Abstract
The solution structure of a new modified thrombin binding aptamer (TBA) containing a 5′–5′ inversion of polarity site, namely d(3′GGT5′-5′TGGTGTGGTTGG3′), is reported. NMR and CD spectroscopy, as well as molecular dynamic and mechanic calculations, have been used to characterize the 3D structure. The modified oligonucleotide is characterized by a chair-like structure consisting of two G-tetrads connected by three edge-wise TT, TGT and TT loops. d(3′GGT5′-5′TGGTGTGGTTGG3′) is characterized by an unusual folding, being three strands parallel to each other and only one strand oriented in opposite manner. This led to an anti-anti-anti-syn and syn-syn-syn-anti arrangement of the Gs in the two tetrads. The thermal stability of the modified oligonucleotide is 4°C higher than the corresponding unmodified TBA. d(3′GGT5′-5′TGGTGTGGTTGG3′) continues to display an anticoagulant activity, even if decreased with respect to the TBA.
INTRODUCTION
SELEX (Systematic Evolution of Ligands by Exponential Enrichment) (1) is a combinatorial chemistry methodology based on oligonucleotide libraries which are screened for high-affinity binding to a given target. High-affinity ligands can be isolated from the library using iterative rounds affinity-based enrichment, alternating with oligonucleotide amplification. Ligands obtained using this methodology have often been referred to as aptamers.
In 1992, Block et al. (2) screened a pool of ∼1013 synthetic oligonucleotides for their interaction with thrombin, which is a serine protease with multiple functions in hemostasis whose roles in coronary heart disease and other thrombotic disorders have promoted efforts toward the identification of specific inhibitors. This study led to the identification of a consensus DNA 15mer, namely 5′GGTTGGTGTGGTTGG3′ (TBA: thrombin binding aptamer), which was found to be a potent inhibitor of thrombin with an EC50 of 20 nM in a fibrinogen clotting assay (3,4). The 3D structure of TBA was solved using NMR techniques (5,6). As Figure 1 shows, TBA is characterized by a chair-like structure consisting of two G-tetrads connected by two TT loops and a single TGT loop. As suggested by others (7), TBA binds at the anion exosite I of thrombin in a conformation little changed from that of the unbound species.
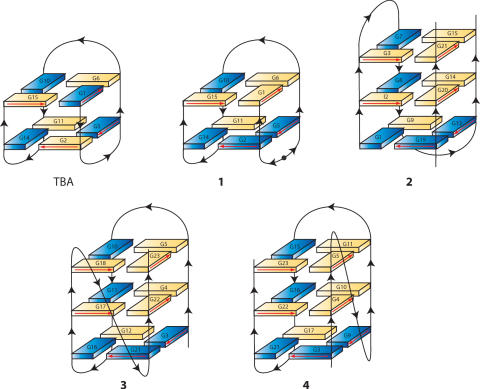
Figure 1.
Schematic illustration of the structures adopted by TBA, modified TBA (1), the three human telomeric repeats sequence into a dimeric quadruplex (22) (2), the four-repeat Tetrahymena d(T2G4)4 sequence (23) (3), and the four-repeat human telomeric d[TAGGG(TTAGGG)3] sequence in K+ solution (24,25). Black arrowheads indicate 5′→3′ polarity of the strands. Black circle in 1 represents the 5′–5′ inversion of polarity site. Anti and syn guanines are depicted as yellow and blue solids, respectively. Red arrows indicate the direction of the proton donors and acceptors in Hoogsteen hydrogen bonds.
In order to improve the properties of TBA, a number of researches (8–10) have been devoted to its structure–activity relationship to post-SELEX modifications. For example, He et al. (9) synthesized numerous TBA analogues containing modified guanosine carrying several substituents at 8 position or at the exocyclic amino group. In a different paper (10), the same authors reported the synthesis and the thrombin inhibiting properties of several TBA-based oligonucleotides containing neutral formacetal linkages. However, although TBA was the first aptamer to be discovered, no studies dealing with the effects of introduction of sites of polarity inversion have been reported yet, even if such interesting backbone alteration has already been utilized in another aptamer, namely Macugen (11) (pegaptanib sodium), that has been recently approved by the US FDA for the treatment of neovascular age-related macular degeneration.
In this frame, we have undertaken a study whose aim is to use a biologically driven approach in order to improve the knowledge of the interactions between thrombin and TBA that are critical for the biological activity. Particularly, we have designed and synthesized a TBA-based oligodeoxynucleotide containing a 5′–5′ site of polarity inversion, namely 3′GGT5′-5′TGGTGTGGTTGG3′ (1). In this article, we report a detailed structural study, along with thermal stability analysis and a structure–activity relationship of this new modified TBA.
MATERIALS AND METHODS
The oligonucleotide 3′GGT5′-5′TGGTGTGGTTGG3′ (1) and the natural counterpart 5′GGTTGGTGTGGTTGG3′ (TBA) were synthesized on a Millipore Cyclon Plus DNA synthesizer, using solid phase β-cyanoethyl phosphoramidite chemistry at 15 μmol scale. The synthesis of 1 was performed by standard methods for the 5′TGGTGTGGTTGG3′ tract, and using 5′-phosphoramidites for the 3′GGT5′ tract. The oligomers were detached from the support and deprotected by treatment with concentrated aqueous ammonia at 55°C for 12 h. The combined filtrates and washings were concentrated under reduced pressure, redissolved in H2O and analyzed and purified by HPLC on a Nucleogel SAX column (Macherey-Nagel, 1000-8/46); using buffer A: 20 mM KH2PO4 aqueous solution, pH 7.0, containing 20% (v/v) CH3CN; buffer B: 1 M KCl, 20 mM KH2PO4 aqueous solution, pH 7.0, containing 20% (v/v) CH3CN; a linear gradient from 0% to 100% B in 30 min and flow rate 1 ml/min were used. The isolated oligomers have the following retention times: 1 = 19.0 min; TBA = 20.5 min. The oligomers were collected and successively desalted by Sep-Pak cartridges (C18). The isolated oligomers resulted to be >99% pure (NMR).
