Abstract
The National Microbial Pathogen Data Resource (NMPDR) (http://www.nmpdr.org) is a National Institute of Allergy and Infections Disease (NIAID)-funded Bioinformatics Resource Center that supports research in selected Category B pathogens. NMPDR contains the complete genomes of ∼50 strains of pathogenic bacteria that are the focus of our curators, as well as >400 other genomes that provide a broad context for comparative analysis across the three phylogenetic Domains. NMPDR integrates complete, public genomes with expertly curated biological subsystems to provide the most consistent genome annotations. Subsystems are sets of functional roles related by a biologically meaningful organizing principle, which are built over large collections of genomes; they provide researchers with consistent functional assignments in a biologically structured context. Investigators can browse subsystems and reactions to develop accurate reconstructions of the metabolic networks of any sequenced organism. NMPDR provides a comprehensive bioinformatics platform, with tools and viewers for genome analysis. Results of precomputed gene clustering analyses can be retrieved in tabular or graphic format with one-click tools. NMPDR tools include Signature Genes, which finds the set of genes in common or that differentiates two groups of organisms. Essentiality data collated from genome-wide studies have been curated. Drug target identification and high-throughput, in silico, compound screening are in development.
INTRODUCTION
The National Microbial Pathogen Data Resource (NMPDR) is one of eight Bioinformatics Resource Centers funded by the National Institute of Allergy and Infectious Disease (NIAID) to provide the comprehensive bioinformatics environment needed to support research in biodefense, emerging infectious diseases, and re-emerging pathogens. The NMPDR focus organisms are NIAID Category B priority pathogens, including the food and water-borne diarrheagenic bacteria Campylobacter jejuni, Vibrio cholerae, Vibrio parahaemolyticus, Vibrio vulnificus, and Listeria monocytogenes. Also included are the nosocomial pathogens Staphylococcus aureus, Streptococcus pneumoniae and Streptococcus pyogenes (Group A Strep). NMPDR is both a central repository for a wide variety of scientific data on these pathogenic microorganisms and a platform for software tools that support investigator-driven data analysis. Its goal is to provide the most accurate functional annotations for its focus organisms in the context of comprehensive comparative analysis. To this end, NMPDR integrates genomic and functional data for hundreds of complete or nearly complete other bacterial, eukaryotic and archaeal genomes, the ‘supporting genomes’, that provide a rich phylogenetic context for data analysis.
Key to the correct interpretation of genomic data is the accuracy and consistency of functional annotations. The annotation environment underlying NMPDR is the SEED (http://www.theseed.org). Developed as an international collaboration, the SEED is a distributed system that organizes genomic data in the form of populated subsystems, rather than on an organism-by-organism basis (1). Subsystems are two-dimensional (2-D) integrations of biological functions with genome sequences, which are represented in spreadsheets as columns of functional roles, rows of genomes, and cells populated by the genes responsible for each function. Subsystems for the NMPDR focus organisms are created in a private instance of the SEED. Functional roles are first manually assigned to a group of proteins in a limited number of microbial genomes, creating a populated subsystem. Tools for comparative analysis, including precomputed sequence similarity and gene co-localization (functional coupling) (2,3), are used to extend the subsystem to additional genomes, resulting in consistent annotation of proteins that play the same functional role in a variety of organisms. Clustering analysis, combined with functional subsystems, is used to identify genes that encode functions known to be present in an organism, but that are performed by divergent proteins not automatically recognized by sequence similarity (4). These tools allow NMPDR to offer the most accurate and consistent annotations to date. To ensure the integrity of the annotations, NMPDR is read-only. Links are provided to the public SEED for community annotation of genes and for the construction of new subsystems by independent investigators.
CONTENT OF DATABASE
Website content and access to data
NMPDR provides access to all publicly available, complete genomes, which are searchable by keyword or alphanumeric identifier from the home page, http://www.nmpdr.org. Advanced search options include sequence search with BLAST (5), and categories for narrowing a keyword search such as taxonomy, alias and subsystem. As a service to investigators who study the Category B pathogens that are the focus of our curators, NMPDR provides organism summary pages as gateways for the analysis of Campylobacter, Listeria, S.aureus, Streptococcus and Vibrio. Keyword searches from the search box on these pages are automatically limited to that group of organisms. The available genomes in each group are presented in a table that lists strain name, genome size, number of annotated RNAs, and number of protein encoding genes (PEGs). The annotated PEGs are categorized into those which have been assigned functional names, and hypothetical genes that have no functional names. These categories are each further divided into genes that have been assigned to subsystems or not. The number of genes in each of the four categories is linked to a detailed list of the genes with links to comparative analysis pages for each PEG.
