Abstract
The Molecular INTeraction database (MINT, http://mint.bio.uniroma2.it/mint/) aims at storing, in a structured format, information about molecular interactions (MIs) by extracting experimental details from work published in peer-reviewed journals. At present the MINT team focuses the curation work on physical interactions between proteins. Genetic or computationally inferred interactions are not included in the database. Over the past four years MINT has undergone extensive revision. The new version of MINT is based on a completely remodeled database structure, which offers more efficient data exploration and analysis, and is characterized by entries with a richer annotation. Over the past few years the number of curated physical interactions has soared to over 95 000. The whole dataset can be freely accessed online in both interactive and batch modes through web-based interfaces and an FTP server. MINT now includes, as an integrated addition, HomoMINT, a database of interactions between human proteins inferred from experiments with ortholog proteins in model organisms (http://mint.bio.uniroma2.it/mint/).
INTRODUCTION
Cells are complex systems whose physiology is governed by an intricate network of molecular interactions (MIs) of which a relevant subset are protein–protein interactions (PPIs). Signal transduction pathways and transcriptional regulation are typical examples of such biological processes mediated by PPI.
The MI database (MINT, http://mint.bio.uniroma2.it/mint/) was designed to collect experimentally verified PPIs in a binary or complex representation.
Over the past four years, MINT has undergone a profound reorganization of both the data model and the database structure, and has dramatically increased the number of stored interactions.
DATABASE STRUCTURE
In January 2006, MINT adopted the IntAct relational model. IntAct is an open source database designed for the storage, presentation and analysis of MIs (1) (schema available at http://intact.sourceforge.net/uml/intactCore.gif). The main advantages of adopting the IntAct model lie in its ability to represent protein complexes and other types of molecules as interaction participants, and in the ease with which new features and toolkits for data storage, representation and analysis can be added. In addition MINT, from now on, will be compatible with all the upgrades and tools developed by the IntAct consortium. MINT is based on the open source PostgreSQL database management system (http://www.postgresql.org). All the data can be accessed as Java objects through the IntAct API by using OJB (http://db.apache.org/ojb/) as the object-relational mapping tool. The web application is based on the Struts framework (http://jakarta.apache.org/struts/) running on the Tomcat servlet container (http://tomcat.apache.org/) and the Apache server (http://www.apache.org/).
DATA GROWTH
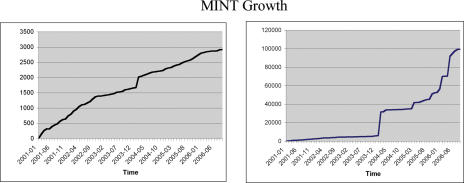
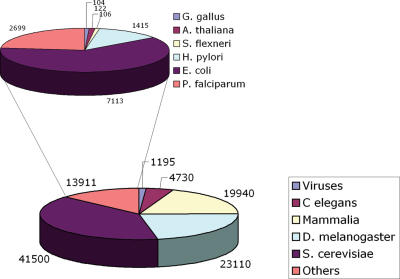
Since MINT was first reported (2) the number of curated entries has grown significantly. Given the new database structure, binary and n-ary interactions can now be represented. The number of binary interactions has increased >20-fold (Figure 1). As of September 2006, more then 95 000 physical interactions involving 27 461 proteins from 325 organisms are stored in MINT (Figure 2). Although, ∼90% of the interactions stored in the database are the result of large scale, genome wide experiments, the value of MINT resides in the high number of curated articles, each of which reports a small number of interactions. This is reflected in the steady increase of curated articles that is mainly due to the curation of publications describing low-throughput experiments (Figure 1). The MINT curation team consists of two full-time and one part-time curators. This is not sufficient to capture all the interaction information steadily reported in research articles. Thus, we have chosen to focus our efforts on consistently curating all the issues of FEBS Letters (since January 2005) and EMBO Journal and EMBO Reports (since January 2006). This choice was made in agreement with the other members of the International Molecular-Interaction Exchange consortium (IMEx http://imex.sourceforge.net/), including DIP (3), IntAct (1), MPact (4) and BIND (5). IMEx aims at avoiding work overlaps, at sharing the curation workload and at exchanging completed records on MI data.
Figure 1.
Mint growth. The graph on the left represents the growth in the number of interactions stored in MINT. The right graph represents the corresponding growth in the number of curated articles.