Nuclear magnetic resonances
NMR samples were prepared at a concentration of ∼2 mM, in 0.6 ml (H2O/D2O 9:1) buffer solution having 10 mM KH2PO4, 70 mM KCl, 0.2 mM EDTA, pH 7.0. For D2O experiments, the H2O was replaced with D2O by drying down the sample, lyophilization and redissolution in D2O alone. NMR spectra were recorded with Varian UnityINOVA 500 MHz and 700 MHz spectrometers. 1H chemical shifts were referenced relative to external sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), whereas 31P chemical shifts were referenced to external phosphoric acid (H3PO4 85% v/v). 1D proton spectra of samples in H2O were recorded using pulsed-field gradient WATERGATE (12) for H2O suppression. A proton-detected 1H–31P heteronuclear COSY was recorded in D2O in the hypercomplex mode with 2048 t2 points and 96 t1 increments, and a spectral width of 500 Hz along the 31P dimension. Phase-sensitive NOESY spectra (13) were recorded with mixing times of 100 and 200 ms (T = 25°C and 5°C). Pulsed-field gradient WATERGATE was used for NOESY and 1H–15N HSQC (14) spectra in H2O. TOCSY spectra (15) with mixing times of 100 ms were recorded with D2O solutions.
All experiments were recorded using STATES-TPPI (16) procedure for quadrature detection. In all 2D experiments, the time domain data consisted of 2048 complex points in t2 and 400–512 fids in t1 dimension. The relaxation delay was kept at 3 s for NOESY experiments used in the structure determination. A relaxation delay of 1.2 s was used for all other experiments. The NMR data were processed on a SGI Octane workstation using FELIX 98 software (Accelrys, San Diego, CA).
Structural calculations
Cross-peak volume integrations were performed with the program FELIX 98, using the NOESY experiment collected at mixing time of 100 ms. The NOE volumes were then converted to distance restraints after they were calibrated using known fixed distances of H2′/H2″ of G1, G2, G5, G8, T9, G10, G11, T12, T13 and G14. Then a NOE restraint file was generated with three distance classifications as follows: strong NOEs (1.8 Å ≤ rij ≤ 3.0 Å, where 1.8 Å is the van der Waals radius and rij is the interproton distance between protons i and j), medium NOEs (2.5 Å ≤ rij ≤ 4.0 Å) and weak NOEs (3.5 Å ≤ rij ≤ 5.5 Å). A total of 198 NOE derived distance restraints were used.
Hydrogen bonds constraints were used: upper and lower distance limits of 2.0 Å and 1.7 Å for hydrogen-acceptor distance, and 3.0 Å and 2.7 Å for donor-acceptor distance, respectively. These constraints for H-bonds did not lead to an increase in residual constraints violation. A total of 54 backbone torsion angles were used in the calculations too. Moreover, according to NMR data, glycosidic torsion angles were kept in a range of −160°/−70° for the G anti, whereas a range of 10°/100° was used for the G syn.
The calculations have been performed using a distance-dependent macroscopic dielectric constant of 4*r and an infinite cut-off for nonbonded interactions to partially compensate for the lack of the solvent (17) have been used. Thus the 3D structures which satisfy NOE and dihedral angle constraints were constructed by simulated annealing calculations. An initial structure of the oligonucleotide was built using a completely random array of atoms. Using the steepest descent followed by quasi-Newton–Raphson method (VA09A), the conformational energy was minimized. Restrained simulations were carried out for 500 ps using the CVFF force field as implemented in Discover software (Accelrys, San Diego, CA). The simulation started at 1000 K, and then the temperature was decreased stepwise until 273 K. The final step was again to energy-minimize to refine the obtained structures, using successively the steepest descent and the quasi-Newton–Raphson (VA09A) algorithms. Both dynamic and mechanic calculations were carried out by using 1 (kcal/mol)/Å2 flatwell distance restraints: 20 structures were generated. RMSD (root mean square deviation) values of 0.75 ± 0.27 Å and 0.90 ± 0.31 Å for the backbone and heavy atoms, respectively, were calculated for the best seven structures.
Illustrations of structures were generated using the INSIGHT II program, version 2005 (Accelrys, San Diego, CA). All the calculations were performed on a PC running Linux WS 4.0.
The final set of coordinates has been deposited in the Protein Data Bank (accession code: 2IDN).
Circular dichroism
CD samples of 1 and TBA were prepared at a concentration of 1 × 10−4 M, by using the buffer solution used for NMR experiments: 10 mM KH2PO4, 70 mM KCl, 0.2 mM EDTA, pH 7.0. CD (circular dichroism) spectra and CD melting curves were registered on a Jasco 715 circular dichroism spectrophotometer in a 0.1 cm pathlength cuvette. For the CD spectra, the wavelength was varied from 220 to 320 nm at 100 nm min−1. The spectra were recorded with a response of 16 s at 2.0 nm bandwidth and normalized by subtraction of the background scan with buffer. The temperature was kept constant at 20°C with a thermoelectrically controlled cell holder (Jasco PTC-348). CD melting curves were registered as a function of temperature from 20 to 90°C at 294 nm with a scan rate of 10°C h−1 for both quadruplexes. The CD melting curves of TBA and modified aptamer showed sigmoidal profiles and were modeled by a two-state transition, using a theoretical equation for an intramolecular association, according to the van't Hoff analysis (18). The Tm and ΔH° values provide the best fit of the experimental melting data. The reported errors for thermodynamic parameters are the SD values of the mean from the multiple determinations. The ΔS° values were calculated by equation ΔS° = ΔH°/Tm and free energy change values by the equation ΔG°(T) = ΔH° − TΔS°.