Organism summary pages are designed to provide user services and a collaborative environment for communities of investigators to share interests beyond bioinformatics. User forums have been established in the iLabs space (http://journals.tdl.org/jodi/article/view/jodi-152/138) (6) at the University of Illinois Graduate School of Library and Information Science for each focus group of pathogens. The user forums are linked from organism summary pages and include bulletin boards, shared document centers, link lists and an inquiry unit builder, which may be used to create interactive problems or case studies for teaching and learning. Email lists are also maintained for each organism group to encourage communication among professionals with common interests.
Organism summary pages contain a wide variety of information about the focus pathogens, from textbook descriptions to the most recently published findings. Technical descriptions of the organisms and genomes are shown as PathInfo, displayed in the style developed for PathPort (7). These descriptions will soon expand to include epidemiological information as well as information concerning organism isolation, identification, culture and safe handling. Access to the most recent developments in the professional literature is provided by a literature aggregator. This tool searches for the organism names in abstracts and keywords of journal articles published by the American Society for Microbiology (ASM) within the past two months, then displays the abstracts on the organism pages with links to the full-text. Because full-text access to the ASM journals requires a subscription until the articles are four months old, a collection of Open Access journals and RSS feeds of abstracts from other subscription journals are also linked. In contrast and just for fun, the most recent news in the popular press regarding these food-borne or flesh-eating pathogens is provided via RSS feed from Google news. Finally, organism-specific resources such as strain collections and other databases are also provided in a link list.
DATA TYPES
Primary data: sequence and features
Complete and essentially complete genomes that have been deposited in RefSeq (8) are the primary data. Most complete genomes are one contiguous length of DNA sequence data, or one ‘contig’. In the absence of other data, genome assemblies that are fragmented into multiple contigs are considered to be ‘essentially complete’ if they satisfy two heuristically derived rules: the sequence data must total more than 300 kbp (to help prevent depositions that are just plasmids from being incorrectly marked as ‘complete genomes’), and at least 70% of the sequence data must be contained in contigs that are longer than 20 kb. Theoretical support for the second rule is provided by Lander-Waterman theory (9). A mean contig length of 20 kbp would imply a conservative lower bound of at least 99.5% coverage for the assembly, and at least 5.4-fold redundancy. If more than 70% of the sequence data are in contigs longer than 20 kb, the mean contig length will certainly be longer than 20 kbp; hence, we may be reasonably confident that the genome is essentially complete. Genome data include the contig sequences, defined sequence features such as genes that encode RNAs or proteins, and annotations associated with each feature. Annotations include accurately determined boundaries of the features, and the assignment of a functional name to encoded proteins. NMPDR curators use comparative tools to correct errors in the start or stop codons of genes, and to change incorrect or ambiguous names in the annotations of PEGs. At least 75% of the total genes in the nearly 50 NMPDR pathogens have been assigned functionally meaningful names. Curators are adding features to our focus genomes, such as prophages or pathogenicity islands, that were not automatically located and identified in the RefSeq data. Curators are also attaching ‘attributes’ to sequence features in the form of key-value pairs (Table 1). The most common attribute is ‘evidence code’, which describes evidence that supports the functional annotation. Protein characteristics are also described as attributes. One important characteristic is essentiality, which has been curated for particular strains based on published experimental evidence. The key for the essentiality attribute is a citation of the data source, which is paired with a value of the essentiality assessment made in that experiment.
Table 1.