Figure 2.
The pie charts represent the number of interactions stored in MINT for the different species.
DATABASE CURATION
Since January 2006, the syntax and the semantics for data representation have been provided by the Proteomics Standards Initiative-Molecular Interaction (PSI-MI 2.5) standards. The PSI workgroup develops and maintains a common data standard allowing users to retrieve all relevant data from different data providers and to perform comparative analysis (6).
Each stored interaction represents, according to the PSI-MI standards, a physical interaction, a direct interaction or a co-localization. All the database records created before January 2005 underwent a drastic quality control performed by the new MINT curation staff composed of PhD level curators. All the previous entries were re-curated according to the new PSI-MI standard. Entries that could not be updated according to PSI-MI standards were deleted from the database in order to maintain a high quality dataset. The MINT Curation Reference Manual is being developed in collaboration with the other IMEx partners and is available at http://imex.sourceforge.net/doc/imex-curationManual.doc. The new data submission HTML forms benefit from an improved automatic check based on curation rules. This ensures that mandatory fields are filled and that annotated ranges or residues are consistent with the protein length reported by Uniprot. In addition, for the purpose of ensuring high fidelity annotation, every new entry derived from low-throughput experiments undergoes a further validation step performed by a different curator before release to the public.
DATA ACCESS AND DISTRIBUTION
The new web-based interface includes a number of improvements and enhancements allowing a more efficient database exploration. Most notably, the query can be based on protein or gene names, UniProt keywords or identifiers of external databases: UniProtKB, PDB, Ensembl, FlyBase, SGD, WormBase, OMIM, HUGE, PubMed and Reactome. It is also possible to query species specific datasets (mammalian, Saccharomyces cerevisiae, Caemorhabaditis elegans, Drosophila melanogaster, viruses). Finally, a sequence similarity search (BLAST) can also be performed to search for proteins which are homologous to the query protein.
The query results are displayed in a table showing in the left frame a summary of the protein features reported in UniProt and, in the right frame, the list of the interaction partners curated in MINT. The interactions can be displayed graphically by an enhanced version of the ‘MINT viewer’, a Java applet derived from the applet Graph (http://java.sun.com). The viewer represents the interactions by lines (edges) and nodes (proteins), and assigns the nodes a size proportional to the protein's molecular weight and a color which depends on the species. The graph displayed by the viewer can be expanded and edited interactively by moving or deleting nodes. Proteins linked to OMIM are now highlighted in red. In order to attribute a reliability index to the reported interactions, we have also assigned each interaction a confidence level, based on the experimental detection method and experimental conditions. The results of the analysis performed in the MINT viewer can be captured in different formats ready for export: PSI1.0-XML, PSI2.5-XML, flat-file, Osprey.
MINT is now also complemented by HomoMINT (7), an inferred human protein interaction network where interactions discovered in model organisms (and collected in MINT) are mapped onto the corresponding human orthologs. Through the MINT web pages it is also possible to search the HomoMINT dataset for inferred interactions.
A web service allows direct computational access so as to freely retrieve interaction networks in different formats: flat-file, PSI1.0-XML, PSI2.5-XML. It is also possible to download specific subsets of the database based on the taxonomy of the interactors.
Acknowledgments
This work is supported by AIRC and by the European Union FP6 Interaction Proteome project and the ENFIN network of excellence. Funding to pay the Open Access publication charges for this article was provided by European Union FP6.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S., Vingron M., Roechert B., Roepstorff P., Valencia A., et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zanzoni A., Montecchi-Palazzi L., Quondam M., Ausiello G., Helmer-Citterich M., Cesareni G. MINT: a Molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 3.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guldener U., Munsterkotter M., Oesterheld M., Pagel P., Ruepp A., Mewes H.W., Stumpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfarano C., Andrade C.E., Anthony K., Bahroos N., Bajec M., Bantoft K., Betel D., Bobechko B., Boutilier K., Burgess E., et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermjakob H., Montecchi-Palazzi L., Bader G., Wojcik J., Salwinski L., Ceol A., Moore S., Orchard S., Sarkans U., von Mering C., et al. The HUPO PSI's molecular interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 7.Persico M., Ceol A., Gavrila C., Hoffmann R., Florio A., Cesareni G. HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics. 2005;6:S21. doi: 10.1186/1471-2105-6-S4-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]