Abstract
Analysis of functional genomics (transcriptomics and proteomics) datasets is hindered in agricultural species because agricultural genome sequences have relatively poor structural and functional annotation. To facilitate systems biology in these species we have established the curated, web-accessible, public resource ‘AgBase’ (www.agbase.msstate.edu). We have improved the structural annotation of agriculturally important genomes by experimentally confirming the in vivo expression of electronically predicted proteins and by proteogenomic mapping. Proteogenomic data are available from the AgBase proteogenomics link. We contribute Gene Ontology (GO) annotations and we provide a two tier system of GO annotations for users. The ‘GO Consortium’ gene association file contains the most rigorous GO annotations based solely on experimental data. The ‘Community’ gene association file contains GO annotations based on expert community knowledge (annotations based directly from author statements and submitted annotations from the community) and annotations for predicted proteins. We have developed two tools for proteomics analysis and these are freely available on request. A suite of tools for analyzing functional genomics datasets using the GO is available online at the AgBase site. We encourage and publicly acknowledge GO annotations from researchers and provide an online mechanism for agricultural researchers to submit requests for GO annotations.
INTRODUCTION
Annotation of agricultural genomes
The complete sequencing of the human genome resulted in improved sequencing technologies and reduced expenses for whole genome sequencing. Consequently many genome sequencing projects are now underway. Among the agriculturally important species the chicken and rice genomes are completed (1–3), the bovine genome has a draft assembly available (4) and other agricultural genome sequencing projects (including pig, maize, wheat, soybean and grape) are in progress (5–8). Effective use of these genome sequences to model biological systems requires both structural annotation (denoting and demarcating genes and other functional elements) and functional annotation (assigning functions to each genomic element) (9,10).
Initial genome structural annotation is done computationally either by homology prediction to known genes in other species (‘predicted’ genes) or by using ab initio prediction algorithms that search for specific patterns indicative of open reading frames (ORFs) (11,12). However, these electronic genome structural-annotation methods produce false positive and false negative predictions as high as 70% (13) and commonly misclassify pseudogenes as functional (14). The use of experimental data for genome annotation is critical for conclusive identification of the functional sequences within genomes, accurate description of intron/exon structures and determination of the potential protein products from each gene in different tissues and cellular states (15). Transcriptome data allow annotation of RNA termini, splice junctions and untranslated RNAs. In addition, quantitative processes at the RNA level are independent of the protein level. However, detection of an mRNA is not conclusive evidence that the gene has a protein product (16). Proteogenomic mapping uses high-throughput mass spectrometry-based methods to provide direct evidence for the existence of proteins in vivo (15,17–19) and their locations in cells (20). Proteomics also more accurately determines the boundaries of functional ORFs and identifies unknown ORFs that cannot be well established on the basis of homology.
In contrast to the structural genome, which is considered to be fixed (at least in the short term), the functional genome rapidly changes both quantitatively and qualitatively in response to the environment. Understanding the functional genome informs us how genes function together. Genome functional annotation describes, interprets and explains the functions of the gene products themselves. The Gene Ontology (GO) project (21) provides the basis for the design, development and implementation of publicly available, expertly curated databases containing comprehensive genome functional annotations using controlled structured vocabularies (ontologies). First-pass electronic GO annotation (‘inferred by electronic annotation’ or IEA) is performed by data mining external sources of gene product information such as InterPro domains and SwissProt keywords. Annotations ‘inferred from sequence or structural similarities’ (ISS; e.g. by BLAST searches) and IEA rapidly provide ‘breadth’ and are especially valuable for organisms with little intensive manual GO curation. All other annotations are performed by expert curators using species-specific literature or (rarely) expert knowledge and these GO annotations are considered to be the ‘gold standard’ (22–24).
To facilitate functional analysis in agriculturally important genomes we launched the AgBase database. The AgBase database is a curated, open-source, web-accessible resource for functional analysis of agricultural plant and animal gene products. Our long-term goal is to serve the needs of the agricultural research communities by facilitating post-genome biology for agriculture researchers and for those researchers primarily using agricultural species as biomedical models.