Prothrombin time (PT) assay
Human plasma samples were collected by venipuncture, in presence of 0.1 volumes 3.8% sodium citrate and fractionated by centrifugation at 2000 g for 5 min. PT times were measured by using Koagulab MJ Coagulation System with a specific kit RecombiPlas Tin HemosIL (Inst. Labs, Lexinton, USA). Briefly, this method relies on the high sensitivity thromboplastin reagent based on recombinant human tissue factor. The addition of recombiplastin to the plasma in the presence of calcium ions initiates the activation of extrinsic pathway. This results ultimately in the conversion of fibrinogen to fibrin, with a formation of solid gel. The procedure was performed according to the manufacturer's instructions. TBA, 1 or vehicle (PBS) were added at different time points in a volume of 2 μl at a final concentration of 20 μM. Data are expressed as mean ± SEM and are representative of at least three different measurements.
RESULTS AND DISCUSSION
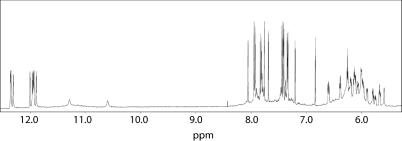
The NMR sample of 1 was heated for 10 min at 80°C and slowly cooled down to room temperature, then its 1H-NMR spectrum was recorded by using pulsed-field gradient WATERGATE (12) for H2O suppression (Figure 2). With the exclusion of some weak resonances due to very minor conformations also present in solution (whose relative intensities turned out to be insensitive to temperature changes), the simple appearance of 1D spectra of 1 indicates that, in the conditions used here, the modified oligomer forms mainly a single well-defined hydrogen-bonded conformation. In fact, the 1H-NMR spectrum of 1 (700 MHz, T = 25°C) shows the presence of eight well-defined signals in the region 11.5–12.5 p.p.m., attributable to imino protons involved in Hoogsteen hydrogen bonds of at least two G-quartets, and of two broader and less intense singlets at 10.6 and 11.29 p.p.m., respectively, probably belonging to two imino protons of two thymines involved in mutual hydrogen bonding (see below). Moreover, fifteen signals in the aromatic region due to the presence of nine guanine H8 and six thymine H6 protons were clearly observable.
Figure 2.
Expanded region of the proton NMR spectrum of 1 (700 MHz, T = 25°C).
A combination of the analysis of 2D NOESY (700 MHz, T = 25°C), TOCSY spectra (700 MHz, T = 25°C), and 31P-NMR spectroscopy (202 MHz, T = 25°C) allowed us to get the almost complete assignment (see Supplementary Material) of both exchangeable and nonexchangeable protons and phosphorus resonances of 1. Particularly, the 1D proton decoupled phosphorus spectrum displays fourteen signals (see Supplementary Material). After assigning the 1H resonances within each deoxyribose by a 2D TOCSY experiment, the 2D proton-detected heteronuclear 1H–31P COSY (see Supplementary Material) allowed us to assign the entire backbone correlating each phosphorus resonance to the respective 5′-coupled H3′ proton and 3′-coupled H5′ and H5′ protons of adjacent ribose. Further, NOEs between base protons and H1′, H2′ and H2″ protons allowed us to assign all aromatic protons to the pertinent base.
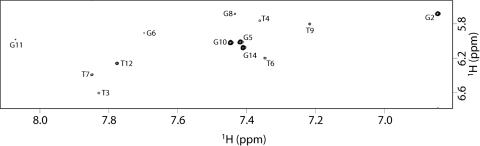
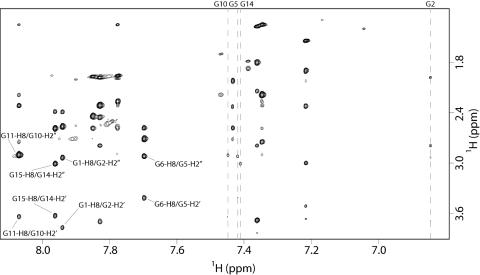
It is interesting to note that the intensities NOESY (700 MHz, T = 25 °C, mixing time 100 ms) cross-peaks between the H8 proton bases and sugar H1′ resonances indicate that four (G2, G5, G10, G14) out of nine Gs of 1 adopt a syn glycosidic conformation (Figure 3), where the H8 resonances of syn G residues are upfield shifted with respect to those of the anti ones (5,19,20). Then, four anti-Gs (G1, G6, G11 and G15) have classical H8/H2′–H2″ sequential connectivities to 5′ neighboring syn-Gs (G2, G5, G10 and G14, respectively) (Figure 4), indicating that the subunits G1–G2, G5–G6, G10–G11 and G14–G15 are involved in the formation of a four-stranded helical structure (underlined residues adopt a syn glycosidic conformation). Moreover, the entire pattern of NOEs observed for all cited Gs indicates that the backbone conformations of these tracts resemble those of the unmodified TBA possessing a right-handed helix structure (Figure 4).
Figure 3.
Expanded region of the NOESY spectrum of 1 (700 MHz, T = 25°C, mt = 200 ms) correlating G–H8 and H1' protons.