Attributes assigned to genomic features
| Key | Value | Usage |
|---|---|---|
| Evidence_code | icw(n) | FIG term: in cluster with; indicates that the PEG occurs in a cluster with n other genes from the same subsystem (very strong evidence) |
| isu | FIG term: in subsystem unique; indicates that the PEG occurs in a subsystem, and it is the only PEG for that genome that has been assigned the functional role (i.e. the cell in the spreadsheet contains a single entry) | |
| idu(n) | FIG term: in subsystem duplicates; indicates that the PEG occurs in a subsystem, but it is in a cell of the spreadsheet containing duplicates | |
| TAS | GO term: traceable author statement | |
| Structure | Stuctural_classification_projected from_homologous_protein_in_E._coli | Link to relevant entry in SCOP: Structural Classification of Proteins (http://scop.mrc-lmb.cam.ac.uk/scop/) |
| PubMed | PMID number | Link to published abstract in PubMed |
| SA_essential_Ji | Essential | Function for S.aureus protein in experiment by Ji et al. (22) |
| Potential_essential | Function for S.aureus protein in experiment by Ji et al. | |
| Undetermined | Function for S.aureus protein in experiment by Ji et al. |
This is a partial list of the most common attributes in the form of key-value pairs. The Usage column gives a brief description of the attribute, or provides a link to the relevant source.
Subsystems of functions
Populated subsystems are a data type unique to the NMPDR and its underlying annotation environment, the SEED (1). Each protein included in a subsystem plays a functional role, which may be enzymatic, signaling, regulatory, structural or other. Subsystems are defined in a table of functional roles, which link to reactions in KEGG (10) when available. Populated subsystems are represented in spreadsheets as columns of functional roles, rows of genomes, and cells populated by the genes assigned the role in the given organism. The spreadsheets permit easy discovery of instances in which genes for a role have not yet been identified. They are also helpful in sorting paralogs by showing cases in which multiple genes in the same organism have been assigned the same role. Many subsystems include a diagram of the relationship among the functional roles and reactions. These often describe a metabolic pathway, but subsystems are not limited to pathways, for example the ribosomal proteins subsystem. A subsystem may exist in all genomes or be present in only a few closely related genomes, for example pathogen-specific virulence factors.
Subsystems may be browsed from the subsystems summaries page. For any selected organism, a comprehensive list of its curated subsystems and functional roles is presented. Populated subsystem spreadsheets are linked to the subsystem headers, while protein context pages are linked from each functional role. Currently, NMPDR includes >300 curated subsystems, including descriptions of central and secondary metabolism, complex structures, virulence factors, and more. At least one third of the genes of the core NMPDR pathogens have been associated with subsystems.
Essential genes
One of the central goals of the NMPDR is to provide a meaningful connection to genomic data for those who study pathogens in the lab. To this end, features such as prophages and pathogenicity islands are being manually curated for our core pathogens, and protein characteristics such as structural classification and essentiality are being annotated as attributes. Like gene sequencing, the study of essential genes has recently been expanded to the scale of whole genomes. NMPDR curators have collated genome-scale essentiality datasets from 14 published studies involving 10 bacterial species, including the NMPDR pathogens S.aureus and S.pneumoniae (11). These data have been annotated as attributes for the respective genes.
Essentiality assessments from these genome-wide studies may also be browsed from the essential genes page. The total number of genes and the number deemed essential in each organism have been graphed to provide a visual summary of the data. The original heterogeneous essentiality assignments have been converted to a consistent (albeit simplified) form for all organisms, ‘essential’, ‘nonessential’, or ‘undetermined’, to facilitate comparative analysis of these data. In ambiguous or conflicting cases, ‘potentially essential’ has been used. Citations and descriptions of the experimental protocols used to generate the data are provided. The bars of the graph are linked to searches for essential genes in each organism. Search results are linked to the respective protein context pages to provide a unique opportunity for detailed comparative analysis of genes demonstrated to be essential in one organism with their homologs in other organisms.
ENVIRONMENT AND TOOLS FOR COMPARATIVE ANALYSIS
Results of keyword and BLAST searches are presented in tables that list a maximum of 50 proteins per page. This default limit may be reset on the advanced search page. If the query term matches records of proteins in NMPDR core organisms, these proteins are returned at the top of the list. For each result, the name of the organism, the functional assignment of the protein, and buttons to open the record are presented. There are two searchable environments for viewing the data record, Generic Genome Browser (GBrowse) and NMPDR.
