Abstract
BindingDB (http://www.bindingdb.org) is a publicly accessible database currently containing ∼20 000 experimentally determined binding affinities of protein–ligand complexes, for 110 protein targets including isoforms and mutational variants, and ∼11 000 small molecule ligands. The data are extracted from the scientific literature, data collection focusing on proteins that are drug-targets or candidate drug-targets and for which structural data are present in the Protein Data Bank. The BindingDB website supports a range of query types, including searches by chemical structure, substructure and similarity; protein sequence; ligand and protein names; affinity ranges and molecular weight. Data sets generated by BindingDB queries can be downloaded in the form of annotated SDfiles for further analysis, or used as the basis for virtual screening of a compound database uploaded by the user. The data in BindingDB are linked both to structural data in the PDB via PDB IDs and chemical and sequence searches, and to the literature in PubMed via PubMed IDs.
INTRODUCTION
The early steps in a modern drug discovery project typically include identifying a biological macromolecule that plays a key role in a disease process, and seeking a low-molecular weight compound that inactivates this macromolecular target by binding it with high affinity. Ligand discovery involves a substantial component of trial and error, despite advances in computer-aided drug-design, so many binding data are generated for each target. Projects directed at ligand discovery therefore generate large quantities of binding data not only for drugs, but also for compounds that do not themselves become drugs. When published, these data become a valuable resource for scientists studying the same macromolecular target, and also for those seeking to develop improved computational models of molecular recognition.
Currently, binding data are published almost exclusively via the scientific journals, which provide an indispensable archival service, are now available in electronic formats, and can be searched in useful ways. However, the journals also impose severe restrictions, as recently emphasized (1). For example, they provide no mechanism for accessing data in numerical form, querying according to chemical structure, downloading computer representations of chemical structure, publishing large datasets in any detail or navigating among binding, structural and sequence data. By providing these missing functionalities, especially to researchers in academia and in small companies who do not have access to the resources of the major pharmaceutical firms, a database of measured binding affinities should accelerate the discovery of targeted ligands. Potential applications of a binding database include:
Analysis of ligands for a specific target to discover chemical features or pharmacophores that correlate with affinity.
Development of quantitative structure–activity relationships.
Interpretation of measured entropies and enthalpies of binding in the context of a receptor's 3D structure.
Parameterization and validation of broadly applicable methods of ligand design.
Identification of candidate lead compounds for a new drug target, by searching for ligands known to bind similar proteins.
Identification of drug candidates with a high risk of side effects, by checking whether similar compounds bind multiple receptors.
Elucidation of the mechanism of a biological effector molecule; e.g. if a naturally occurring compound inhibits cellular proliferation, a search of the database for chemically similar compounds may reveal that a similar compound binds a protein known to be involved in regulation of the cell cycle.
A binding database also offers the possibility of publishing data that are not amenable to journal publication, such as very large data sets, and raw experimental data which can be useful in the assessment of data quality.
BindingDB (http://www.bindingdb.org) was created to address these needs. It currently holds ∼20 000 measurements, making it one of the most extensive public databases of protein–ligand binding affinities, and it is continuing to grow. The present paper summarizes these data holdings as well as new website features and capabilities; basic technical aspects of BindingDB have been described previously (2–4).
CONTENTS OF THE DATABASE
Data collection currently focuses on targets whose three-dimensional structures are available in the Protein Data Bank (5,6) (PDB) or can be accurately modeled. Such data are of particular interest because they are amenable to structural analysis and are suitable for the development and validation of computational models of binding. Statistical sampling of the PDB in 2003 revealed that ∼150 of the non-redundant proteins therein were considered current or potential drug-targets (unpublished data) and were thus suitable for data collection by BindingDB. This analysis omits additional drug-targets whose structures could be built by comparative modeling. Restricting attention to proteins of known structure allows BindingDB to complement, rather than overlap, other binding databases collecting data for membrane proteins whose 3D structures are, in the main, unavailable; e.g. GPCRDB [www.gpcr.org (7)], the IUPHAR receptor database (www.iuphar-db.org) and GLIDA [http://gdds.pharm.kyoto-u.ac.jp/services/glida/index.php (8)].