Figure 4.
Expanded region of NOESY spectrum of 1 (700 MHz, T = 25°C, mt = 100 ms) correlating G–H8 and H2′–H2″ protons. Classical G–H8/H2′–H2″ NOEs (nuclear Overhauser effect) correlations for the four-stranded part of the complex are reported. Pattern of NOEs for the Gs in syn glycosidic conformation are highlighted by vertical dashed lines.
The alternation of syn and anti G residues within each strand suggests that, as TBA, 1 folds into a monomolecular foldback quadruplex, characterized by two G tetrads. Further, a number of unusual NOE connectivities were observed between syn-Gs and Ts, indicating that 5′TG3′ and 5′GT3′ tracts do not adopt a helical winding, thus suggesting that, most probably, the TT and TGT tracts form loops.
In order to assign exchangeable protons to their respective G or T residues, a 1H–15N HSQC experiment (700 MHz, T = 5°C) has been acquired (see Supplementary Material). Eight exchangeable protons (the eight at lower field) correlate with 15N in the region between 143 and 147 p.p.m. indicating that they are attributable to imino protons of G residues (21), whereas the signals at 10.65 and 11.35 p.p.m. correlate with nitrogens at 155.7 and 155.6 p.p.m., respectively, thus indicating that the latter belong to T residues (21). Interestingly, these two signals are characterized by strong NOE effects between each other, thus suggesting that two thymines are in close proximity and, verisimilarly, interacting by a mutual hydrogen bond, exactly as observed in the unmodified TBA. Moreover, NOEs between H1′ proton of G8, methyl protons of T9 and three G imino protons (δH 12.03, 11.97 and 11.91 p.p.m.), and NOEs between other four imino protons (δH 12.37, 12.35, 12.30 and 11.95 p.p.m.) and H6, methyl, H3′ and H1′ protons of T4 and H3′, H2′, H4′ and H1′ of T13, led us to assign the exchangeable protons of two tetrads (see Supplementary Material).
The whole of data indicates that the structure of 1 is a foldback quadruplex characterized by two G tetrads formed by the residues G1, G6, G10, G15 and G2, G5, G11, G14, respectively, where the underlined residues possess a syn glycosidic conformation. Interestingly, the two tetrads assume an anti-anti-anti-syn and syn-syn-syn-anti arrangement of the bases. It is noteworthy that even the T imino protons (δH 10.65 and 11.35 p.p.m., T = 5°C) are characterized by several NOE connectivities with a set of G imino protons belonging to only one tetrad (δH 11.95, 12.80, 12.35 and 12.37 p.p.m.). Moreover, the pattern of NOEs attributable to the loop T12–T13 and T3–T4 let us to identify the T4 and T13 as the residues that are H-bonded to each other. Interestingly, T13 and T4 are mutually hydrogen bonded in TBA as well. Then, the facts that TGT loop possesses some NOEs with G imino protons of one tetrad and that the two TT loops are characterized by NOEs with G imino protons of the other tetrad indicate that no loop assumes a dog-eared conformation. All this means that the 1 folds into a chair-like quadruplex structure possessing three strands parallel to each other and only one strand oriented in reverse manner (Figure 1).
In order to get the 3D structure of 1 at atomic level, an estimation of proton–proton distances has been retrieved from cross-peak intensities in 2D NOESY experiments acquired at 700 MHz, both at T = 25°C and T = 5°C (mixing time 100 ms for the experiment acquired in D2O and 200 ms for the experiments acquired in H2O). Pseudo-atoms were introduced where needed. A total of 198 distances were used for the calculations and, as suggested by the presence of 8 G and 2 T imino protons in the 1D 1H-NMR spectrum, 32 supplementary distance restraints (HN1–O6, N1–O6, HN2–N7, N2–N7) for 16 hydrogen bonds corresponding to the 2 G-quartets, and 4 distance restraints (HN3–O4, N3–O4) for 2 hydrogen bonds between T13 and T4 were also incorporated during the computations (Table 1). The backbone torsion angles β and ɛ were estimated from the scalar coupling measured in the 2D proton-detected heteronuclear 1H–31P COSY by using the semi-empirical Karplus equation (22). Thus, since for all sugars in 1 all JP,H5′ and JP,H5″ were <8 Hz, β backbone torsion angles were restricted to 180 ± 30° (22). For residues 3, 4, 5, 6, 7, 8, 9 and 11, JP,H3′ was >8 Hz, and the corresponding ɛ angles were restricted to −120 ± 30° (21). Sugar ring conformations were obtained from the analysis of the PE-COSY spectrum. From the ΣJH4′ estimation (<10 Hz), no dihedral restraints for γ were applied to the four G nucleotides in the syn glycosidic conformation, whereas γ backbone torsion angles for residues G6 and G11 (possessing an anti conformation) were restricted to 60 ± 30° (21). All measurable JH1′,H2′ were reasonably large, indicating a predominant S-type nature of sugar ring conformations: δ and ν2 angle constraints have been added consistently. Further, in agreement with NMR data, glycosidic torsion angles for four out of eight guanines involved in the formation of the two G-tetrads were fixed in the anti-domain (−160°/−70°), whereas the χ angle was kept in a range of 10°/100° (syn-conformation) for the remaining four G residues.
Table 1.