ACCESSING INFORMATION FROM AgBase
The AgBase database provides experimentally derived structural annotations, GO annotations and tools for analyzing functional genomics data. Structural annotations based on proteogenomic mapping are provided as expressed peptide sequence tags or ‘ePSTs’ (20) and the proteogenomic pipeline used to generate these ePSTs from proteomic data is freely available upon request. AgBase curators provide manually curated GO annotations. We work in collaboration with the European Bioinformatics Institute GOA project [EBI-GOA (25)] and specifically focus our efforts on providing GO annotations for gene products EBI-GOA has not automatically annotated using IEA. The AgBase gene detail page displays GO annotations from both AgBase and EBI-GOA and the group responsible for each GO annotation is attributed. Gene association files of gene products annotated by AgBase are available for download in a tab-delimited format. Through EBI-GOA our GO annotations are added to the GO and UniProtKB databases (21,26). Tools developed by the AgBase consortium are made freely available upon request and where possible are web-based. Tools are designed to analyze large datasets generated by functional genomics based approaches. The AgBase database can be accessed directly via http://www.agbase.msstate.edu, or where appropriate the NCBI Genome Resources pages provide links to GO databases at AgBase.
Users can access information by text searches of protein or gene names, text searches of GO terms, searching via a variety of accession numbers or via BLAST searches. The AgBase tools also access the AgBase database. Users can choose to search the entire AgBase database or narrow their search by choosing an organism-specific database. The text search performs an exact substring search and multiple queries are also supported. Searches based on UniProtKB accessions and identifiers, UniParc identifiers, EMBL accessions, InterPro identifiers and NCBI non-redundant protein database GenBank gi numbers are supported. A BLAST based search is also available in instances where the user has sequences not represented in these databases or for which there is no database accession. To facilitate data mining the user has the option of searching the AgBase database by taxon ID or using either GO term names or identifiers. The proteogenomic database may also be searched using either text or BLAST based searches.
A TWO TIER SYSTEM OF GO ANNOTATIONS
Our AgBase download page provides a ‘GO Consortium’ gene association file containing fully quality checked annotations supported by the GO Consortium and a ‘Community’ gene association file containing annotations checked only for formatting errors by the AgBase biocurators following GO Consortium guidelines (http://www.geneontology.org/GO.annotation.shtml#script). This two-tiered system allows users to choose the breadth of GO coverage most appropriate for their experimental needs.
The community gene association file contains three kinds of annotations:
GO annotations for ‘predicted proteins’ without UniProtKB identifiers that until 10 July 2006, were not supported by EBI-GOA.
ISS annotations to evidence codes that stopped being accepted as of April 2006. For example, we recently completed ISS annotation of 1609 sheep proteins in UniProtKB that had no GO annotation but fewer than half will be released into the UniProtKB database following the newest GO Consortium guidelines.
GO annotations from community researchers that have not yet been quality checked by a trained GO curator. This type of annotation will be transferred to the GO Consortium gene association file after quality checks by AgBase biocurators and the original submitter be acknowledged in this gene association file.
Documentation is provided at the AgBase download site and via a link from the AgBase gene detail page informing users about these different gene association files and the checks that have been employed for each file. We envision that, just as for the species with much more advanced GO annotation, the GO annotations in the community gene association file will be superseded as more manually curated GO annotations become available for agricultural species.
ANALYZING FUNCTIONAL DATASETS
The tools currently available at the AgBase database can be divided into two categories: tools designed to assist with analysis of proteomics data and tools to evaluate experimental datasets using the GO. Two tools are available at AgBase to assist with analyzing proteomics data: the proteogenomic pipeline and the ProtIDer tool. The GOProfiler tool is designed to give a statistical summary of existing GO annotations. The GO suite of tools developed for functional analysis of large datasets includes GOProfiler, GORetriever, GOanna and GOSlimViewer. This suite of tools is designed so that users can work online to analyze their large-scale datasets using GO.
The proteogenomic pipeline is used to provide experimentally based structural annotations at a complete genome level, and the data we have obtained from proteogenomic mapping are available from the proteogenomics link at AgBase. The ProtIDer tool can be used to create a database of highly homologous proteins from ESTs and EST assemblies and is designed to assist with proteomic analysis in species which do not have genomic sequence available. In addition to providing the ProtIDer tool upon request, we will also provide organism-specific databases for proteomic analysis upon request.
GOProfiler provides an overview of current GO annotations available for particular species. The user provides a species taxonomy ID number (a link to a taxonomy browser is provided to assist users) and GOProfiler returns the number of GO associations and the number of annotated proteins for that species. A separate list of unannotated proteins is also provided.
