of comparing the presence or absence of protein-coding genes among various genomes. In fact, in MBGD, only one genome is chosen from each species in the default setting. On the other hand, comparisons between the nucleotide sequences of closely related genomes are often useful to observe evolutionary changes in each genome directly, or to identify conserved regions such as regulatory sequences. To utilize information on closely related genomes, we implemented the CGAT interface for pairwise genome comparisons.
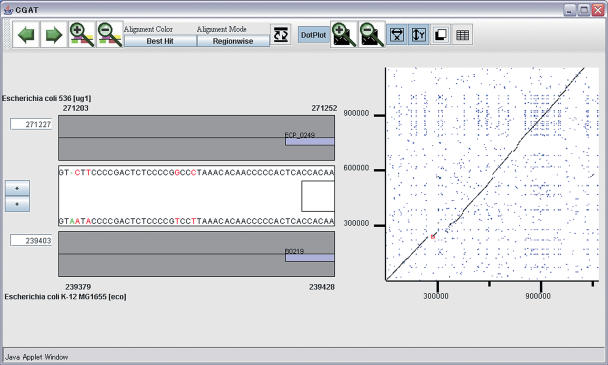
Originally, CGAT is a standalone program that employs client-server architecture (7) (available at http://mbgd.genome.ad.jp/CGAT/). In MBGD, we implemented a customized applet version of the client program (AlignmentViewer), which reads the data from the MBGD database. AlignmentViewer consists of an alignment display and a dotplot display with scrolling and zooming facilities (Figure 4), which are updated in a coherent fashion by user operations. They display precomputed alignments, which are, in this case, obtained by mapping the protein sequence alignments, which are calculated according to the protocol in Figure 3, onto each genome. Nevertheless, users can see on the AlignmentViewer a consecutive long nucleotide sequence alignment extended to non-coding regions (Figure 4), due to its navigation functionality of the alignment space along the orthologous alignments, as well as its functionality to calculate sequence alignments dynamically between specified regions.
Figure 4.
The CGAT interface showing the nucleotide sequence alignment between the genomes of two strains of Escherichia coli, 536 (‘ug1’) and K-12 (‘eco’). Here, the 536 genome is registered via the My MBGD functionality.
The CGAT interface is invoked from the page for pairwise comparisons, which can be accessed from the top page (Figure 2). After selecting a taxonomy group to compare, a pairwise comparison table is displayed, where the number in each cell (i, j) indicates the percentage ratio of the orthologous clusters shared between genomes i and j to those in genome i. Clicking each cell invokes the CGAT interface to compare genomes i and j. However, as mentioned, only one genome is selected from each species in the default setting. Therefore, users must choose a set of genomes on the taxonomy browser before accessing the pairwise comparison page, if they want to compare genomes between different strains of the same species; in this case, a clustering process is invoked to create a new cluster table containing the specified set of genomes.
FUTURE PLANS
Comparative genome analysis on the basis of ortholog identification is the most powerful approach for consistent and robust functional annotation. MBGD has used a unique protocol to assign a function category to each gene, which is based on a majority vote of the assignments made by some early genome projects. Although this protocol appears to give a robust assignment, there are several established function classification systems such as Gene Ontology (12) and COG categories (2). We have a plan to incorporate such information in order to improve the functional characterization of each orthologous group, whereby one can use MBGD as a resource for robust genome annotation, especially in combination with the My MBGD functionality.
The number of genome sequences continues to grow. However, the current interface of MBGD is not effective for simultaneous comparisons of a large number of genomes. Users must choose an appropriate number of genomes to compare for the effective use of the MBGD functionalities. Thus, another important task is to develop an interface suitable for simultaneous comparisons of a huge number of genomes.
Acknowledgments
The author thanks Toshio Higuchi for his programing support. The construction of the database is supported by a Grant-in-Aid for Publication of Scientific Research Results from the Japan Society for the Promotion of Science (JSPS). This work was also supported by the Institute for Bioinformatics Research Development, Japan Science Technology Agency (BIRD-JST) and by a Grant-in-Aid for Scientific Research on Priority Areas ‘Comparative Genomics’ from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. Funding to pay the Open Access publication charges for this article was provided by BIRD-JST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Uchiyama I. MBGD: microbial genome database for comparative analysis. Nucleic Acids Res. 2003;31:58–62. doi: 10.1093/nar/gkg109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haft D.H., Loftus B.J., Richardson D.L., Yang F., Eisen J.A., Paulsen I.T., White O. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen F., Mackey A.J., Stoeckert C.J., Jr, Roos D.S. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markowitz V.M., Korzeniewski F., Palaniappan K., Szeto E., Werner G., Padki A., Zhao X., Dubchak I., Hugenholtz P., Anderson I., et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchiyama I. Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes. Nucleic Acids Res. 2006;34:647–658. doi: 10.1093/nar/gkj448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchiyama I., Higuchi T., Kobayashi I. CGAT: a comparative genome analysis tool for visualizing alignments in the analysis of complex evolutionary changes between closely related genomes. BMC Bioinformatics. 2006;7:472. doi: 10.1186/1471-2105-7-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith T.F., Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 10.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 11.Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liolios K., Tavernarakis N., Hugenholtz P., Kyrpides N.C. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]