Experimental constraints and structure statistics of the best 20 structures
| Experimental constraints | |
|---|---|
| Total NOEs | 198 |
| NOEs from non-exchangeable protons | 185 |
| NOEs from exchangeable protons | 13 |
| Hydrogen bonds constraints | 36 |
| Dihedral angle constraints | 62 |
| CVFF energy (kcal mol−1) of the minimized structures | |
| Total | 370.158 ± 4.529 |
| nonbond | 40.783 ± 5.145 |
| restraint | 13.213 ± 1.417 |
| NOEs violations | |
| Number > 0.2 Å | 3.40 ± 1.63 |
| Maximum (Å) | 0.560 ± 0.263 |
| Sum (Å) | 3.821 ± 0.693 |
| Average violation (Å) | 0.016 |
| r.m.s. deviations from the mean structure (Å) | |
| All backbone heavy atoms | 0.75 ± 0.27 |
| All heavy atoms | 0.90 ± 0.31 |
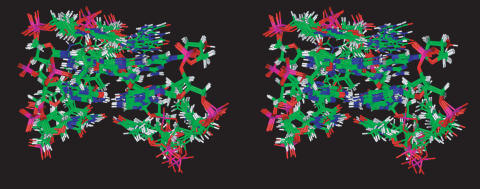
Therefore, 3D structures which satisfy NOEs were constructed by simulated annealing (SA) calculations. An initial structure of the oligonucleotide was constructed possessing a random conformation and minimized, in order to eliminate any possible source of initial bias in the folding pathway. Restrained simulations were carried out for 500 ps using the CVFF (consistent valence force field). The restrained SA (simulated annealing) calculations started at 1000 K, and, thereafter, the temperature was decreased stepwise down to 273 K. The aim was to energy-minimize and refine the structures. A total of 20 structures were generated. Average RMSD values of 0.75 ± 0.27 Å and 0.90 ± 0.31 Å for the backbone and all heavy atoms, respectively, were obtained from the superimposition of the best seven structures (Figure 5). These data, along with the lack of significant violations of the experimental restraints (see Table 1), suggest that the obtained structures are representative of the structure actually adopted in solution by 1.
Figure 5.
Stereoview representation of the superimposed seven best structures of 1. Backbones and bases are depicted in colored ‘stick’ (carbons, green; nitrogens, blue; oxygens, red; hydrogens, white).
As expected, 1 shows a right-handed helical backbone geometry and three edge-wise connecting TT, TGT and TT loops. As the unmodified TBA, 1 involves two stacked G-tetrads, but differs in guanine syn/anti distribution around the GGGG tetrads, being characterized by the unusual anti-anti-anti-syn and syn-syn-syn-anti arrangement of the two tetrads respectively, and in the relative strand orientations, with three strands parallel to each other and with only one oriented in opposite manner.
Interestingly, a similar strand orientation and glycosidic angle arrangement of the bases of the tetrads have already been observed for the assembly of three human telomeric repeats into a dimeric G-quadruplex (23) (2), the four-repeat Tetrahymena d(T2G4)4 sequence (24) (3), and the four-repeat human telomeric d(TAGGG(TTAGGG)3] sequence in K+ solution (25,26) (4) (Figure 1). Even though sequences 2–4 differ each other in both molecularity and folding topology, when their structures are represented in such a way that the four strands possess the same orientation (Figure 1), a certain resemblance among the three models is clearly apparent. Particularly, all the corresponding guanines are characterized by the same glycosidic angle arrangements. It is noteworthy that the only two tetrads in the modified TBA (1) show the same glycosidic angle arrangements as those in the lower two planes in 2–4. A further structural element joining the four complexes is the presence in all of them of one narrow, one wide and two medium width grooves.
Furthermore, a comparison of the most representative structures of 1 (the one with lowest energy after minimization), and the already reported NMR structure of the unmodified TBA (PDB code: 148D) has been accomplished by analyzing the helix parameters by CURVES (27,28) (Tables 2 and 3).
Table 2.
Rise, tilt, roll and twist of TBA and of the best-minimized structure of 1
| TBA | Rise | Tilt | Roll | Twist | 1 |
|---|---|---|---|---|---|
| G1/G2 | 3.44 | −10.60 | −179.75 | 50.93 | |
| 3.25 | 7.84 | 160.55 | −45.79 | G1/G2 | |
| G5/G6 | 2.67 | 6.22 | −161.77 | 48.90 | |
| 3.64 | −1.25 | −178.12 | −49.88 | G5/G6 | |
| G10/G11 | 2.80 | 10.45 | −178.83 | 48.50 | |
| 3.92 | −4.04 | 151.25 | −52.77 | G10/G11 | |
| G14/G15 | 4.67 | −17.19 | −177.35 | 55.65 | |
| 3.24 | −12.25 | −156.98 | −49.22 | G14/G15 |
Table 3.
