Abstract
The conserved domain database (CDD) is part of NCBI's Entrez database system and serves as a primary resource for the annotation of conserved domain footprints on protein sequences in Entrez. Entrez's global query interface can be accessed at http://www.ncbi.nlm.nih.gov/Entrez and will search CDD and many other databases. Domain annotation for proteins in Entrez has been pre-computed and is readily available in the form of ‘Conserved Domain’ links. Novel protein sequences can be scanned against CDD using the CD-Search service; this service searches databases of CDD-derived profile models with protein sequence queries using BLAST heuristics, at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. Protein query sequences submitted to NCBI's protein BLAST search service are scanned for conserved domain signatures by default. The CDD collection contains models imported from Pfam, SMART and COG, as well as domain models curated at NCBI. NCBI curated models are organized into hierarchies of domains related by common descent. Here we report on the status of the curation effort and present a novel helper application, CDTree, which enables users of the CDD resource to examine curated hierarchies. More importantly, CDD and CDTree used in concert, serve as a powerful tool in protein classification, as they allow users to analyze protein sequences in the context of domain family hierarchies.
INTRODUCTION
The annotation of conserved domain footprints on protein sequences often serves as the first step toward characterizing protein function in silico. Protein domains may be viewed as units in the molecular evolution of proteins and can be organized into an evolutionary classification. The set of protein domains characterized so far appears to describe no more than a few thousand superfamilies, where members of each superfamily are related to each other by common descent. NCBI's conserved domain database (CDD) attempts to collate that set and to organize related domain models in a hierarchical fashion, meant to reflect major ancient gene duplication events and subsequent functional diversification.
Computational annotation of protein function is generally obtained via sequence similarity: once a close neighbor with known function has been identified, its annotation is copied to the sequence with unknown function. This strategy may work very well in functionally homogeneous families and when applied only for very close neighbors or suspected orthologs, but it is doomed to fail often when domain or protein families are sufficiently diverse and when no close neighbors with known function are available.
To this end, the CDD (1) provides a strategy toward a more accurate assessment of such neighbor relationships, similar to approaches termed ‘phylogenomic inference’ (2). CDD acknowledges that protein domain families may be very diverse and that they may contain sets of related subfamilies. Of these, only few may have been characterized experimentally, and within this set function may have diverged considerably. While it may be possible, and certainly efficient, to represent such a set of subfamilies with just a single family model, that model could only provide very generic annotation. In CDD curation, we attempt to detect evidence for duplication and functional divergence in domain families by means of phylogenetic analysis. We record the resulting subfamily structure as a set of explicit models, but limit the analysis to ancient duplication events—several hundred million years in the past, as judged by the taxonomic distribution of protein sequences with particular domain subfamily footprints.
CDD provides a search tool employing reverse position-specific BLAST (RPS–BLAST), where query sequences are compared to databases of position-specific score matrices (PSSMs), and E-values are obtained in much the same way as in the widely used PSI-BLAST application (3). When CDD is scanned with protein query sequences, a region on a query may pick up more than one overlapping footprint from a set of related models. One of those models provides the best score or lowest E-value, but that alone may not be sufficient to indicate that the query sequence is a bona fide member of the corresponding subfamily. Since the CDD collection also contains imported models, which have not been curated at NCBI, search results may present a mixture of curated models (accessions starting with ‘cd..’) and un-curated models (accessions starting with ‘pfam’, ‘smart’ or ‘COG’). By default, overlapping domain hits are sorted by E-value, but curated models are listed first, if their E-values exceed a secondary significance threshold of 1e-05. Default displays are presented in a concise fashion, where domain hits that overlap with the top-ranked domain hits are hidden.
We have started to distribute CDTree, a helper application for the web browser. CDTree allows users to examine the results of simple phylogenetic analysis on the sequences from a curated domain hierarchy, and view their query sequence in the context of such a phylogenetic sequence tree.
ASSESSING DOMAIN FAMILY MEMBERSHIP
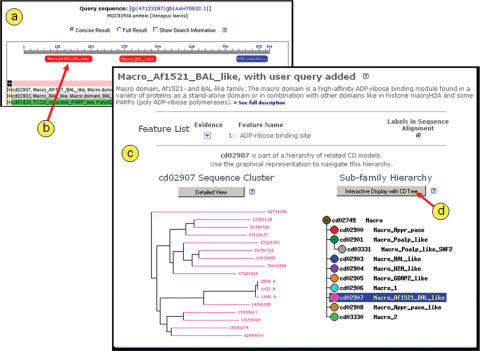
Figures 1 and 2 demonstrate how one might use a web browser, the curated CDD resource and the CDTree application to obtain confidence for the transfer of annotation from a domain model to a particular protein sequence. A cartoon depicting domain model footprints on a protein sequence can be obtained by following the ‘Conserved Domains’ link from an Entrez protein search results page, or by submitting a live CD-search (4) request (Figure 1a). A particular domain annotation is examined in detail by clicking on the corresponding item in the graphical display (Figure 1b). This generates a CD summary page (Figure 1c) which lists descriptive information and a multiple sequence alignment including the user query sequence (not shown). The summary page contains a description of the particular family. If the domain model has been curated at NCBI (CDD accessions starting with ‘cd..’), it also lists conserved features that have been recorded by NCBI curators, displays a sequence cluster tree diagram for the particular family and indicates its position in a hierarchy of related domains. A button labeled ‘Interactive Display with CDTree’ (Figure 1d) launches CDTree.