The GBrowse environment
The GBrowse from the Generic Software Components for Model Organisms (GMOD) project was implemented in NMPDR to provide a familiar, easy-to-use tool for exploring genome context (12). GBrowse provides both a bird's eye and a detailed view of a genomic region, be it a chromosome, a sequence contig or an extra chromosomal element. It allows the user to zoom in and out, slide up and downstream along a sequence, and recenter the viewing area. The detailed view provides a variety of data tracks. For the core pathogens, tracks are available to view annotated prophages and pathogenicity islands, as well as the displayed region in closely related genomes, allowing visual comparison of genome similarity. Tracks available for all genomes will display GC content or functionally coupled genes. The CDS:curated track provides links for all the displayed genes to NMPDR protein context pages for further browsing in the NMPDR environment.
THE NMPDR ENVIRONMENT
The heart of NMPDR and the gateway to most of the analysis tools and capabilities of the system reside in protein context pages. These pages show a graphical display of the genomic context of the PEG of interest (the ‘focus PEG’), highlighted in green, centered in a 16 kb region. Neighboring genes that are conserved in other genomes (and are presumably functionally clustered) are drawn in blue, while neighboring genes whose relative proximity is not conserved in at least four other species are shown in red. Clicking on any gene arrow will link to that PEG's context page. Each page lists the organism name, with links to the NCBI Taxonomy ID, and the current functional assignment for the focus PEG. Convenient buttons allow immediate display of the protein sequence, DNA sequence, and DNA sequence including 500 nt of flanking sequence on either side; all sequences are provided in FASTA format for easy copy and paste into other programs. Another button shows a table of the functional coupling scores and identities of the blue genes.
Homologous regions and proteins
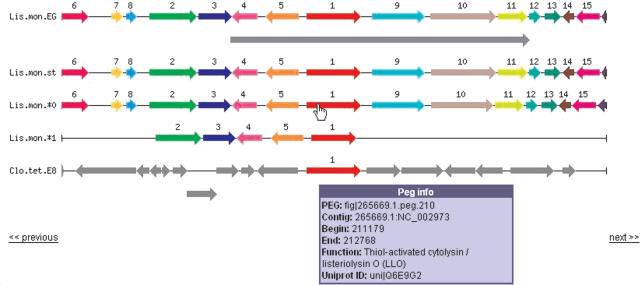
The ‘Compare Regions’ tool shows the displayed region with corresponding regions in other genomes (Figure 1). The size of the compared region and the number of compared genomes are adjustable. Navigation links allow the user to scroll upstream and downstream, effectively providing a means for ‘chromosome walking’. Each set of homologous genes is displayed in a common color and assigned a common number. Non-homologous proteins and non-protein features are shown in gray. Mousing over the arrows will display information for that gene, and clicking ‘show’ on any arrow will open that protein page with the compare regions display refocused on that gene.
Figure 1.
Compare regions shows a graphical display of homologous chromosomal regions that opens showing the five genomes with the highest score, based on similar proteins in this region, and phylogenetic distance. The display is centered on this focus PEG, which is shown in red and numbered 1. Sets of homologous genes share a color and a numerical label, which are ordered by frequency of co-localization with the focus PEG. The size of the region and the number of genomes may be reset. Clicking on any arrow in the display will refocus the comparison on that gene. The focus PEG always points to the right, even if it is located on the minus strand. The next and previous links allow you to walk the contig. This example shows a large, gray, pathogenecity island annotated in one strain of L.monocytogenes at the top, while at the bottom, Clostridium tetani shares only a homologous cytolysin with the Listeria. The gray proteins in the region of the C.tetani cytolysin are not homologous with the listerial proteins. The Listeria strain that seems to be lacking proteins represents the full length of a very short contig.
The ‘Bidirectional Best Hits’ tool returns a table of homologous proteins in other organisms precomputed using BLASTP and ranked by similarity. These are presumed to be orthologs because they are reciprocal best hits between two proteins in two genomes. Homologous proteins within one genome, or paralogs, cannot be identified by this method and are not listed in this table. Sequences selected from this list may be aligned with ClustalW (13). Phylogenetic relationships of the aligned sequences are shown in a neighbor-joining tree.