Shear, stretch, stagger, buckle, propeller, opening of TBA and of the best-minimized structure of 1
| TBA | Shear | Stretch | Stagger | Buckle | Propeller | Opening | 1 |
|---|---|---|---|---|---|---|---|
| G1/G6 | −6.15 | 1.72 | −0.88 | 11.32 | 176.45 | −89.48 | |
| −6.67 | 2.57 | −0.38 | 3.51 | 174.13 | 91.17 | G1/G6 | |
| G1/G10 | 4.82 | 2.32 | −2.69 | 3.47 | −176.25 | −0.95 | |
| −6.59 | 2.09 | −0.71 | 38.43 | 164.49 | 6.02 | G1/G10 | |
| G1/G15 | −6.02 | 1.87 | −0.59 | −5.12 | −162.35 | 86.32 | |
| −6.95 | 2.01 | −0.37 | 28.42 | −172.46 | −86.37 | G1/G15 | |
| G2/G5 | 6.42 | 2.77 | −0.11 | −5.50 | 157.97 | −91.51 | |
| 6.23 | 2.08 | 0.02 | −5.58 | −168.29 | 87.08 | G2/G5 | |
| G2/G11 | −6.04 | 1.75 | −2.05 | 24.52 | −177.68 | −3.39 | |
| 7.00 | 2.51 | −0.04 | 26.55 | −147.31 | −0.96 | G2/G11 | |
| G2/G14 | 5.41 | 2.07 | −1.82 | −5.12 | −162.35 | 86.32 | |
| 6.59 | 1.88 | −0.37 | 8.34 | −176.04 | −89.79 | G2/G14 |
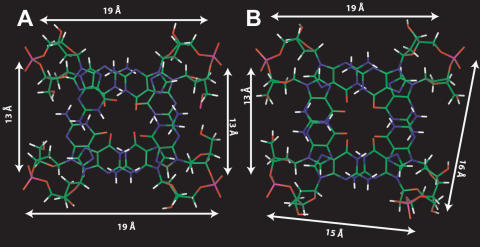
These data clearly suggest that the overall structure adopted by 1 is similar to that of TBA. The base stacking of the two quadruplexes is quite similar, being the five-membered rings of the top guanines overlapped to the five-membered rings of the underneath guanine bases (Figure 6). Furthermore, as suggested by Tilt and Stagger values, the stacking between G pairs is slightly better in 1 than in TBA. The groove widths for 1 are just a little bit different from that of unmodified TBA. Particularly, TBA is characterized by two wide grooves (∼19 Å) between the strands G5–G6 and G10–G11 and between the strands G1–G2 and G14–G15 respectively, and other two narrow grooves that are calculated to be ∼13 Å. On the other hand, 1 possesses the groove between the strands G1–G2 and G5–G6 just a little bit wider (∼16 Å) and the groove between the strands G1–G2 and G14–G15 just a little bit narrower (∼15 Å) (Figure 6).
Figure 6.
Top view of the two G-tetrads of TBA (A) and 1 (B). Backbones and bases are depicted in colored ‘stick’ (carbons, green; nitrogens, blue; oxygens, red; hydrogens, white). White arrows indicate groove widths.
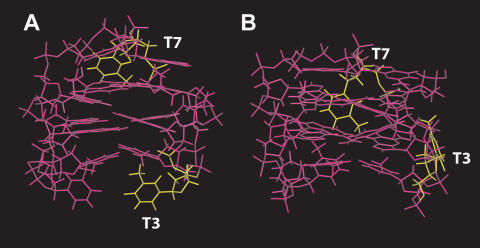
As for the sugar puckering, the two molecules possess the same sugar ring conformations, being in S-type conformation. It is interesting to note that the main structural difference rely on the orientation of T3. In fact, as indicated by the diagnostic NOE connectivities between T3–Me and G2–H8, and between T3–H6 and G2–H8, the base of T3 folds back into the groove between the strands G1–G2 and G5–G6 in 1, and faces closely the G2–H8, thus justifying the very low chemical shift value of T3–H6 (δH 6.84) (Figure 7). Furthermore, another slight structural difference is to be ascribed to the orientation of T7. In fact, T7 folds back into the groove formed by the strands G5–G6 and G10–G11. Actually, this latter structural change seems due to a different folding of the backbone of this part of the molecule, where the sugar of T7 is closer to the bases G8 and G6 than the unmodified TBA (Figure 7).
Figure 7.
Side view of TBA (A) and 1 (B). All residues are depicted in magenta, except T3 and T7 (yellow). The different orientations of T3 and T7 are clearly observable.
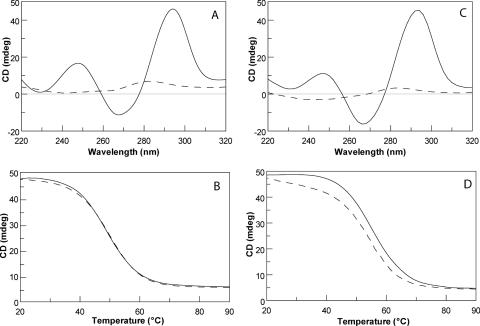
In order to determine the effects of the inversion of polarity site on the CD profile and the thermal stability of the resulting quadruplex structure, CD spectra and CD melting and annealing experiments were acquired for 1 and its natural counterpart (Figure 8). In particular, the CD spectra of 1 and TBA, performed at 20°C, are almost superimposable, both exhibiting two positive bands at 248 and 294 nm and a negative one at 267 nm. As expected, the TBA spectrum is typical of anti-parallel quadruplex structures (29–31). The spectrum of 1 is also typical of antiparallel quadruplex structures because 1 possesses two G-tetrads with an alternating anti and syn glycosidic conformation along each strand. However, this spectrum differs from CD spectra previously reported for the same ‘3+1’ strand arrangements (26), where the presence of an additional peak at 260 nm was attributed to the presence of third G-tetrad that implies a non-alternating guanine glycosidic conformation between the middle and bottom G-tetrads.
Figure 8.
CD spectra of TBA (A) and 1 (C) at 20°C (continuous lines) and 90°C (dashed lines). Melting (continuous lines) and annealing (dashed lines) CD experiments of TBA (B) and 1 (D).
As for melting and annealing CD experiments, taking into account that the rates of quadruplex formation/dissociation are very slow, we collected the data at 10°C/h. No significant hysteresis emerged for each oligonucleotide comparing annealing and melting curves thereby indicating that, at the scan rate used, both systems were at equilibrium. The melting curves were analyzed using van't Hoff analysis, and thermodynamic parameters are shown in Table 4. The values for TBA are in good agreement with those recently obtained by UV technique (32,33). The transition for the modified aptamer occurs 5°C above the transition of TBA and the enthalpy change values are consistent with the opening of two G-tetrads (34,35). The entropy values suggest that the modified aptamer possesses a more rigid structure with respect to TBA, according to a better stacking between G pairs found in the NMR study. The whole set of thermodynamic parameters shows that the quadruplex formed by 1 is more stable than its unmodified counterpart.