Figure 1.
(a) Pre-computed domain annotation is retrieved for a protein sequence in the Entrez database, gi|47123187 from Xenopus laevis. The graphical annotation can be obtained by following the ‘Conserved Domains’ link on the Entrez document summary for gi|47123187. By default, graphical domain summaries hide redundant information from the user. By clicking on the red balloon (b), representing a conserved domain footprint for the model cd02907, the user launches a summary view of that domain model, which also preserves information about the (query) sequence of interest (c). The CD summary page displays details about the actual model and its hierarchy. A section labeled ‘Links’ (data not shown), for example, provides links to all protein sequences in Entrez that match the current domain model, to references in PubMed and Entrez Books, and to the original source of the curated family, which may be a model imported from outside databases such as Pfam. Clicking on the button labeled ‘Interactive Display with CDTree’ (d) launches CDTree on the user's computer as a local application, which retrieves its data via the web-browser. The CDTree view corresponding to this example is shown in Figure 2. CDTree launching is not enabled for alignment models imported from outside sources.
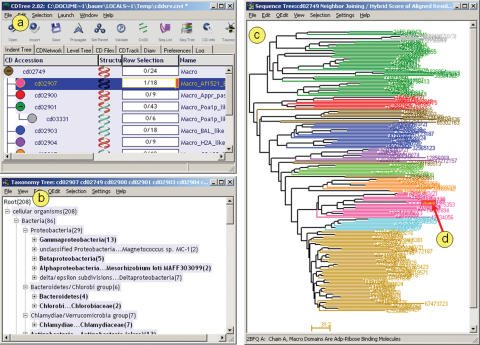
Figure 2.
CDTree default display, as launched from a web browser, showing a curated domain hierarchy with an embedded user query sequence. A protein sequence found in NCBI's Entrez database was inspected for the presence of conserved domains. The user has followed links from one of the domain footprints annotated on the model, and inspected a particular domain model, cd02907. From the CD summary page, as shown in Figure 1, CDTree was launched as a helper application. The main window (a) presents the organization of the conserved domain hierarchy, as already visible on the CD summary page (Figure 1). In this case, cd02749 or ‘Macro’ is a generic ‘parent’ model, which has been split up into several more specific ‘children’. The sequence tree shown in panel (c) provides evidence for this particular subfamily structure. Groups of branches rendered in the same color correspond to alignment rows that have been assigned to a particular subgroup. Sequence trees are always calculated from the curated CD alignments; in this particular example, the distance data have been obtained from pair-wise alignment scores. Aligned residue pairs are scored with the BLOSUM62 matrix. Pair-wise scores are subtracted from the highest observed pair-wise score to yield distances, and the distance units plotted here correspond to BLOSUM62 scores. By default, a taxonomy viewer window is opened as well (b). Users may select and highlight whole branches in the sequence tree view and examine corresponding highlights in the taxonomy viewer, to understand the taxonomic scope of particular subfamilies, or select/highlight taxa in the taxonomy viewer and examine their distribution in the sequence tree. In this example, a user query sequence has been added to one of the models, cd02907. cd02907 gave the best scoring hit in a database search for a particular region of the user's query sequence. In the sequence tree display, the user query is highlighted by default (d). It appears that the user query is a typical member of this particular subfamily, as it clusters tightly with all the other members, and therefore transfer of annotation from the model to the sequence - or functional inference - may be appropriate.
CDTree is a helper application for the web browser and must be downloaded and installed on the user's computer. Instructions for installing CDTree are found at http://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml. CDTree functions as a viewer for curated protein domain hierarchies; it retrieves data for display via the web browser. CDTree is a combined domain hierarchy viewer and editor. It uses a separate program, Cn3D (5), to view 3D structure and to display and edit multiple alignments of protein structure and sequence. Cn3D is distributed, installed and configured along with CDTree. CDTree requires a recent version of Cn3D, version v4.2, which is contained in the CDTree installation package. The installation package also contains a stand-alone application, ‘fa2cd’, which can be used to convert FASTA-formatted multiple sequence alignments into models stored in the ‘CD’ format, so that they can be imported into CDTree. CDTree also allows the de novo buildup of alignment models starting from single protein sequences. More details can be found on the CDTree home page (see Table 1). A manuscript detailing CDTree and its various uses is in preparation.