Proteins are selected for data collection based upon their importance as drug-targets or model systems, as well as the availability of suitable data. Once a protein is selected, relevant scientific articles are identified and their data are extracted and deposited into BindingDB. Data from multiple laboratories and companies are sought in order to obtain a wide range of chemotypes for the targeted protein. The journals from which data are drawn include J. Med. Chem., Bioorg. Med. Chem. Lett. and Biochem. Web-accessible forms also allow direct deposition by experimentalists, but this route has not generated a significant number of entries. The majority of the data are based upon enzyme inhibition studies (>19 000 measurements), but a smaller number of data from the more informative method of isothermal titration calorimetry also are included (416 measurements). Each data entry includes detailed experimental conditions, such as solution composition, pH and temperature, because these can affect the measured affinities.
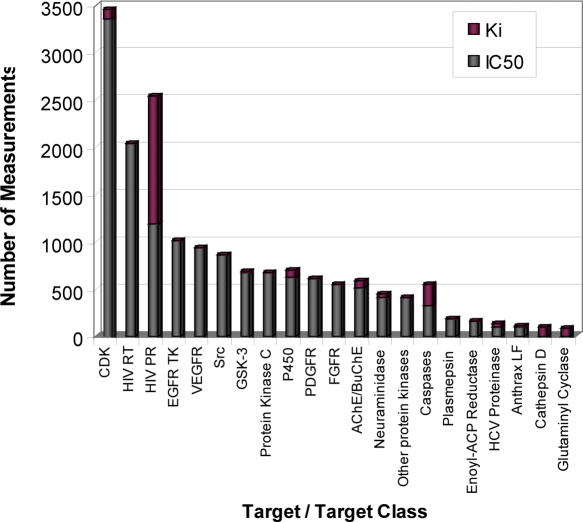
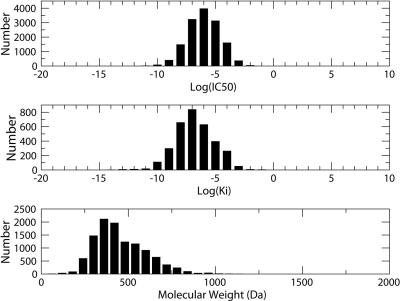
BindingDB currently holds ∼20 000 binding data for ∼11 000 different small molecule ligands and 110 different drug-targets; or 74 targets when mutants and isoforms are not counted separately. Examples include anthrax lethal factor, various caspases and kinases and HIV protease and reverse transcriptase. Perhaps the most similar public effort is KiBank (9), which provides a sparser user-interface to a substantial data set of ∼16 000 Ki data for 5900 small molecule ligands and 50 protein targets, apparently including proteins for which no structural data are available. For a perspective on BindingDB's current data holdings, Figure 1 shows the number of binding measurements for various targets and target classes, and Figure 2 provides histograms of Ki and IC50 values, and of the molecular weights of the small molecules across all entries. Although structural data are available for every protein target included in BindingDB, BindingDB collects data for many ligands that are not represented in the PDB. For example, the PDB has ∼50 structures of acetylcholinesterases, while BindingDB has affinity data for acetylcholinesterase with ∼250 different ligands. More generally, ∼2% of ligands in BindingDB have an exact match in the PDB and ∼15% of ligands in BindingDB have 90% similarity to a ligand in the PDB based upon the search criterion of the PDB. Thus, BindingDB's data collection differs significantly from those of databases which only collect affinities for protein–ligand complexes in the PDB, notably BindingMOAD (10) which holds ∼1400 data, PDBBind (11,12) with ∼1600 data, and AffinDB (13) with ∼750 data.
Figure 1.
Number of measurements in BindingDB for various targets and target classes.
Figure 2.
Histograms of binding affinities (1 M standard concentration), and molecular weights of ligands in BindingDB.
