Abstract
We formerly released the porcine expressed sequence tag (EST) database Pig EST Data Explorer (PEDE; http://pede.dna.affrc.go.jp/), which comprised 68 076 high-quality ESTs obtained by using full-length-enriched cDNA libraries derived from seven tissues. We have added eight tissues and cell types to the EST analysis and have integrated 94 555 additional high-quality ESTs into the database. We also fully sequenced the inserts of 10 147 of the cDNA clones that had undergone EST analysis; the sequences and annotation of the cDNA clones were stored in the database. Further, we constructed an interface that can be used to perform various searches in the database. The PEDE database is the primary resource of expressed pig genes that are supported by full-length cDNA sequences. This resource not only enables us to pick cDNA clones of interest for a particular analysis, but it also confirms and thus contributes to the sequencing integrity of the pig genome, which is now being compiled by an international consortium (http://www.piggenome.org/). PEDE has therefore evolved into what we now call ‘Pig Expression Data Explorer’.
INTRODUCTION
The pig is not only a type of livestock that occupies a large proportion of the meat market—it is also a possible candidate animal model for biomedical research addressing regenerative medicine or preclinical investigations in pharmacology (1). The great usefulness of pigs in agriculture and in experimental and applied medicine demands that we have a sound knowledge of the molecular biology of the pig, as represented by genome sequences and gene expression data.
Many groups, including ours, have contributed to the recent rapid accumulation of pig expression data through expressed sequence tag (EST) analysis, and >1 600 000 ESTs are available in public databases such as the DDBJ/EMBL/GenBank nucleotide databases and Ensembl Trace Server/NCBI Trace Archive. However, the availability of porcine ESTs from full-length-enriched cDNA libraries has been quite limited. In addition, there have been few large-scale attempts to sequence and characterize broad collections of full-length pig cDNA sequences.
We have previously constructed and described the Pig EST Data Explorer (PEDE) database (http://pede.dna.affrc.go.jp/), which comprises more than 68 076 high-quality ESTs based on full-length-enriched cDNA libraries (i.e. those that were enriched in clones whose inserts contained full-length gene coding sequences) and which offers Internet-based search interfaces (2). Here, we describe the more than 100 000 porcine ESTs we have collected from additional libraries, the majority of which were constructed as full-length-enriched libraries. The new ESTs obtained were assembled into contigs, and we have picked representative cDNA clones for each contig and for each singlet that was highly similar to a known gene in other mammals. We have fully sequenced these representative cDNA clones and added these data to our porcine gene sequence collection. In addition, we have annotated these sequences in light of the results of sequence similarity searches and stored this information in the PEDE database. Finally, we have added various features to the PEDE search interface to increase the usefulness of the database.
CONSTRUCTION OF PORCINE FULL-LENGTH cDNA LIBRARIES AND EST ANALYSIS
To add to the ESTs we reported previously (2), we prepared cDNA libraries from the adrenal gland, alveolar macrophages, intestine, mesenteric lymph nodes, trachea and testis from crossbred pigs and skin from a Berkshire pig. Dendritic cells were induced from an adherent population of peripheral blood mononuclear cells from a Landrace pig, as described previously (3). We used the oligo-capped method as described in the previous studies (2,4) to generate porcine full-length cDNA libraries from all of the previously listed tissues or types of alveolar macrophages and dendritic cells. Libraries from those samples were constructed by using a SMART cDNA library construction kit, as described previously (5) (Clontech, Mountain View, CA), because of the small amounts of RNA these cells yielded. The cDNAs were cloned unidirectionally into pCMVFL3 (Invitrogen, Carlsbad, CA; Toyobo, Osaka, Japan) or pME18SFL3 (Toyobo) for oligo-capped cDNA clones and into pDNR-LIB (Clontech) for cDNA clones by the SMART method. The latest details of the sequencing status and assemblies generated by the procedure described (2) (PEDE assemblies) are shown in Supplementary Table S1 (http://pede.dna.affrc.go.jp/suppl_2007/suppl_table1.php).