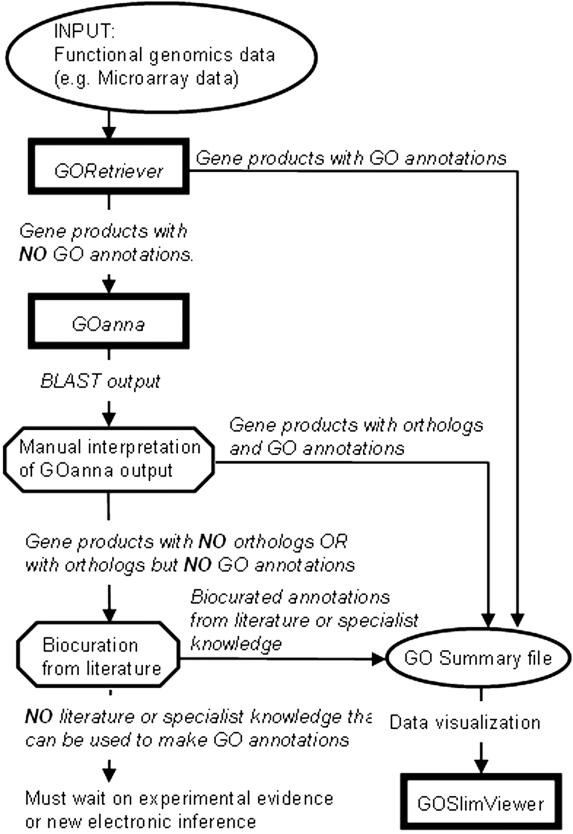
GORetriever, GOanna and GOSlimViewer are designed to be used as a pipeline for analyzing functional datasets using the GO (Figure 1). GORetriever inputs a list of database identifiers and searches a set of annotated databases for existing GO annotations of protein sequences. Annotation information from designated remote databases is stored in a local database to allow fast processing of large protein datasets. GORetriever returns the data online, as a downloadable Excel file and as a simplified text file (the GO Summary file) which can subsequently be used in GOSlimViewer. A list of queries without GO annotations is also provided and the user can add annotation to this list using GOanna. The GOanna tool differs from the GORetriever tool in that it does BLAST searches against a user-defined local database. The GOanna tool accepts a range of inputs, including fasta files. GOanna returns up to six proteins that exceed an operator-defined E-value threshold in an Excel format containing HTML links to each BLAST alignment. Users manually inspect the output and, in cases where they are satisfied that the query is orthologous to an annotated protein, transfer the GO annotation from the annotated protein to their query. These data can be added to their GO Summary file. If there are no orthologous proteins with GO annotation available the user may add GO annotation by curating published literature or from expert knowledge. The final GO Summary file is used as an input file for the GOSlimViewer tool. This tool is used to provide a high-level summary of the GO terms for a dataset and the output is a simple text file which can be charted in Excel to obtain publication quality figures.
Figure 1.
Analysis of functional datasets using the AgBase GO tools. Together the AgBase GO suite of tools form a pipeline for using GO to analyze microarray and other functional genomics datasets. Square boxes represent the tools, octagonal boxes are points in the pipeline that require human interpretation and ovals represent data files. GORetriever searches a set of annotated databases for existing GO annotations; sequences without annotations from GORetriever are entered into GOanna which BLAST searches a local GO database. GOanna returns up to six proteins that exceed an operator-defined E-value threshold. The user determines if the GO annotations from any of these matches can be used. Users may then choose to add literature-based or expert knowledge GO annotations to produce the final GO Summary file. GOSlimViewer can then be used to generate a high level view of the GO annotation for the proteins in a dataset using GO Slim sets.
Where these tools have modest computational requirements they are designed for online use, but all tools are freely available upon request. Each tool has comprehensive online help, including worked examples, and users are encouraged to contact us directly should they require additional information.
COMMUNITY REQUESTS FOR GO ANNOTATIONS
There are numerous tools available for functional analysis of datasets (27–32) but these tools assume that a significant number of the gene products of interest have GO annotation available. Since this is not the case for agriculturally important organisms we designed the GOanna tool so that a user can leverage data from closely related organisms to add GO annotations to their experimental dataset. However, even highly homologous genes may have different functions in different organisms and direct experimental evidence is the best data for assigning function to a particular gene product. Typically (but not always) individual groups within the GO Consortium are responsible for annotating particular species. In species where there is a dedicated GO annotation effort, researchers can contact the responsible database directly to request further annotations but there is currently no mechanism for requesting annotations in the many other species that researchers may be studying.