Table 4.
Thermodynamic parameters for the unfolding of 1 and TBA
| Tm (°C) (±1) | ΔH° (kJ mol−1) (±20) | ΔS° (kJ mol−1 K−1) (±0.05) | ΔG°(298 K) (kJ mol−1) (±1) | |
|---|---|---|---|---|
| 1 | 57 | 180 | 0.54 | 17 |
| TBA | 52 | 160 | 0.49 | 13 |
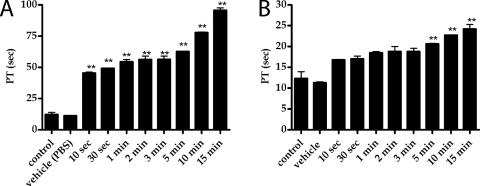
In order to study the effects of the structural changes on the biological activity of 1, if any, we performed a PT assay. PT assay on 1 was performed on human plasma in strict comparison with TBA. In order to eliminate the variation caused by the measurements performed on different days, the samples of 1 and TBA were prepared at the same time and folded together by heating the samples for 10 min at 80°C and slowly cooling them down to room temperature. The assays have been conducted after one week from the preparation procedure. As shown in Figure 9, overall, TBA showed a statistically significant and a more marked effect over 1, both in terms of the onset of activity and endurance. Indeed, incubation studies demonstrated that TBA effect is already present after 10 s of incubation and lasts up to 15 min of incubation. On the other hand, 1 showed a slower onset of the effect, indeed PT time started to be significantly prolonged following 5 min of incubation. Also the magnitude of the activity of 1 is reduced compared with TBA. The biological data reported here imply that the modifications of the structure adopted by 1 affect sensibly the biological activity, and therefore its interaction with the thrombin. Looking at the overall folding of the modified aptamer, such a reduction in the biological activity was not expected. However, it is interesting to note that the TBA inhibits the thrombin activity when interacting with its fibrinogen exosite (36). In particular, it has been proposed that TBA could associate with thrombin in two different ways: through the T7-G8-T9 loop (as suggested by X-ray data) or through the T3 and T12 residues (as suggested by NMR models) (36). As far as the X-ray data are concerned, among all the observed interactions, T7 is buried in a pocket of hydrophobic residues of the fibrinogen exosite of thrombin made by Ile24, His71, Ile79 and Tyr117, whereas, according to NMR data, the T3 residue interacts with the thrombin residues Tyr76 and Ile82. Therefore, the reduced biological activity of 1 could be ascribed to the different orientation adopted by T3, and, to a less extent, by T7 in 1 with respect to the unmodified TBA (Figure 7). The longer onset displayed by 1 could be tentatively explained considering that 1 needs to properly orientate T3 and T7 upon its binding to thrombin. Nevertheless, the anticoagulant activity of 1 could not be simply ascribed to these changes, but could even be due to a combination of all structural changes observed such as: (i) relative strand orientation; (ii) glycosidic arrangements of the bases (iii) groove widths.
Figure 9.
Prothrombin time assessed by using human plasma. On the x-axis the incubation time is reported, while PT time in seconds is reported on y-axis. (A) Refers to TBA, while (B) to 1.
The present data give a further insight into the understanding of the variables involved in the formation and the stability of quadruplex structures. In fact, 1 clearly prefers to adopt a ‘3+1’ strand orientation instead of a classical ‘2+2’ typical of this kind of structure (see TBA, Figure 1), in spite of the fact that both arrangements are equally possible. Thus, these findings further enforce the idea that the ‘3+1’ core topology is not to be considered an anomaly, but is rather a robust G-quadruplex scaffold (23–26). The structure reported here represents also the first example of a ‘3+1’ core topology quadruplex structure containing only two G-tetrads. Furthermore, our findings may contribute to the comprehension of the folding of telomeric DNA (23–26), and could shed more light on the understanding of the thrombin/TBA interaction phenomenon. Finally, considering that the ‘3+1’ arrangement increases the thermal stability of the quadruplex and that 1 is not devoid of biological activity, the scaffold described in the present paper can be seen as a unique platform for structure-based anticoagulant drug design. Further investigations concerning oligodeoxynucleotides (ODNs) containing either 3′–3′ or 5′–5′ inversion of polarity sites and their thermodynamic study are in progress in our laboratories.
Supplementary Material
Acknowledgments
This work is supported by Italian M.U.R.S.T. (P.R.I.N. 2004 and 2005) and Regione Campania (L.41, L.5). The authors are grateful to ‘Centro di Servizio Interdipartimentale di Analisi Strumentale’, C.S.I.A.S., for supplying NMR facilities. Funding to pay the Open Access publication charges for this article was provided by Italian M.U.R.S.T. (P.R.I.N. 2004 and 2005).
Conflict of interest statement. None declared.