SEQUENCING OF FULL-LENGTH cDNA INSERTS
For each contig, we picked a representative cDNA clone that included the initiation start site, and we determined the complete sequence of the insert. We also picked clones corresponding to singlets that were estimated to encode full-length coding sequences (CDSs) of orthologs of human genes (Table 1). The selected clones were sequenced by primer walking from both the 3′ and 5′ ends; sequence reads derived from each clone were assembled by using Phred and Phrap (6,7). Remaining regions of low-quality sequence data (Phred quality value, ≤25) were resequenced by using custom-designed primers. The resultant assembled sequence for each clone was inspected manually, and errors in the sequence were corrected by using Consed (8). To date, of the 15 000 clones picked, 10 147 have been sequenced completely.
Table 1.
Clones that were picked from the EST analysis for determination of their full-length cDNA sequences
| Clone set | EST accumulated until | Description | Clone number | Completely sequenced |
|---|---|---|---|---|
| A | March 2003 | Representative clones in contigs | 5546 | 4119 |
| B | May 2005 | Representative clones in contigs | 4769 | 4092 |
| C | May 2005 | Singlets apparently corresponding to human genes | 5079 | 1936 |
| Total | 15 394 | 10 147 |
The authors thus far have picked two sets of cDNA clones (A and B) as representative clones for contigs. We also have picked the clones of singlets that appeared to contain full-length CDSs of the orthologs of known human genes and which did not overlap any contig (C).
CHARACTERISTICS OF THE FULL-LENGTH cDNA CLONES
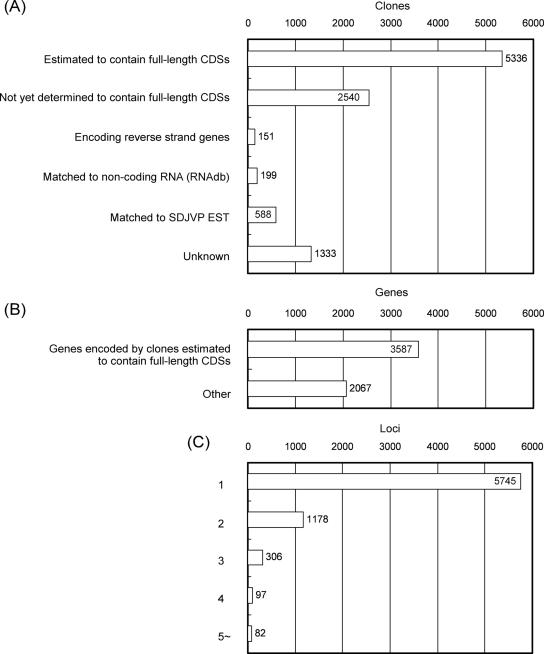
Sequences of full-length pig cDNA inserts were used in BLAST similarity searches (9) with translated human, mouse, dog, cattle and pig RefSeq sequences; human, mouse, cattle and pig UniGene clusters; and human genome sequences from the National Center for Biotechnology Information (10), as done for the EST assemblies in the PEDE database (2). We considered the cDNA clones to contain full-length CDSs if the length from the head to the tail of the match region (BLAST score, >50) in the ORF of the cDNA clone was 67–150% of the length of the CDS of the matched reference gene, although this criterion may exclude some cDNA clones that encode functional ORFs in pigs. As shown in Figure 1A, the similarity search demonstrated that 5336 cDNA clones were estimated to encode full-length CDSs of pig genes (corresponding to 3587 genes, Figure 1B), whereas 2540 failed to meet our criterion for full-length CDS-encoding clones but nevertheless showed high similarity to human or other mammalian genes. The cDNA clones we have finished sequencing correspond to at least 5654 genes (Figure 1B), and additional transcripts that failed to show marked similarity to known genes may in fact contain functional genes in pigs. Approximately, 200 clones showed high similarity to the reverse strands of known genes or sequences registered in RNAdb, which is a database of non-coding RNA sequences (11). Transcripts encoded by these clones may exert a regulatory function in gene expression (Figure 1A).