WEB INTERFACE: QUERY, DOWNLOAD AND VIRTUAL COMPOUND SCREENING
The BindingDB website provides an increasingly rich set of tools for query, analysis and download of binding data. Search capabilities include queries by target name; ligand name; affinity range; chemical structure, substructure and similarity; and target sequence, via BLAST (14). Query results are presented in a summary table, with the option to drill down to more detail on a given measurement. Available details include citation data, with links to PubMed and the option to retrieve all binding data from the same publication; sequence data and SMILES strings (15,16) and chemical structures. Hyperlinks to the PDB allow easy navigation to structural data for a given ligand, protein or complex. Additional tools also allow the user to build a ‘data set’ which can be downloaded in the form of an MDL SDfile containing chemical structures, target information and affinities.
The website also provides web-accessible tools for virtual screening of candidate ligands; we are not aware of any other public website that provides this functionality. The user provides a training set of ligands active against a given target or class of targets, either by using queries to form a BindingDB data set, or by uploading an SDfile from disk. The user then uploads his or her own SDfile of candidate ligands, selects one of three machine-learning methods installed on the BindingDB server, and starts the calculation. The software returns a ranking of the user's candidate ligands, where the top-ranked compounds are most likely to share the activity of the training set of active compounds. The results can be downloaded in the form of an SDfile containing the score of each compound; optionally, the compounds in the SDfile can be ranked according to their scores. The three machine-learning methods are as follows.
Maximum similarity
JChem (17) chemical fingerprints are computed with default parameters for each active compound and for each candidate ligand. The software computes the Tanimoto similarity [see, e.g. (18)] of each candidate compound to each active, and ranks the candidate compounds according to their maximal similarity to any active.
Binary kernel discrimination
JChem chemical fingerprints are computed with default parameters for each active compound and for a set of decoy compounds that are presumed to be inactive. The decoy compounds can be supplied by the user, or BindingDB can supply a random set of drug-like compounds drawn from the Zinc compound database (19). The BKD method (20) is then trained on a subset of the known actives and decoys, and tested on the remainder of the actives and decoys. The results of the test are reported to the user in terms of the fold enrichment of the known actives among the top 2% and top 10% of the ranked test-set compounds. If a high degree of enrichment is obtained (e.g. 10-fold enrichment) then it is reasonable to screen the user's candidate ligands with the trained model. When the user uploads these compounds, JChem fingerprints are computed for them, and the compounds are scored and ranked. The scores of the candidate ligands can be compared with those of the test-set actives and decoys, which are also provided as part of the output.
Support vector machine
As for the BKD, a set of active compounds and a set of decoys is established. The user is then presented with a list of quantitative molecular descriptors that can be used for the screening process; a reasonable default set of these is suggested by the website in order to aid the user. Descriptors are computed for all the compounds with Molconn-Z (eduSoft LC), and the descriptor set is then refined to avoid using highly correlated, and therefore redundant, descriptors (21). The LibSVM software (22) is then trained with a subset of the actives and decoys, and applied to the remaining active and decoy compounds to generate training set and test-set rankings, as previously described (21). The quality of these results are reported as enrichment factors, as for the BKD, and the user can then upload an SDfile of compounds to be ranked with the trained SVM model.
Maximum similarity is the fastest of the three methods and thus may be most convenient for very large screening sets. The BKD method is slower, but can recover more diverse actives. The SVM method also is slower than maximum similarity, but is arguably the best at finding actives that differ significantly from the known actives used to train the algorithm.
AVAILABILITY AND CITATION
BindingDB is freely accessible at http://www.bindingdb.org, and also may be accessed by following links from compounds at PubChem. To download SDfiles, users must complete a simple registration process and agree not to republish the data without explicit permission. Users are invited to contact us through the ‘Email us’ link and to participate in the user-forum at http://www.bindingdb.org/forum/forum.jsp. Suggestions regarding data sets to be extracted and deposited in BindingDB, and for web site features, are welcomed. Works using BindingDB should cite references (2–4) in this paper.