Genomic context
Two especially powerful analysis tools are linked as buttons in the protein context table, ‘Find Best Clusters’ and ‘Pins.’ The ‘Pins’ button opens a new window showing a graphical display of homologous chromosomal regions, centered on the focus PEG. This is an expansion of the ‘Compare Regions’ function, listing all genomes that have a homologous region, arranged in phylogenetic order. Here again, the focus PEG and its homologs (in red, and labeled 1) are aligned in the center of the page; all the genes within 8 kb of this central ‘pin’ are shown. Sets of homologous genes are depicted in a common color and share a common numerical label. The numerical labels are ordered by frequency of co-localization with the focus PEG. Mousing over the genes reveals their identity, and clicking on them will open the corresponding NMPDR protein context page. Similarity thresholds for the search are customizable. The ‘Commentary’ button at the foot of the Pins page displays a comprehensive information table for all sets of homologous genes.
The ‘Find Best Clusters’ (CL) button addresses the issue that not all clusters occur in all genomes. Even though the protein being examined might not be functionally clustered in the current genome, homologs in other genomes might be in clusters. The ‘Find Best Clusters’ function examines all proteins with highly significant similarity to the focus protein and displays them in order from those with the most to the fewest number of functionally clustered proteins. Each entry includes a link to the corresponding protein context page, from which the details of the cluster can be explored. This is a powerful analysis tool that provides insight into the functionality of genes by examining the genomic context of corresponding genes in other organisms.
NMPDR has precomputed instances in which genes appear to be functionally coupled, displayed in the fc-sc column in the protein context table. If this cell contains a number, there is evidence based on clustering that the genes are functionally coupled to the focus gene. The numbers are scores of functional relatedness that take into account the number of genomes in which the two genes are neighbors, as well as the phylogenetic distance between the genomes. The strength of evidence that two genes are functionally related is greater when the genes are clustered in a large number of organisms over a wide phylogenetic space, and this fact is reflected in the score. This link will return a table of homologous pairs with links to their NMPDR protein context pages.
Biological context
If the focus PEG has been assigned to one (or more) subsystem(s), they will be listed below the context table. Subsystem names link to the specific subsystem page, where functional data can be accessed. The ‘Protein Families’ function will show a table listing the protein families of which the focus PEG is a member. These are not restricted to NMPDR and SEED-curated families, but include families in other databases, like TIGRfam (14), Pfam (15) and KEGG (10).
Annotation details
Gene annotation is an ongoing process whereby current annotations are modified, changed, and expanded as new data become available. Therefore, viewing annotation details can greatly aid in the discernment of functional roles. Links are provided to the history of the annotation for the focus PEG, and ‘View All Related Annotations’ will display a list of the annotation details for proteins in other organisms that are homologous to the focus PEG, providing a test for the consistency of annotations. The source of the annotation is provided in each instance. ‘Assignments for Essentially Identical Proteins’ returns a table displaying the functional assignment for the focus PEG in the NMPDR/SEED and other databases, such as UniProt (16), KEGG (10), SwissProt (17), etc. This also provides a consistency check and is useful in identifying potentially erroneous annotations. Links to related entries (the focus PEG) in other databases are also provided. Evidence supporting the current functional assignment, such as structural and/or functional information, and pertinent literature links, are provided in the ‘Attributes’ table.
External analysis tools
Links to useful analytical tools at other sites are provided at the bottom of each protein page. Among others, these tools include TMpred (18), PSORT (19) and PPSearch (a service of the European Bioinformatics Institute; http://www.ebi.ac.uk), which locate transmembrane regions, predict cellular localization, and find signature amino acid patterns, respectively. There is also a link to NCBI's Position Specific Iterated BLAST (Psi-BLAST) (20), which may discover protein family relationships among proteins with fairly low overall sequence similarity. These tools launch with the focus PEG's information already entered, obviating the need to copy and paste sequences or other identifiers.
SIGNATURE GENES TOOL
Of special interest to researchers working on NMPDR core organisms is the ‘Signature Genes Tool.’ In essence this is a ‘compare and contrast’ function that allows the researcher to find genes in common in a set of organisms (the ‘compare’ side of the equation), and/or the genes that distinguish two sets of organisms (the ‘contrast’ part of the equation). This function is useful to look for genes that might be involved in the generation of a specific phenotype. The tool is not limited to pair-wise, genome-to-genome comparisons; genes from one organism (the ‘given’ organism) can be compared to the genes from any number of other organisms to find the shared genes or the genes that distinguish them. It is also possible to combine sets of organisms, so that a number of organisms can be contrasted in toto to another set of organisms.