Figure 1.
(A and B) Results of BLAST similarity searches of fully sequenced cDNA clones against translated RefSeq sequences. (A) A group of 7876 clones shows high similarity (BLAST score >50) to corresponding sequences in the human, mouse, dog, cattle or pig RefSeq and 5336 of the 7876 clones were estimated to contain full-length CDSs. Non-coding RNA sequences were derived from sequences stored in RNAdb (11). SDJVP: EST sequences performed by Sino–Danish Joint Venture Project. (B) The pig cDNA clones estimated to encode full-length CDSs correspond to 3587 human, mouse, dog or cattle genes or already-known pig genes. This decreased number of corresponding genes compared with the estimated number of full-length encoding clones in (A) likely reflects alternative splicing. (C) Transcripts estimated to be ‘alternative transcripts’ among the fully sequenced cDNA clones. We estimated that the 10 147 cDNA clones represent transcripts derived from 7408 independent loci; 5745 transcripts are derived from independent loci, and 2739 cDNA clones seem to be alternative transcripts.
We also estimated the number of loci from which the transcripts encoded in the cDNA clones were derived. We considered that two transcripts were derived from the same locus if they shared >98% match over at least 200 bp. According to this criterion, the 10 147 cDNA clones were derived from ∼7400 independent loci, and 5745 loci each gave rise to a single clone (Figure 1C).
DATABASE AND SEARCH INTERFACES OF THE FULL-LENGTH cDNA CLONES
Into the PEDE database, we integrated the additional full-length sequences of cDNA clones and the results of the similarity searches with known genic sequences of the pig and other mammals. We updated the search interfaces that we had prepared for the EST assemblies, such as keyword and locus searches for the assemblies (http://pede.dna.affrc.go.jp/seq_search/seq_viewer.php) and a BLAST similarity search with the PEDE assemblies (http://pede.dna.affrc.go.jp/pedeblast/pedeblast_main.html), to include the full-length cDNA sequences. In the keyword and locus search views, cDNA clones and EST assemblies can be selected by gene symbol, keywords, or chromosomes according to their similarity to human, mouse, dog, cattle and pig RefSeq genic sequences. In the result view, the identified sequences can be downloaded in multi-FastA format. This result view also links to a list showing the clones and assemblies that match to a particular gene. Details of each clone, including its nucleotide sequence, a summary of the BLAST results, and identified single nucleotide polymorphisms (SNPs), are summarized in a page linked to the search result page as previously describe for the EST assemblies (2).
We also prepared an interface to search cDNA clones or EST assemblies according to gene ontology (GO) (12) terms. The full-length cDNA clones and EST assemblies that are related to target GO identifiers, which are selected from a tree structure, can be viewed as a list on the database (http://pede.dna.affrc.go.jp/seq_search/go_viewer.php).
IMPLEMENTATION
The PEDE database was developed on the PostgreSQL relational database system, and its interfaces were constructed using PHP script language on the Apache Internet Web server. The PEDE database is provided as one of the resources in the Animal Genome Database (http://animal.dna.affrc.go.jp/). It is accessible freely and directly at http://pede.dna.affrc.go.jp/.
CONCLUSIONS AND PERSPECTIVES
The PEDE database provides a catalog of porcine expressed genes as ESTs and full-length cDNA clones. The full-length cDNA sequences enable us to design oligoprobes for use with microarrays to reflect the expression patterns in pigs with increased accuracy. Furthermore, the full-length cDNA clones promote direct functional analyses of porcine genes because they can be introduced into cells as expression vectors. However, the benefits of this collection of porcine full-length cDNA sequences are not limited to analyses of gene function and expression. The availability of these full-length sequences will increase the utility of the porcine genome sequences being generated by the international Swine Genome Sequencing Consortium (13) (http://www.piggenome.org/). The reliable cDNA sequences stored in the PEDE database can be used to validate the integrity of assembled genome sequences. We are now developing a collection of SNPs from the 3′ ends of the full-length pig cDNA sequences, with the goal of constructing a reliable linkage map, further contributing to the reliability of the pig genome assembly. Alignment of the full-length cDNA sequences with the genomic sequences will yield clues for determining the transcriptional elements adjacent to CDSs in pigs. Through comparison with the pig genome sequences, the PEDE database will increase the number of full-length pig cDNA sequences and the volume of other information related to porcine genes.