Acknowledgments
The authors gratefully acknowledge prior support by the National Institute of Standards and Technology and the National Science Foundation (Grant Number 9808318), and current support by the National Institute of General Medical Sciences, NIH (Grant Number GM070064). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the sponsors. Funding to pay the Open Access publication charges for this article was provided by NIH Grant GM070064.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bourne P. Will a biological database be different from a biological journal? PLoS Comput. Biol. 2005;1:179–181. doi: 10.1371/journal.pcbi.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X., Liu M., Gilson M.K. BindingDB: A Web-accessible molecular recognition database. Comb. Chem. High Throughput Screen. 2002;4:719–725. doi: 10.2174/1386207013330670. [DOI] [PubMed] [Google Scholar]
- 3.Chen X., Lin Y., Liu M., Gilson M.K. The binding database: Data management and interface design. Bioinformatics. 2002;18:130–139. doi: 10.1093/bioinformatics/18.1.130. [DOI] [PubMed] [Google Scholar]
- 4.Chen X., Liu M., Gilson M.K. The binding database: Overview and user's guide. Biopolymers/Nucleic Acid Sci. 2002;61:127–141. doi: 10.1002/1097-0282(2002)61:2<127::AID-BIP10076>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein F.C., Koetzle T.F., Williams T.F., Meyer G.J.B., Jr, Brice M.D., Rodgers J.R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 6.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horn F., Weare J., Beukers M., Horsch S., Bairoch A., Chen W., Edvardsen O., Campagne F., Vriend G. GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res. 1998;26:275–279. doi: 10.1093/nar/26.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okuno Y., Yang J., Taneishi K., Yabuuchi H., Tsujimoto G. GLIDA: GPCR-ligand database for chemical genomic drug discovery. Nucleic Acids Res. 2006;34:D673–D677. doi: 10.1093/nar/gkj028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhanga J., Aizawaa M., Amaria S., Iwasawab Y., Nakanoc T., Nakata K. Development of KiBank, a database supporting structure-based drug design. Comput. Biol. Chem. 2004;28:401–407. doi: 10.1016/j.compbiolchem.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Hu L., Benson M.L., Smith R.D., Lerner M.G., Carlson H.A. Binding moad (mother of all databases) Prot. Struct. Func. Bioinf. 2006;60:333–340. doi: 10.1002/prot.20512. [DOI] [PubMed] [Google Scholar]
- 11.Wang R., Fang X., Lu Y., Wang S. The PDBBind database: collection of binding affinities for protein–ligand complexes with known three-dimensional structures. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 12.Wang R., Fang X., Lu Y., Yang C., Wang W. The PDBBind database: Methodologies and updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 13.Block P., Sotriffer C.A., Dramburg I., Klebe G. AffinDB: A freely accessible database of affinities for protein–ligand complexes from the PDB. Nucleic Acids Res. 2006;34:D522–D526. doi: 10.1093/nar/gkj039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;214:1–8. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 15.Weininger D. SMILES, a chemical language and information-system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comp. Sci. 1988;28:31–36. [Google Scholar]
- 16.Weininger D., Weininger A., Weininger J.L. SMILES 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Comp. Sci. 1989;29:97–101. [Google Scholar]
- 17.Csizmadia F. JChem: Java applets and modules supporting chemical database handling from web browsers. J. Chem. Inf. Comput. Sci. 2000;40:323–324. doi: 10.1021/ci9902696. [DOI] [PubMed] [Google Scholar]
- 18.Willett P., Barnard J.M., Downs G.M. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998;38:983–996. [Google Scholar]
- 19.Irwin J.J., Shoichet B.K. ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harper G., Bradshaw J., Gittins J.C., Green D.V., Leach A.R.J. Prediction of biological activity for high-throughput screening using binary kernel discrimination. J. Chem. Inf. Comput. Sci. 2001;41:1295–1300. doi: 10.1021/ci000397q. [DOI] [PubMed] [Google Scholar]
- 21.Jorissen R.N., Gilson M.K. Virtual screening of molecular databases using a Support Vector Machine. J. Chem. Inf. Model. 2005;45:569–561. doi: 10.1021/ci049641u. [DOI] [PubMed] [Google Scholar]
- 22.Chang C.-C., Lin C.-J. LIBSVM: a library for support vector machines 2001. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm. [Google Scholar]