At AgBase we have developed a Community Requests and Submission form to meet the GO annotation needs of researchers interested in agriculturally important species. The type of data that a user must complete to make a request is scalable, allowing both users new to GO and users who have had GO biocuration training to enter requests. The basic request for GO annotations requires that the researcher identify the gene product of interest using a unique database accession identifier, the species the gene product comes from and nominate functional literature about the gene product (using PubMed ID). Researchers with expert knowledge about particular gene products have the option of supplying additional information about the types of experimental data available and the GO terms associated with the gene product. Users with biocuration training can upload their data as a gene association directly to be sanity checked and assimilated into GO databases. Researchers who submit their own gene association files will be provided with a unique AgBase annotator ID so that their annotations can be acknowledged. All users are asked to register and provide a valid email address so that they may be notified about the progress of their requests and submissions.
Requests are prioritized based on the number of community requests for each gene product and when the request was received. Gene product priority lists are species specific (i.e. chicken and cow gene products will not compete) and time spent on annotating each species is split proportionally based on the number of requests for each species. Annotations that have been submitted as a gene association file receive the highest priority as they are quality checked and submitted to the GO Consortium. Requests are acknowledged with a return email stating their rank in the request queue and researchers are notified by email when their request has been processed and the GO annotation added to the AgBase database; they are also notified if there is no functional data available for GO annotation.
AVAILABILITY OF AGBASE DATA AND TOOLS
Access to the AgBase databases is via http://www.agbase.msstate.edu and access to data is unrestricted. The tools we have developed are either freely available online at AgBase or by contacting us at agbase@cse.msstate.edu.
OBTAINING HELP FROM AgBase
Extensive online help, including worked examples, is available at AgBase by clicking on the help link in the top right corner of the site. All of our computational tools are freely available via AgBase, and technical support can be obtained by contacting us. Our biocurators make every effort to maintain data integrity by linking data with researchers, references and methods. However, similar to all databases, AgBase is an on-going project and interaction with the user community is vital for its success. We encourage the submission of data, correction of errors and suggestions for making AgBase of greater use including ideas for new computational tools (email: agbase@cse.msstate.edu).
FUTURE DIRECTIONS
The primary focus of the AgBase databases is to provide a resource that facilitates functional analysis in agriculturally important species. We will continue to work with GO Consortium members (particularly EBI-GOA), other agricultural based groups [including the Roslin Institute and Gramene (33)] and community groups to provide improved functional annotations for agricultural organisms. We are also using experimental-based approaches for improving genomic structural annotation and future work will focus on improving the proteogenomic pipeline, visualizing ePST data and integrating this structural annotation data with functional data.
Acknowledgments
We would like to thank MGI and EBI-GOA for their continued help and support with the Gene Ontology aspects of this manuscript. Financial support for our projects has come from the USDA NRI, MSU Office of Research (MAFES contribution number J11020), MSU Bagley College of Engineering, MSU College of Veterinary Medicine and the MSU Life Science and Biotechnology institute. Funding to pay the Open Access publication charges for this article was provided by the Office of Research and Graduate Studies, College of Veterinary Medicine, MSU.
Conflict of interest statement. None declared.