REFERENCES
- 1.Gold L., Polisky B., Uhlenbeck O., Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 2.Block L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 3.Griffin L.C., Tidmarsh G.F., Bock L.C., Toole J.J., Leung L.K. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulant in extracorporeal circuits. Bloods. 1993;81:3271–3276. [PubMed] [Google Scholar]
- 4.Li W.X., Kaplan A.V., Grant G.F., Toole J.J., Leung L.K. A novel nucleotide-based thrombin inhibitor inhibits clot-bound thrombin and reduces arterial platelet thrombus formation. Blood. 1994;83:677–682. [PubMed] [Google Scholar]
- 5.Wang K.Y., McCurdy S., Shea R.G., Swaminathan S., Bolton P.H. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 6.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paborsky L.R., McCurdy S.N., Griffin L.C., Toole J.J., Leung L.K. The single-stranded DNA aptamer-binding site of human thrombin. J. Biol. Chem. 1993;268:20808–20811. [PubMed] [Google Scholar]
- 8.Dias E., Battiste J.L., Williamson J.R. Chemical probe for glycosidic conformation in telomeric DNAs. J. Am. Chem. Soc. 1994;116:4479–4480. [Google Scholar]
- 9.He G.-X., Krawczyk S.H., Swaminathan S., Regan S.G., Dougherty J.P., Terhorst T., Law V.S., Griffin L.C., Coutrè S., Bischofberger N. N2- and C8-substituted oligodeoxynucleotides with enhanced thrombin inhibitory activity in vitro and in vivo. J. Med. Chem. 1998;41:2234–2242. doi: 10.1021/jm970434d. [DOI] [PubMed] [Google Scholar]
- 10.He G.-X., Williams J.P., Postich M.J., Swaminathan S., Regan S.G., Terhorst T., Law V.S., Griffin L.C., Cheri M.T., Coutrè S., Bischofberger N. In vitro and in vivo activities of oligonucleotide-based thrombin inhibitors containing neutral formacetal linkages. J. Med. Chem. 1998;41:4224–4231. doi: 10.1021/jm970766i. [DOI] [PubMed] [Google Scholar]
- 11.Nimjee S.M., Rusconi C.P., Sullenger B.A. Aptamers: an emerging class of therapeutics. Ann. Rev. Medicine. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 12.Piotto M., Saudek V., Sklenar V.J. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 13.Jeener J., Meier B., Bachmann H.P., Ernst R.R. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979;71:4546–4553. [Google Scholar]
- 14.Sklenar V., Piotto M., Leppik R., Saudek V. Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. 1993;102:241–245. [Google Scholar]
- 15.Braunschweiler L., Ernst R.R. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 16.Marion D., Ikura M., Tschudin R., Bax A. Rapid recording of 2D NMR spectra without phase cycling: application to the study of hydrogen exchange in proteins. J. Magn. Reson. 1989;85:393–399. [Google Scholar]
- 17.Weiner S.J., Kollman P.A., Case D.A., Singh U.C., Ghio C., Alagona G., Profeta S., Weiner P.J. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- 18.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 19.Smith F.W., Feigon J. Strand orientation in the DNA quadruplex formed from the Oxytricha telomere repeat oligonucleotide d(G4T4G4) in solution. Biochemistry. 1993;32:8682–8692. doi: 10.1021/bi00084a040. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Patel D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3]G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez C., Szyperski T., Ono A., Iwai H., Tate S., Kainosho M., Wuthrich K. NMR with 13C,15N-doubly-labeled DNA: the Antennapedia Homeodomain Complex with a 14mer DNA Duplex. J. Biomol. NMR. 1998;12:25–37. [PubMed] [Google Scholar]
- 22.Kim S, Lin L., Reid B.R. Determination of nucleic acid backbone conformation by 1H-NMR. Biochemistry. 1992;31:3564–3574. doi: 10.1021/bi00129a003. [DOI] [PubMed] [Google Scholar]
- 23.Zhang N., Phan A.T., Patel D.J. (3+1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Am. Chem. Soc. 2005;127:17277–17285. doi: 10.1021/ja0543090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Patel D.J. Solution structure of the Tetrahymena telomeric repeat d(T2G4) G tetraplex. Structure. 1994;2:1141–1155. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 25.Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128 doi: 10.1021/ja062791w. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 28.Lavery R., Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 29.Lu M., Guo Q., Kallenbach N.R. Thermodynamics of G-tetraplex formation by telomeric DNAs. Biochemistry. 1993;32:598–601. doi: 10.1021/bi00053a027. [DOI] [PubMed] [Google Scholar]
- 30.Lu M., Guo Q., Kallenbach N.R. Effect of thymine tract length on the structure and stability of model telomeric sequences. Biochemistry. 1993;32:3596–3603. doi: 10.1021/bi00065a010. [DOI] [PubMed] [Google Scholar]
- 31.Smirnov I., Shafer R.H. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 32.Miyoshi D., Karimata H., Sugimoto N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006;128:7957–7963. doi: 10.1021/ja061267m. [DOI] [PubMed] [Google Scholar]
- 33.Olsen C.M., Gmeiner W.H., Marky L.A. Unfolding of G-quadruplexes: energetic, and ion and water contributions of G-Quartet Stacking. J. Phys. Chem B. 2006;110:6962–6969. doi: 10.1021/jp0574697. [DOI] [PubMed] [Google Scholar]
- 34.Shafer R.H. Stability and structure of model DNA triplexes and quadruplexes and their interactions with small ligands. Prog. Nucleic Acids Res. Mol. Biol. 1998;59:55–94. doi: 10.1016/s0079-6603(08)61029-6. [DOI] [PubMed] [Google Scholar]
- 35.Petraccone L., Erra E., Nasti L., Galeone A., Randazzo A., Mayol L., Barone G., Giancola C. Effect of modified thymine on the structure and stability of [d(TGGGGT)]4 quadruplex. J. Biol. Macromol. 2002;31:131–137. doi: 10.1016/s0141-8130(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 36.Padmanachan K., Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Cryst. 1996;D52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.