When the user launches CDTree from a CD summary page, CDTree displays the contents of the curated domain or domain hierarchy, and serves as a viewer for the evidence supporting a particular subfamily structure. By default, CDTree displays a sequence tree view, a taxonomy view and a hierarchy overview. The sequence tree has been pre-computed and is contained in the data sent by the server. When a query sequence has been submitted to CD-Search and a matching CDD model has been identified, launching CDTree from its CD summary page will cause the user's query sequence to be added to the model and CDTree will recalculate a sequence tree. This tree will now contain the query sequence as well (Figure 2c).
Potentially, this allows users to distinguish between several cases: (i) Query sequences may be bona fide members of clusters which curators have explicitly declared subfamilies, and which may carry specific functional annotation, as in the case illustrated in Figures 1 and 2; (ii) query sequences may be bona fide members of subfamilies which do not (yet) carry specific functional annotation; (iii) query sequences may be members of clusters, which curators have not declared subfamilies, as they may not have seen enough evidence for a subfamily at the time of curation; (iv) query sequences may be outliers and not cluster with any particular group of sequences in the curated hierarchy. Only the first scenario listed above may allow the transfer of specific functional annotation from the model to the query. In all the other cases, annotation transfer from the hierarchy's generic ‘parent’ model may be more appropriate, assuming that the ‘parent’ model provides annotation that is valid for all or the majority of members in a superfamily.
CDD CONTENTS AND AVAILABILITY
CDD can be searched directly as part of NCBI's Entrez database and query system (6), where it is listed as the ‘Domains’ database. Entries in CDD are cross-linked reciprocally to NCBI taxonomy, citations in PubMed®, and to protein sequences in Entrez. Links to protein sequences reflect the results of pre-computed RPS–BLAST searches and are updated on a daily basis and stored in the CDART database (7).
The current version of CDD, version v2.09, contains a total of 12 422 models, of which 2494 have been curated at NCBI. Of these curated models, <300 are solitary domain models, while the rest are organized into hierarchies. The largest hierarchies contain well over 100 individual models each. 5252 models have been obtained from Pfam (version 11) (8), 575 have been obtained from SMART (9) and 4101 have been derived from the COG collection (10). Together, these models cover about 69% of non-identical protein sequences in NCBI's Entrez protein database. The full set of models as imported from Pfam, SMART, COG and KOG(9), are available in separate search databases, although not all of them have been indexed in Entrez, since lineage-specific models with limited taxonomic scope, as well as largely redundant models, have been filtered out.
The size of the CDD model collection, details with respect to versions of external databases mirrored in CDD, and the control of redundancy may change over time, as we attempt to provide a resource that is more comprehensive as well as efficient. Expert curation of CDD is an ongoing effort, and we plan to eventually replace a majority of imported models with hierarchies curated at NCBI. Table 1 lists URLs and FTP site addresses for tools and services mentioned above.
Table 1.
URLs and FTP-site addresses for CDD and CDD-related services
| CDD | Conserved Domain Database home page | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml |
|---|---|---|
| CDD FTP | CD alignment models, pre-built search databases | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd |
| CD-Search | Live and pre-computed RPS-BLAST results | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| CDTree | Domain hierarchy viewer and editor | http://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml |
| Cn3D | 3D structure and alignment viewer | http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml |
| rpsblast | Stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution | ftp://ftp.ncbi.nlm.nih.gov/toolbox executables can be obtained from: http://www.ncbi.nlm.nih.gov/BLAST/download.shtml |
Acknowledgments
We thank the authors of the Pfam, SMART and COG resources. Development of CDTree would not have been possible without Paul Thiessen and his work on Cn3D, Lewis Geer, Jane He, Naigong Zhang and Praveen Cherukuri and their work on the CDART resource, the NCBI BLAST group, the NCBI IEB and the NCBI C++ toolkit developers. This work was supported by the Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Comments, suggestions and questions are welcome and should be directed to: info@ncbi.nlm.nih.gov. Funding to pay the Open Access publication charges for this article was provided by Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Marchler-Bauer A., Anderson J.B., Cherukuri P.F., DeWeese-Scott C., Geer L.Y., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown D., Sjolander K. Functional classification using phylogenomic inference. PLoS Comput. Biol. 2006;2:e77. doi: 10.1371/journal.pcbi.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchler-Bauer A., Bryant S.H. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Geer L.Y., Chappey C., Kans J.A., Bryant S.H. Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 6.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center of Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geer L.Y., Domrachev M., Lipman D.J., Bryant S.H. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn R.D., Mistry I., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letunic I., Copley R.R., Pils B., Pinkert S., Schultz J., Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatusov R.L., Fedorova N.D., Jackson J.D., Jacobs A.R., Kiryutin B., Koonin E.V., Krylov D.M., Mazumder R., Mekhedov S.L., Nikolskaya A.N., et al. The COG database: and updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]