In conclusion, the PEDE database, which now includes >10 000 full-length cDNA sequences and covers much broader porcine gene expression data than the former version, will help users to explore pig genes that may affect economic traits in the livestock industry. This useful resource also will enable scientists to prepare a catalog of genes likely to be of interest when pigs are used as animal models in research applications, such as preclinical pharmacology investigations and studies of transplantation biology.
Acknowledgments
We thank Yasumichi Sakai, Kazuyoshi Makino, and Akira Irako (Mitsubishi Space Software) for their assistance in the computational analysis and Takako Suzuki and Hiromi Sakata for their technical assistance. We are indebted to Yoshihiro Muneta (National Institute of Animal Health) and Katsuhiro Aikawa (National Institute of Livestock and Grassland Science) for preparation of the porcine cells and tissues. This work was supported by the Animal Genome Research Project and the Food Traceability Research Project of the Ministry of Agriculture, Forestry and Fisheries of Japan and by a Grant-in-Aid from the Japan Racing Association. The sequence data described in this paper have been submitted to the DDBJ/EMBL/GenBank database under accession nos BW954997–BW985219, CJ000001–CJ039835, DB781565–DB806061 and AK230469–AK240615. Funding to pay the Open Access publication charges for this article was provided by the Ministry of Agriculture, Forestry and Fisheries of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vodicka P., Smetana K., Jr, Dvorankova B., Emerick T., Xu Y.Z., Ourednik J., Ourednik V., Motlik J. The miniature pig as an animal model in biomedical research. Ann. N. Y. Acad. Sci. 2005;1049:161–171. doi: 10.1196/annals.1334.015. [DOI] [PubMed] [Google Scholar]
- 2.Uenishi H., Eguchi T., Suzuki K., Sawazaki T., Toki D., Shinkai H., Okumura N., Hamasima N., Awata T. PEDE (Pig EST Data Explorer): construction of a database for ESTs derived from porcine full-length cDNA libraries. Nucleic Acids Res. 2004;32:D484–D488. doi: 10.1093/nar/gkh037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paillot R., Laval F., Audonnet J.C., Andreoni C., Juillard V. Functional and phenotypic characterization of distinct porcine dendritic cells derived from peripheral blood monocytes. Immunology. 2001;102:396–404. doi: 10.1046/j.1365-2567.2001.01200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki Y., Yoshitomo-Nakagawa K., Maruyama K., Suyama A., Sugano S. Construction and characterization of a full length-enriched and a 5′ end-enriched cDNA library. Gene. 1997;200:149–156. doi: 10.1016/s0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- 5.Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewing B., Hillier L., Wendl M.C., Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 7.Ewing B., Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 8.Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler D.L., Barrett T., Benson D.A., Bryant S.H., Canese K., Chetvernin V., Church D.M., DiCuccio M., Edgar R., Federhen S., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006;34:D173–D180. doi: 10.1093/nar/gkj158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang K.C., Stephen S., Engstrom P.G., Tajul-Arifin K., Chen W., Wahlestedt C., Lenhard B., Hayashizaki Y., Mattick J.S. RNAdb—a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schook L.B., Beever J.E., Rogers J., Humphray S., Archibald A., Chardon P., Milan D., Rohrer G., Eversole K. Swine Genome Sequencing Consortium (SGSC): a strategic roadmap for sequencing the pig genome. Comp. Func. Genomics. 2005;6:251–255. doi: 10.1002/cfg.479. [DOI] [PMC free article] [PubMed] [Google Scholar]