The similarity threshold for the comparisons is set at an expectation value of 1e−10 by default, but this can be reset by the user. The results are displayed as a list of genes, with links to their protein context pages for further exploration.
DRUG TARGETS INITIATIVE
In support of research on organisms considered to be potential agents of biowarfare or bioterrorism or to cause emerging or re-emerging diseases, NMPDR is developing a pipeline for identification of candidate drug targets and virtual screening of potential drugs. The first draft of the table of candidate drug targets represents a very limited set of proteins that have been determined to be essential in at least one of the NMPDR pathogens, have been included in subsystems by our curators, have orthologs with experimentally determined structures in the Protein Data Bank (21), and have orthologs in a substantial number of the bacterial pathogens curated in the BRC system. These proteins will be used to optimize the virtual screening process. The table of candidates will grow as characteristics such as known antibiotic target, known antibiotic inhibitor, or associated with virulence are included.
Taking advantage of the high performance computing capabilities at Argonne National Laboratory and the Universiy of Chicago, we have begun to to do in silico screens of millions of ligand structures against the 3-D structures of selected drug targets. In silico screening is the computational molecular docking of a library of ligands against a protein structure using an algorithm to compute the binding energy. This screening has been performed for a few likely targets to refine ligand selection and screening procedures. Once the protocol has been fully optimized, in silico screening will be performed for ∼70 targets annually. Results of the computational docking will be provided as they become available. The ultimate goal of this drug targets initiative is to use computational resources to winnow millions of potential drug compounds to hundreds for in vitro biochemical analysis and target validation.
SUMMARY
NMPDR provides the most consistent functional annotations available in a structured biological context with a wealth of powerful tools for comparative analysis. This web-based environment for comparative genomics provides more than the standard bioinformatics resources to the scientific community conducting research on NIAID Priority Category B pathogens. Tools such as ‘Signature Genes’ and ‘Functional Clusters’ have been designed to help experimental researchers gain biological insight. A new interactive subsystems display tool is being developed that will allow scientists to visualize data, such as essentiality attributes and microarray results, in the context of functional diagrams like metabolic networks. Resources will expand to suit the needs and requests of the user community.
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400042C. Funding to pay the Open Access publication charges for this article was provided by NIH Contract No. HHSN266200400042C.
Conflict of interest statement. None declared.
REFERENCES
- 1.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M., de Crecy-Lagard V., Diaz N., Disz T., Edwards R., et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. Use of contiguity on the chromosome to predict functional coupling. In Silico Biol. 1999;1:93–108. [PubMed] [Google Scholar]
- 3.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osterman A., Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr. Opin. Chem. Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Bishop A.P., Bruce B.C., Lunsford K.J., Jones M.C., Nazarova M., Linderman D., Won M., Heidorn P.B., Ramprakash R., Brock A. Supporting community inquiry with digital resources. J. Digit. Info. 2004;5 Article no. 308. [Google Scholar]
- 7.He Y., Vines R.R., Wattam A.R., Abramochkin G.V., Dickerman A.W., Eckart J.D., Sobral B.W. PIML: the Pathogen Information Markup Language. Bioinformatics. 2005;21:116–121. doi: 10.1093/bioinformatics/bth462. [DOI] [PubMed] [Google Scholar]
- 8.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lander E.S., Waterman M.S. Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- 10.Kanehisha M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes S., Edwards R., Kubal M., Fonstein M., Stevens R., Osterman A. Essential genes on metabolic maps. Curr. Opin. Chem. Biol. 2006;17:448–456. doi: 10.1016/j.copbio.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J.D., Higgins D.G., Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:239–241. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeckmann B., Bairoch A., Apweiler R., Blatter M.-C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann K., Stoffel W. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe Seyler. 1993;374:166. [Google Scholar]
- 19.Gardy J.L., Laird M.R., Chen F., Rey S., Walsh C.J., Ester M., Brinkman F.S.L. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji Y.D., Zhang B., Van Horn S.F., Warren P., Woodnutt G., Burnham M.K.R., Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]