REFERENCES
- 1.Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 2.Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 3.Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 4.Spencer G., Tomlin R. NIH News Advisory. 2006. [Google Scholar]
- 5.Messing J., Dooner H.K. Organization and variability of the maize genome. Curr. Opin. Plant Biol. 2006;9:157–163. doi: 10.1016/j.pbi.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Meyers S.N., Rogatcheva M.B., Larkin D.M., Yerle M., Milan D., Hawken R.J., Schook L.B., Beever J.E. Piggy-BACing the human genome II. A high-resolution, physically anchored, comparative map of the porcine autosomes. Genomics. 2005;86:739–752. doi: 10.1016/j.ygeno.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Riaz S., Dangl G.S., Edwards K.J., Meredith C.P. A microsatellite marker based framework linkage map of Vitis vinifera L. Theor. Appl. Genet. 2004;108:864–872. doi: 10.1007/s00122-003-1488-5. [DOI] [PubMed] [Google Scholar]
- 8.Stacey G., Vodkin L., Parrott W.A., Shoemaker R.C. National Science Foundation-sponsored workshop report. Draft plan for soybean genomics. Plant Physiol. 2004;135:59–70. doi: 10.1104/pp.103.037903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orchard S., Hermjakob H., Apweiler R. Annotating the human proteome. Mol. Cell Proteomics. 2005;4:435–440. doi: 10.1074/mcp.R500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Reeves G.A., Thornton J.M. Integrating biological data through the genome. Hum. Mol. Genet. 2006;15:R81–R87. doi: 10.1093/hmg/ddl086. [DOI] [PubMed] [Google Scholar]
- 11.Delcher A.L., Harmon D., Kasif S., White O., Salzberg S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukashin A.V., Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennetzen J.L., Coleman C., Liu R., Ma J., Ramakrishna W. Consistent over-estimation of gene number in complex plant genomes. Curr. Opin. Plant Biol. 2004;7:732–736. doi: 10.1016/j.pbi.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Eyras E., Reymond A., Castelo R., Bye J.M., Camara F., Flicek P., Huckle E.J., Parra G., Shteynberg D.D., Wyss C., et al. Gene finding in the chicken genome. BMC Bioinformatics. 2005;6:131. doi: 10.1186/1471-2105-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desiere F., Deutsch E.W., Nesvizhskii A.I., Mallick P., King N.L., Eng J.K., Aderem A., Boyle R., Brunner E., Donohoe S., et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 2005;6:R9. doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gygi S.P., Rochon Y., Franza B.R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaffe J.D., Berg H.C., Church G.M. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- 18.Jaffe J.D., Stange-Thomann N., Smith C., DeCaprio D., Fisher S., Butler J., Calvo S., Elkins T., FitzGerald M.G., Hafez N., et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 2004;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalume D.E., Peri S., Reddy R., Zhong J., Okulate M., Kumar N., Pandey A. Genome annotation of Anopheles gambiae using mass spectrometry-derived data. BMC Genomics. 2005;6:128. doi: 10.1186/1471-2164-6-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarthy F.M., Cooksey A.M., Wang N., Bridges S.M., Pharr G.T., Burgess S.C. Modeling a whole organ using proteomics: the avian bursa of Fabricius. Proteomics. 2006;6:2759–2771. doi: 10.1002/pmic.200500648. [DOI] [PubMed] [Google Scholar]
- 21.The Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berardini T.Z., Mundodi S., Reiser L., Huala E., Garcia-Hernandez M., Zhang P., Mueller L.A., Yoon J., Doyle A., Lander G., et al. Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 2004;135:745–755. doi: 10.1104/pp.104.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couto F.M., Silva M.J., Coutinho P.M. Finding genomic ontology terms in text using evidence content. BMC Bioinformatics. 2005;6:S21. doi: 10.1186/1471-2105-6-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doms A., Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005;33:W783–W786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camon E., Magrane M., Barrell D., Lee V., Dimmer E., Maslen J., Binns D., Harte N., Lopez R., Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben-Shaul Y., Bergman H., Soreq H. Identifying subtle interrelated changes in functional gene categories using continuous measures of gene expression. Bioinformatics. 2005;21:1129–1137. doi: 10.1093/bioinformatics/bti149. [DOI] [PubMed] [Google Scholar]
- 28.Berriz G.F., King O.D., Bryant B., Sander C., Roth F.P. Characterizing gene sets with FuncAssociate. Bioinformatics. 2003;19:2502–2504. doi: 10.1093/bioinformatics/btg363. [DOI] [PubMed] [Google Scholar]
- 29.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 30.Khatri P., Bhavsar P., Bawa G., Draghici S. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 2004;32:W449–W456. doi: 10.1093/nar/gkh409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah N.H., Fedoroff N.V. CLENCH: a program for calculating Cluster ENriCHment using the Gene Ontology. Bioinformatics. 2004;20:1196–1197. doi: 10.1093/bioinformatics/bth056. [DOI] [PubMed] [Google Scholar]
- 32.Zeeberg B.R., Qin H., Narasimhan S., Sunshine M., Cao H., Kane D.W., Reimers M., Stephens R.M., Bryant D., Burt S.K., et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaiswal P., Ni J., Yap I., Ware D., Spooner W., Youens-Clark K., Ren L., Liang C., Zhao W., Ratnapu K., et al. Gramene: a bird's eye view of cereal genomes. Nucleic Acids Res. 2006;34:D717–D723. doi: 10.1093/nar/gkj154. [DOI] [PMC free article] [PubMed] [Google Scholar]